Abstract
The dataset investigates the neural correlates of appetite in people living with motor neuron disease (plwMND) compared to non-neurodegenerative disease controls. Thirty-six plwMND and twenty-three controls underwent two fMRI sessions: one in a fasted state and one postprandial. Participants viewed visual stimuli of non-food items, low-calorie foods, and high-calorie foods in a randomised block design. Imaging data included T1w, T2w, and task-based and resting-state fMRI scans, and measures are complemented by subjective appetite questionnaires and anthropometric measures. This dataset is unique for its inclusion of functional imaging across prandial states, offering insights into the neural mechanisms of appetite regulation in patients with MND. Researchers can explore various aspects of the data, including the functional responses to food stimuli and their associations with clinical and appetite measures. The data, deposited in OpenNeuro, follows the Brain Imaging Data Structure (BIDS) standard, ensuring compatibility and reproducibility for future research. This comprehensive dataset provides a resource for studying the central mechanisms of appetite regulation in MND.
Similar content being viewed by others
Background & Summary
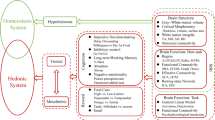
The loss of appetite is a common feature in people living with MND (plwMND), leading to negative energy balance, which is associated with poorer disease outcomes and prognosis1. Impairments in appetite and metabolism are associated with decreases in fat mass and body weight, leading to faster disease progression and earlier death2. The loss of appetite appears to worsen over time and is associated with declining functional outcomes3. The loss of appetite in plwMND is thought to occur due to several factors, including hypothalamic deficits, anxiety and depression, bulbar symptoms, and respiratory dysfunction4. The provision of appropriate clinical care for patients with a loss of appetite requires a complete understanding of the underlying etiological mechanisms that lead to impaired appetite control. Extra motor structural deficits and disease pathology have been observed throughout the brains of plwMND, including regions involved in appetite regulation5,6,7,8. For example, proteinopathy6 and atrophy9 of the hypothalamus, a critical appetite regulation brain structure, is documented in the literature. Neural activity in the brain has not been directly studied in relation to visual food stimuli and appetite in MND, and so additional research is needed to better understand how disease may impact perceptions of and reward for food in MND.
Appetitive behaviours are controlled by brain regions involved in visual and sensory perception, reward, memory, and decision-making. To better understand their functional roles in food-related behaviours, studies have utilised functional MRI (fMRI) to investigate the neural correlates to visual food stimuli10. Imaging sessions are often conducted during fasting and after the consumption of a meal (postprandial) to incorporate subjective changes in appetite into the study. Such fMRI study designs capture differential neural reward responses to food under different nutritional states. For instance, the activation of the basolateral amygdala was identified to be associated with food addiction under the fasted condition only, and not the postprandial condition11.
Additionally, these imaging sessions often include images of high-calorie foods, low-calorie foods and non-food items10. High-calorie foods often elicit differential hedonic sensations in pleasure and satisfaction due to their greater capacity to supplement energy reserves12. Indeed, disease-specific food stimuli neural correlates have been identified in obesity-prone individuals, Prader-Willi syndrome, and anorexia nervosa13,14,15.
For the present dataset16, thirty-six plwMND and twenty-three non-neurodegenerative disease control participants underwent two sessions of imaging, one under a fasted condition and one under a postprandial condition. Visual stimuli of non-food items, low-calorie foods, and high-calorie foods were presented to participants during scanning in a randomised block design. For each participant, we collected T1w and T2w anatomical scans, and two task-based fMRI scans, seven minutes each. The first scan was taken under a 12-hour overnight fasted-state, after which, participants were offered a liquid mixed meal, and then proceeded to undergo a second session of imaging. Questionnaires on subjective appetite were administered before, between and after the imaging sessions. Each participant has corresponding anthropometric, appetite and disease measures.
This dataset is unique due to its inclusion of functional imaging across prandial states, complemented by matching appetite, anthropometric and clinical measures. This data set is of value as it provides researchers with essential data for the investigation of neural correlates related to nutritional status and appetite in a patient population where loss of appetite is commonly observed and of clinical importance. Several research questions can be asked when interrogating this dataset. For example, this dataset can contribute to studies that contrast loss versus gain of appetite across the spectrum of patients with MND. Researchers can analyse many aspects of the fMRI paradigm, including discriminating functional responses between visual stimuli of food based on their caloric content. Researchers can also integrate clinical and appetite data into the fMRI design matrix to identify other biologically and clinically meaningful associations. The resting state data can be used to identify and analyse default mode networks with patient data, providing greater insights into disease impacts on a large network scale. Finally, our data can be integrated with other imaging datasets to increase statistical power for analysis in the broader MND scientific community.
