Abstract
The elemental content of life is a key trait shaping ecology and evolution, yet organismal stoichiometry has largely been studied on a case-by-case basis. This limitation has hindered our ability to identify broad patterns and mechanisms across taxa and ecosystems. To address this, we present StoichLife, a global dataset of 28,049 records from 5,876 species spanning terrestrial, freshwater, and marine realms. Compiled from published and unpublished sources, StoichLife documents elemental content and stoichiometric ratios (%C, %N, %P, C:N, C:P, and N:P) for individual plants and animals. The dataset is standardized and, where available, includes information on taxonomy, habitat, body mass (for animals), geography, and environmental conditions such as temperature, solar radiation, and nutrient availability. By providing an unprecedented breadth of organismal stoichiometry, StoichLife enables the exploration of global patterns, ecological and evolutionary drivers, and context-dependent variations. This resource advances our understanding of the chemical makeup of life and its responses to environmental change, supporting progress in ecological stoichiometry and related fields.
Similar content being viewed by others
Background & Summary
Owing to their shared evolutionary history, all living organisms possess a common biochemistry of 28 naturally occurring chemical elements1,2. These elements play essential roles in core biological processes, including the storage of genetic information, metabolic regulation, mechanical support, and protective mechanisms3,4,5. Despite this universal elemental composition, the relative proportions of these elements —referred to as stoichiometry—vary both within and among species3,6. Such stoichiometric variation underlies key functional traits related to resource uptake, assimilation, storage, and release7,8,9, reflecting the evolution of diverse life-history strategies shaped by organismal morphology and function. Furthermore, stoichiometric variation may reflect ecological adaptations to the environmental conditions in which different organisms have evolved3,4,10,11.
The framework of ecological stoichiometry3 has driven extensive research into how organisms acquire, store, and transfer nutrients essential for growth and reproduction across diverse environments. Despite significant advancements, progress has been hindered by the limited availability and synthesis of elemental content data for plants and animals across broad spatial and taxonomic scales, as well as insufficient integration between ecological and evolutionary mechanisms (but see6,12). Therefore, a comprehensive understanding of spatiotemporal patterns and underlying mechanisms governing stoichiometric diversity is crucial. Beyond explaining observed patterns in organismal elemental content, such insights would enhance our ability to predict how organisms will respond to ongoing and future environmental changes, particularly under intensifying global change drivers.
To address this critical gap, we introduce StoichLife — the first global dataset for biogeographical and macroevolutionary patterns in organismal stoichiometry, developed within the sBIOMAPS working group at iDiv in Germany13. StoichLife provides openly accessible data without restrictions, though we kindly ask users to acknowledge this paper when using the database. The database aims to advance research in ecological stoichiometry, functional biogeography, macroecology, and macroevolution. Additionally, StoichLife remains open to integrating new data and future updates.
Methods
Data compilation
We developed the StoichLife template structure, which contains data pertaining to elemental content and their ratios (%C, %N, %P, C:N, C:P, and N:P) alongside body size measurements, and information regarding sampling locality, country, and taxonomic affiliation (i.e., phylum, class, order, family, species or morphospecies). This template was distributed among prospective data contributors actively engaged in sampling and analyzing plant and animal elemental content across diverse regions worldwide (between 2014 and 2022). As part of this effort, we engaged 24 researchers who contributed 50 datasets on animals (both invertebrates and vertebrates), including 15 previously published datasets14,15,16,17,18,19,20,21,22,23,24,25,26,27,28, one dataset from two sources29,30, and 34 unpublished datasets (details of collection outlined in Supplementary Text 1; refs. 31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55). Additionally, we incorporated data from six large databases used in prior studies, comprising one zooplankton dataset56, 20 aquatic animal datasets57, five datasets for both animals and plants10, three datasets for coral reef macroalgae (Pangaea database58), 80 datasets for green leaves59, and 35 datasets for plants60. Datasets were included based on the following criteria: (i) elemental analyses were conducted on individual organisms under natural conditions, excluding those subjected to experimental manipulations such as nutrient enrichment; (ii) for animals, analyses were performed on whole-body (bulk) tissue, while for plants, stoichiometry was primarily assessed through leaf or shoot elemental composition; and (iii) georeferenced coordinates of sampling sites were available to facilitate spatial analyses.