Overall, this imaging data, integrated with rich clinical data, which would otherwise be expensive and resource-intensive to generate, provides a platform to begin to explore alterations in central mechanisms for appetite regulation in plwMND. The research output from this dataset will provide insights into the non-motor aspects of the disease and inform future directions for targeted studies investigating the central mechanisms of appetite control in plwMND. It is hoped that these studies will guide the development of clinical management strategies for plwMND experiencing a loss of appetite, improving patient care and quality of life.
Methods
Participants
The present dataset includes thirty-six participants with a diagnosis of possible, probable, or definite Amyotrophic Lateral Sclerosis (ALS; the most common subtype of MND), as defined by the El Escorial criteria17. Twenty-three non-neurodegenerative (NND) controls were recruited as a convenience sample of spouses, friends, or colleagues of plwMND. We note that convenience sampling itself brings potential biases and limitations to the dataset, including confounding factors such as similar living conditions, diets, and genetic backgrounds. Studies utilising this dataset will need to consider the potential bias this brings to the interpretation of results. Additionally, the relatively lower sample size may impact the power of this dataset, and so researchers will need to consider this when integrating this dataset into future studies.
Participants underwent imaging at the Herston Imaging Research Facility (HIRF) at the Royal Brisbane and Women’s Hospital (RBWH) campus in Australia. To control for factors that could directly impact appetite and to minimise participant burden during imaging, individuals undergoing gastrostomy care, with a history of diabetes, and/or with respiratory impairment where forced vital capacity was <60% of the predicted forced expiratory volume were excluded. Two participants received an updated diagnosis of primary lateral sclerosis (PLS), one received an eventual diagnosis of ALS-bvFTD, and one had a diagnosis of MND that is undefined.
Ethics
This study was approved by the University of Queensland, the Royal Brisbane and Women’s Hospital (RBWH, HREC/17/QRBW/616), and Uniting Care Health Human Research Ethics (Ref no. 1801) Committees. All participants, 34 to 74 years of age, provided written and informed consent to participate in the research and for deidentified data to be made available through publication and for other research purposes. To protect the identities of participants, the software mri_reface (v0.3.3) was used to reface all anatomical images18. Additionally, any identifiable information in the metadata has also been removed.
Imaging and assessments
Imaging and assessments for this study were conducted over the course of 32 months, between July 2018 and February 2021. Participants fasted for 12 hours (overnight) before attending the research visit (nil by mouth, other than water). Participants completed two imaging sessions, each approximately 45 minutes in duration, during this research visit. Following arrival at the imaging facility at 0800 hours, and prior to their first MRI session, participants were asked to complete the Council on Nutrition Appetite Questionnaire (CNAQ), an eight-item questionnaire designed to assess appetite19, and part one of a Visual Analogue Scale (VAS) to assess subjective sensations of appetite20.
Participants were positioned in the MRI machine and instructed to observe visual stimuli during scanning. At completion of the first MRI scan, participants consumed a liquid meal (Sustagen®, Nestle; 15 kJ/kg.Bw; Protein - 0.18 g/kg.Bw, Fat - 0.05 g/kg.Bw, Carbohydrates - 0.61 g/kg.Bw), before again completing the VAS for repeat assessment of subjective measures of appetite (part one) and palatability of the test meal (part two)20. The elapsed time between imaging sessions was 33 ± 8 minutes, and the elapsed time between the consumption of the liquid meal and the start of the second imaging session was 29 ± 12 minutes. After the second imaging session, participants completed the VAS (part one) for the third time.
The task during each session involved displaying three categories of images on a projector screen for the participant to view: high-calorie food, low-calorie food, and non-food items. During each functional scan, there were eighteen blocks of images, with six blocks for each category presented in a random order. Each block contained six randomly chosen images from the same category, and each image was displayed for three seconds. After each block, a fixation cross was shown on the screen, and participants were instructed to respond by pressing a button to ensure attentiveness in the scanner. The fixation cross was displayed between blocks for 14–17 seconds as a baseline. During scans, participants were also equipped with a pulse oximeter, ECG electrodes, and a breathing belt to record physiological activity.
Image acquisition
All scans were acquired using a 3-Tesla (3 T) Siemens Prisma scanner (Siemens Healthcare, Erlangen, Germany). T1-weighted (T1w) structural scans were obtained in the first session from a 3-dimensional 1 mm3 isotropic MP2RAGE sequence (TR/TE/TIs/Flip Angles/FoV/Acquisition Time = 5000 ms/2.98 ms/701 ms,2500 ms/4°,5°/256 mm × 240 mm/9 m:02 s)21. Scans were conducted as a single slab with 192 slices, positioned isocentre and oriented in the sagittal plane, with the phase encoding direction anterior to posterior and a distance factor of 50%. Images were acquired using the GRAPPA parallel imaging technique with a phase encoding acceleration factor of three and thirty-two phase encoding lines used as reference. The first imaging session also included T2-weighted (T2w) fluid-attenuated inversion recovery (FLAIR) scans at 1 mm3 isotropic resolution (TR/TE/TI/FoV/Acquisition Time = 5000 ms/386 ms/1800ms/256 mm × 256 mm/5m52s)22. T2w scans were conducted as a single slab with 176 slices, positioned isocentre and oriented in the sagittal plane, with the phase encoding direction anterior to posterior, and a slice thickness of 1 mm. Images were acquired using the GRAPPA parallel imaging technique, with a phase encoding acceleration factor of twenty-four.
Functional scans were obtained from both imaging sessions through a 2D gradient echo planar imaging (EPI) sequence at 2.4 mm3 isotropic resolution (TR/TE/FoV/Flip Angle/Acquisition Time = 820 ms/33 ms/206 mm × 206 mm/53°/11 m:12 s), with sixty slices covering a 206 mm field of view, which includes the entire cortex and cerebellum. The acquisition was conducted in one slice group, positioned in the isocentre, transverse orientation, with a distance factor of 0% and phase encoding direction anterior to posterior. The functional scan was conducted with a simultaneous multi-slice acceleration, with an acceleration factor of six.
Stimulus material
The stimulus set consisted of three hundred colour pictures containing two hundred food stimuli and one hundred pictures of non-food items. All images were obtained from the FoodPics Database23. Food stimuli contained images of hot and cold meals, including meals with high and low-calorie contents (Supplementary table 1). Non-food items included general household items, animals, insects, etc. Image sets used for fasting and postprandial response testing were matched for complexity, size, brightness, calorie content, and macronutrient balance (Table 1).
fMRIPrep
Anatomical and functional preprocessing was conducted in fMRIPrep 23.1.424. The full methods can be found in the Supplementary methods.
PhysIO
To account for physiological noise originating from respiration and cardiac activity, the PhysIO toolbox was used to process breathing belt, ECG and pulse oximetry data, creating eight regressors for respiration and six regressors for cardiac activity25. Additionally, to account for excessive movement, any volumes exceeding a framewise displacement of 0.5 mm were censored in the resulting design matrix.
Data Records
This dataset ds005874 is deposited in the online neuroimaging repository, OpenNeuro16. The platform is an open platform that features thousands of MRI datasets for various natural science applications. Per the requirements for an OpenNeuro dataset16, the files are organised into the Brain Imaging Data Structure (BIDS) v1.10.0. BIDS is a standard that prescribes a structure for organising neuroimaging data to ensure clarity of the data, compatibility and reproducibility for future research26.
In compliance with the BIDS specification, images and their json sidecar files are stored under their respective modality (i.e., func, anat), session (i.e., ses-01, ses-02) and subject (i.e., sub-01, sub-02, etc.) directories. Sidecar files consist of modality-specific information as outlined in the specification. Here, every directory and recursive directories consist of a tsv file describing the contents of the folder, along with a json sidecar file defining any non-standard terminologies. Raw physiological data are stored under the sourcedata directory under their respective session and subject subdirectories. Phenotypic information is stored under the phenotype folder, and derivative datasets are stored under the derivatives folder. Custom code used to generate data are stored under the code directory. All files within the BIDS specification follow the key-value pair entities convention. Data records are summarised in Fig. 1.