Data search
To complement data contributors, we conducted a systematic literature review to identify ecological stoichiometry studies published before 2021. Using Clarivate Analytics’ Web of Science Core database, we employed a set of search terms: “nutrient content” OR “nutrient composition” OR “elemental content” OR “elemental composition” OR “chemical composition” OR “nitrogen content” OR “nitrogen composition” OR “phosphorus content” OR “phosphorus composition” OR “percent nitrogen” OR “percent phosphorus” OR “N:P” OR “nitrogen-to-phosphorus” OR “ecological stoichiometry”.
This rigorous search yielded a total of 2,620 papers, which were further filtered to exclude: (i) microbial data, which typically represent analyses of entire microbial communities rather than individual cells; (ii) studies involving laboratory or field experiments; and (iii) literature reviews and opinion papers. Following this refinement, 110 eligible papers remained. After thoroughly examining these papers, we narrowed the selection down to 33 papers that met our criteria8,41,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91. In cases where archived datasets lacked essential details (e.g., species-mean resolution), we contacted authors to request raw data. Additionally, we cross-referenced all identified datasets within Andrieux et al.92, the most comprehensive synthesis study on animal stoichiometry to date. However, no additional datasets meeting our criteria were found through this process.
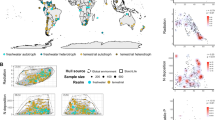
StoichLife consolidates 227 datasets, comprising an unprecedented 28,049 individual records from both published and unpublished sources13. At the time of data compilation (2022/09/19): 14% of datasets (n = 31) and 26% of records (n = 7,361) were unpublished. StoichLife contains data on plant and animal elemental content (%C, %N, and %P; Fig. 1) from terrestrial, freshwater, and marine realms. Spanning a broad geographical extent from 68°S (Antarctica) to 81°N (Arctic) and from 160°W (Hawaii, USA) to 177°E (New Zealand), StoichLife encompasses a diverse array of taxonomic groups, including plants and animals, across realms (Fig. 1). Despite its inherent limitations, the StoichLife dataset is the most comprehensive compilation of animal and plant data from terrestrial, freshwater, and marine realms across the globe to date (Figs. 1 and 2).
Workflow and structure of the StoichLife dataset. The database includes 227 plant and animal elemental content datasets across terrestrial, freshwater, and marine realms. StoichLife comprises 38 variables, including environmental descriptors related to temperature, solar radiation, and environmental N and P availability. These were compiled from the literature using bioinformatic approaches to allow for reproducibility, consistency, and efficiency. Each data type was checked and validated using the R programming language (see Methods - Data processing section).
Data Processing
Dataset checking, cleaning, and formatting
Three distinct data types were processed: quantitative, taxonomic, and spatial. These data underwent rigorous validation and quality assurance procedures using the R software93. The quantitative data include elements such as individual %C and element ratios (e.g., C:N) and individual body mass measurements (dry mass). Elemental content values were verified to represent the percentage of each element in dry body mass, while elemental ratios were checked to ensure they accurately reflected both mass and molar ratios.
The taxonomic data encompass classifications ranging from species and morphospecies to higher taxonomic ranks, including families, orders, classes, phylum, and kingdoms. Data validation involved both automated and manual inspection to correct spelling errors, complete missing taxonomic information where feasible, address ambiguously identified morphospecies (e.g., “Geophilidae,” “Psychodidae sp.1”), and ensure the accuracy of currently accepted names. In cases where two distinct morphospecies with identical names appeared in different datasets (e.g., “Psychodidae sp.1”), we assigned unique identifiers to distinguish them (e.g., “Psychodidae sp.1_A” and “Psychodidae sp.1_B” in the two datasets). Taxonomic affiliations were validated using the Global Biodiversity Information Facility (GBIF), Integrated Taxonomic Information System (ITIS), and Catalogue of Life (COL) databases via the taxadb-package (version 0.1.594) in R. Additionally, plant taxonomy was verified using the Plants of the World online website (https://powo.science.kew.org/; Fig. 1).
To standardize taxonomic names across datasets, we applied a set of harmonization criteria. First, when missing taxonomic information was identified and multiple synonyms were available, we retained the accepted name based on established taxonomic databases (GBIF, ITIS, and COL). Second, taxonomic synonyms found between original publications and taxonomic databases were standardized to their accepted names. Third, in cases where taxonomic information in original sources aligned with taxonomic information from GBIF, ITIS, or COL, we prioritized GBIF as the primary reference unless substantial discrepancies were found. Finally, when inconsistencies remained unresolved after validation (e.g., differing taxonomic information between the original publication and taxonomic sources), we deferred to the taxonomic information provided by the original publications or data contributors. The StoichLife dataset preserves both the initial taxonomic classifications provided by contributors and the revised taxonomy to maintain transparency and facilitate future updates.
The spatial data include latitude and longitude coordinates of sampling locations. These coordinates underwent extensive validation through visual inspection, where they were plotted onto a global map to identify and correct any spatial errors. Common discrepancies, such as marine data mistakenly recorded as inland or vice versa, were rectified using geographical details provided in the original publications (Fig. 1). These steps ensured that spatial data were accurate and georeferenced correctly, enhancing their utility for ecological and biogeographical analyses.
StoichLife comprises 38 variables (columns) and 28,049 individual records (rows; Supplementary Table 1). Each record corresponds to at least one measurement of one elemental content or ratio taken at the level (i.e., %C, %N, %P, C:N, C:P, and N:P) of an individual organism. Records in the StoichLife dataset are distributed across 1,120 locations, each associated with both latitude and longitude coordinates (n = 23,290 records; Fig. 2; Supplementary Table 1).
In addition, 3,616 records contain only latitudinal information, while 1,143 records lack spatial information entirely. The dataset spans terrestrial (n = 16,832), freshwater (n = 8,935), and marine (n = 2,282) realms. Most records originate from the northern hemisphere (n = 19,268) compared to the southern hemisphere (n = 7,638; Fig. 2). Geographically, the dataset exhibits extensive coverage in Europe, the Americas, East Asia, and Eastern Australia, with notable gaps observed in Africa (excluding South Africa), the Middle East, Central and Southeast Asia, Western Australia, and Russia (Fig. 2). In the marine realm, Oceania (Western Pacific) and the Eastern Indian Ocean are underrepresented.
The revised taxonomy within StoichLife comprises 5,876 species (65.4%) and morphospecies (34.6%), spanning 837 families, 208 orders, 50 classes, 16 phyla, and 2 kingdoms. Animalia is the most extensively documented kingdom (n = 19,664 entries), followed by Plantae (n = 8,385 entries; Fig. 3; Supplementary Table 1). The percentage of taxa identified at the species level is higher in plants (92.4% of all plant taxa) than in animals (27.7% of all animal taxa). Two major classes, Insecta (Animalia) and Magnoliopsida (Plantae), account for approximately 44% of all entries in StoichLife, with 7,205 individual records for Insecta and 5,130 for Magnoliopsida (Fig. 3). Among these, Magnoliopsida has the highest number of species or morphospecies, (n = 2,420), followed by Insecta (n = 1,366) (Fig. 3).
Number of observations within taxonomic classes. Classes are sorted by decreasing number of observations. Numbers beside bars indicate the number of species or morphospecies within each class. Number of observations is given in the thousands. Classes with <10 observations are not shown (n = 13). Species or morphospecies for which the class was not identified are not shown (n = 600).
However, 30 out of 51 classes contain 50 or fewer individual records. The dataset includes 10,322 individual animal records (1,570 species or morphospecies) and 6,510 individual plant records (3,048 species or morphospecies) from the terrestrial realm. In freshwater habitats, there are 7,882 individual animal records (751 species or morphospecies) and 1,053 individual plant records (206 species or morphospecies). Meanwhile, marine data contain 1,460 individual animal records (133 species or morphospecies) and 822 individual plant records (172 species or morphospecies; primarily algae). Given that certain species inhabit several environments due to their life cycles, habitat classifications were preserved as indicated in the original sources. For example, terrestrial insects with aquatic larval stages were classified according to their primary feeding environment, distinguishing between aquatic larvae and terrestrial adults.