Summary of the dataset. The dataset is formatted into the brain imaging data structure (BIDS) standard. Each subject includes two imaging sessions; anatomical T1-weighted (T1w) and T2-weighted (T2w) scans, and resting and task functional scans with corresponding task files that contain onset and duration information. Each functional scan has associated physiological files, which are stored under ‘sourcedata’ in the Siemens VB proprietary format. Derivative data consists of outputs from fMRIPrep, and PhysIO. Anthropometric and relevant clinical data is stored under ‘phenotype’.
Imaging data
Under each subject directory are data from the two imaging sessions, with the first session under a fasted condition and the second session after the consumption of a liquid mixed meal (postprandial). The first session consists of anatomical T1w and T2w images.
Task-based and resting fMRI data was collected for both sessions. Task fMRI files had their associated event categories, onsets, and durations described in their corresponding events tab-separated value (TSV) file. To account for physiological noise during the functional scans, respiratory, cardio, pulmonary and external data was recorded. As the format is not currently covered under the current BIDS specification, physiological data is stored under the sourcedata folder.
fMRIPrep
The output from the fMRIPrep pipeline is found under the derivatives folder. Any anatomical data that may be identifiable has been removed. The output consists of segmentations, probability maps, and masks for the anatomical data; and pre-processed functional volumes that have undergone slice-timing correction, realignment, co-registration and segmentation (denoted with the entity, desc-preproc). A summary of the processing is automatically generated by fMRIPrep as an HTML file in each subject’s base directory.
PhysIO
The physiological regressors generated by PhysIO are found under the derivatives folder. The output includes a series of graphs that summarise the regressors, as well as the final regressor matrix, saved as a comma-separated value (CSV) file, that is compatible with SPM12 for first-level fMRI analysis.
Appetite and clinical measures
Our dataset includes demographics, appetite and clinical measures, stored under the phenotype folder16. The dataset consists of thirty-six plwMND, accounting for 61% of the participants (Fig. 2a). Case-control comparisons are conducted using a two-tailed t-test with Welch’s correction for continuous variables and Fisher’s exact test for categorical variables. The handedness measure is missing for one case and one control participant. Overall, there are no differences between cases and controls for age, height and weight (Fig. 2b–d). CNAQ is significantly lower in cases, however (p = 0.005; Fig. 2e), suggesting loss of appetite in this cohort of plwMND when compared to NND Controls. There are no differences in sex and handedness between the case and control groups (Fig. 2f,g).
In plwMND, additional clinical measures were also collected. Most patients presented with spinal onset disease (81%), followed by bulbar onset disease (17%). One individual presented with both spinal and bulbar onset disease (Fig. 3a). Out of thirty-six cases, thirty-two (86%) have ALS, three individuals (8%) had an eventual diagnosis of PLS, and one individual was identified as having an undefined MND. One individual participant received an eventual diagnosis of ALS-bvFTD (Fig. 3b). Our dataset also includes a wide range of ages at first assessment, ranging from 34 to 74 years (Fig. 3c). Diagnostic delay and onset to assessment show a positive skew, with most cases falling below 27 months and 20 months, respectively (Fig. 3d,e).
Technical Validation
Characteristics of visual stimuli shown to participants are well balanced between image sets, ensuring inherent image quality is controlled under the fasting and postprandial state (Table 1). Moreover, image blocks and individual images are randomised to reduce sources of order-based bias in functional activations.
The quality of the anatomical images was assessed using MRIQC v24.0.0. In Fig. 4, the quality assurance metrics of interest included those indicative of head motion – entropy focus criterion (EFC) and coefficient of joint variation (CJV) between grey and white matter. Additionally, the signal-to-noise ratio (SNR) in the cerebral spinal fluid (CSF), grey matter (GM) and white matter (WM) were also analysed to evaluate the quality of signal in each tissue type. The spread of EFC and CJV for T1w and T2w images indicates a wide range of head movements across the cohort. On the other hand, SNR for all three tissue types demonstrates consistent signal quality in grey matter and white matter. Following a manual inspection of each anatomical volume, the T1w and T2w images were found to be suitable for further analysis.
Quality of functional images were also assessed using fMRIPrep24. Significant head movement within fMRI images were identified for nearly all participants, which can significantly impact the signal and hence the interpretability of a given volume. The median percentage of volumes exceeding a framewise displacement of 0.5 mm per participants is 5.76% with an interquartile range of 9.34%. Four participants (sub-23, sub-41, sub-42, sub-54) had more than 50% of volumes exceeding 0.5 mm framewise displacement. We note that these are all plwMND, which makes sense given the difficulty and uncomfortable nature of staying still with MND in an MRI machine. To address excessive movement, functional image nuisance regressors, including movement, framewise displacement and global signal are stored under the fMRIPrep output. Coupled with physiological data, nuisance regressors are processed and transformed using the PhysIO toolbox25 to create SPM12-compatible files in a fMRI first-level analysis. As part of the regressors are censored volumes where framewise displacement exceeds 0.5 mm. Altogether, this enables researchers to account for confounding physiological and excessive movements in their fMRI analysis of this dataset.
Code availability
The code used to generate derivative datasets can be found under the Code folder of the repository on OpenNeuro16.
References
*Ngo, S. T. et al. Loss of appetite is associated with a loss of weight and fat mass in patients with amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener, 1–9 https://doi.org/10.1080/21678421.2019.1621346 (2019).
Ludolph, A. et al. Nutritional and metabolic factors in amyotrophic lateral sclerosis. Nat Rev Neurol 19, 511–524, https://doi.org/10.1038/s41582-023-00845-8 (2023).
Sarmet, M., Kabani, A., Maragakis, N. J. & Mehta, A. K. Appetite and quality of life in amyotrophic lateral sclerosis: A scoping review. Muscle Nerve 66, 653–660, https://doi.org/10.1002/mus.27694 (2022).
Ngo, S. T., Mi, J. D., Henderson, R. D., McCombe, P. A. & Steyn, F. J. Exploring targets and therapies for amyotrophic lateral sclerosis: current insights into dietary interventions. Degener Neurol Neuromuscul Dis 7, 95–108, https://doi.org/10.2147/dnnd.S120607 (2017).
Chang, J. et al. Lower hypothalamic volume with lower body mass index is associated with shorter survival in patients with amyotrophic lateral sclerosis. Eur J Neurol 30, 57–68, https://doi.org/10.1111/ene.15589 (2023).
Gabery, S. et al. Loss of the metabolism and sleep regulating neuronal populations expressing orexin and oxytocin in the hypothalamus in amyotrophic lateral sclerosis. Neuropathol Appl Neurobiol 47, 979–989, https://doi.org/10.1111/nan.12709 (2021).
Bolborea, M. et al. Loss of hypothalamic MCH decreases food intake in amyotrophic lateral sclerosis. Acta Neuropathol 145, 773–791, https://doi.org/10.1007/s00401-023-02569-x (2023).
Bede, P. et al. Hippocampal, Fornix, Uncinate, and Amygdala Degeneration in Amyotrophic Lateral Sclerosis: the Pathological Substrate of Memory Impairment (4916). Neurology 94, 4916, https://doi.org/10.1212/WNL.94.15_supplement.4916 (2020).
Michielsen, A. et al. Association Between Hypothalamic Volume and Metabolism, Cognition, and Behavior in Patients With Amyotrophic Lateral Sclerosis. Neurology 103, e209603, https://doi.org/10.1212/wnl.0000000000209603 (2024).
Zheng, L., Miao, M. & Gan, Y. A systematic and meta-analytic review on the neural correlates of viewing high- and low-calorie foods among normal-weight adults. Neurosci Biobehav Rev 138, 104721, https://doi.org/10.1016/j.neubiorev.2022.104721 (2022).
Pursey, K. M., Contreras-Rodriguez, O., Collins, C. E., Stanwell, P. & Burrows, T. L. Food Addiction Symptoms and Amygdala Response in Fasted and Fed States. Nutrients 11 https://doi.org/10.3390/nu11061285 (2019).
Berthoud, H. R., Münzberg, H. & Morrison, C. D. Blaming the Brain for Obesity: Integration of Hedonic and Homeostatic Mechanisms. Gastroenterology 152, 1728–1738, https://doi.org/10.1053/j.gastro.2016.12.050 (2017).
Santel, S., Baving, L., Krauel, K., Münte, T. F. & Rotte, M. Hunger and satiety in anorexia nervosa: fMRI during cognitive processing of food pictures. Brain Res 1114, 138–148, https://doi.org/10.1016/j.brainres.2006.07.045 (2006).
Holsen, L. M. et al. Neural mechanisms underlying hyperphagia in Prader-Willi syndrome. Obesity (Silver Spring) 14, 1028–1037, https://doi.org/10.1038/oby.2006.118 (2006).
Cornier, M. A. et al. Differences in the neuronal response to food in obesity-resistant as compared to obesity-prone individuals. Physiol Behav 110-111, 122–128, https://doi.org/10.1016/j.physbeh.2013.01.002 (2013).
Chang, J. et al. An fMRI Dataset for Appetite Neural Correlates in People Living with Motor Neuron Disease OpenNeuro ds005874 https://doi.org/10.18112/openneuro.ds005874.v1.0.0 (2025).
Brooks, B. R., Miller, R. G., Swash, M. & Munsat, T. L. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord 1, 293–299, https://doi.org/10.1080/146608200300079536 (2000).
Schwarz, C. G. et al. Changing the face of neuroimaging research: Comparing a new MRI de-facing technique with popular alternatives. Neuroimage 231, 117845, https://doi.org/10.1016/j.neuroimage.2021.117845 (2021).
Wilson, M. M. et al. Appetite assessment: simple appetite questionnaire predicts weight loss in community-dwelling adults and nursing home residents. Am J Clin Nutr 82, 1074–1081, https://doi.org/10.1093/ajcn/82.5.1074 (2005).
Flint, A., Raben, A., Blundell, J. E. & Astrup, A. Reproducibility, power and validity of visual analogue scales in assessment of appetite sensations in single test meal studies. Int J Obes Relat Metab Disord 24, 38–48, https://doi.org/10.1038/sj.ijo.0801083 (2000).
Marques, J. P. et al. MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. Neuroimage 49, 1271–1281, https://doi.org/10.1016/j.neuroimage.2009.10.002 (2010).
Hajnal, J. V. et al. Use of fluid attenuated inversion recovery (FLAIR) pulse sequences in MRI of the brain. J Comput Assist Tomogr 16, 841–844, https://doi.org/10.1097/00004728-199211000-00001 (1992).
Blechert, J., Meule, A., Busch, N. A. & Ohla, K. Food-pics: an image database for experimental research on eating and appetite. Front Psychol 5, 617, https://doi.org/10.3389/fpsyg.2014.00617 (2014).
Esteban, O. et al. fMRIPrep: a robust preprocessing pipeline for functional MRI. Nat Methods 16, 111–116, https://doi.org/10.1038/s41592-018-0235-4 (2019).
Kasper, L. et al. The PhysIO Toolbox for Modeling Physiological Noise in fMRI Data. J Neurosci Methods 276, 56–72, https://doi.org/10.1016/j.jneumeth.2016.10.019 (2017).
Gorgolewski, K. J. et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci Data 3, 160044, https://doi.org/10.1038/sdata.2016.44 (2016).
Acknowledgements
We thank all participants for their contributions. We thank Dr Fleur Garton for advice on the statistical analysis and selection of visual stimuli. We also thank the radiology and professional staff at the Herston Imaging Research Facility, National Imaging Facility, UQ Centre for Clinical Research, and UQ Research Computing Centre. The authors acknowledge the valuable funding of Wesley Medical Research (project No. 2017-07) and The Faculty of Medicine (The University of Queensland, Brisbane). JC is supported by the UQ Graduate School Scholarship (RTP) and the MND Research Australia PhD Scholarship Top-up Grant. STN acknowledges support through the Scott Sullivan MND Research Fellowship (University of Queensland, RBWH Foundation, and the MND and Me Foundation) and a FightMND Mid-Career Fellowship. TS is supported by a Motor Neurone Disease Research Australia (MNDRA) Postdoctoral Research Fellowship (PDF2112).
Author information
Authors and Affiliations
Contributions
F.J.S., S.T.N., R.D.H., P.A.M., and C.C.G. obtained funding for the study. F.J.S., S.T.N., J.L. and C.C.G. designed the study. F.J.S. and S.T.N. managed the project and oversaw study ethics. F.J.S., S.T.N., R.D.H., P.A.M. and D.L. conducted data collection, and J.C., T.B.S. and K.G. conducted data analysis. J.C. and T.B.S. wrote the code. J.C. wrote the manuscript and generated all figures. T.B.S., J.C., F.J.S. and S.T.N. edited the manuscript. All authors revised and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chang, J., Lv, J., Guo, C.C. et al. An fMRI dataset for appetite neural correlates in people living with Motor Neuron Disease. Sci Data 12, 466 (2025). https://doi.org/10.1038/s41597-025-04828-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-04828-w