The number of individual records varies by element and stoichiometric ratio, with %N (n = 25,652) being the most common, followed by %C (n = 18,558), C:N (n = 18,373), %P (n = 14,457), N:P (n = 12,244), and C:P (n = 7,092). Only 7,091 individual records (25.3% of all entries) contain values for all three elements (i.e., %C, %N, and %P), with the majority belonging to animals (n = 5,635; 20.1% of all records) rather than plants (n = 1,456; 5.2% of all records). Overall, StoichLife provides a wide range of elemental values and their ratios (Figs. 4 and 5; Supplementary Table 1): %C [0.6–78.1], %N [0.06–19.5], %P [0.004–8.9], C:N molar [2.07–223.3], C:P molar [7.4–9154.7], and N:P molar [0.2–789.4].
Distribution of elemental ratios for the 16 phyla included in the database. Figures on top of each boxplot indicate the number of observations. Species or morphospecies for which the phylum was not identified are not shown (n = 7). For the sake of visual clarity, values above 100, 3000, and 400 were removed for C:N (n = 8), C:P (n = 17), and N:P (n = 5), respectively.
These values exhibit substantial variations across taxonomic and trophic groups (i.e., phyla; Figs. 4 and 5) and realms (i.e., freshwater, marine, and terrestrial; Fig. 6).
Body mass data, measured as whole-organismal dry mass in g, were recorded for 9,942 individual animals. The two most represented taxa were Arthropoda (n = 6,552) and Chordata (n = 2,868; Fig. 7), with smaller contributions from Mollusca (n = 149), Annelida (n = 132), Nematoda (n = 62), Platyhelminthes (n = 58), Chaetognatha (n = 48), Cnidaria (n = 35), Acanthocephala (n = 27), and Ctenophora (n = 11). Terrestrial animals accounted for 3,885 individual records, while 5,347 originated from freshwater and 710 from marine realms. Body dry mass values ranged from <0.001 g (copepod nauplii) to over 800 g (Salmo salar Linnaeus, 1758; Supplementary Table 1), demonstrating substantial variation across taxa (Fig. 7).
To facilitate investigations into environmental drivers of organism elemental content, StoichLife integrates information on environmental factors such as air temperature, solar radiation, as well as environmental nitrogen and phosphorus availability. Mean annual air temperature at 10 m above ground or sea surface (T10M; °C; 0.5° × 0.5° resolution) and solar radiation data (ALLSKY_SFC_SW_DWN: All Sky Insolation Incident on a Horizontal Surface; W/m2; 1° × 1° resolution) from each sampling site were extracted from the National Aeronautics and Space Administration Prediction of Worldwide Energy Resources project (NASA POWER; https://power.larc.nasa.gov). Both temperature and solar data have temporal coverage from 1981 to 2022/09.
Global nitrogen (N) availability at each sampling site, represented by inorganic N deposition (kg N/km2/ year1; resolution of 2° × 2.5°); was retrieved from a published database95 covering the period from 1984 to 2016). Soil phosphorus (P) data (P labile; g P/m2; resolution 0.5° × 0.5° resolution) were extracted from the Oak Ridge National Laboratory Distributed Active Archive Center for Biogeochemical Dynamics (ORNL DAAC; https://daac.ornl.gov). While these soil P data lack specific temporal coverage, they are nominally representative of pre-industrial conditions ca. 1850. Additionally, marine total P data (sea surface P measurement; micromoles P/kg; resolution 1° × 1° resolution were sourced from the National Center for Atmospheric Research World Ocean Atlas (NCAR WOA; https://climatedataguide.ucar.edu; WOA13), covering the period from 1955 to 2012. To ensure consistency across datasets, we computed the average values for each of these four environmental factors across the respective temporal windows. This approach was chosen for several reasons: (1) our primary objective was to examine spatial rather than temporal variations in organismal elemental content; (2) samples were collected at different times by various researchers, and specific sampling dates were often unavailable; and (3) the environmental data were measured or modeled over different temporal timeframes, making direct temporal alignment impractical.
Data Record
The dataset is provided as an Excel file at Dryad13, which includes key information such as elemental content in % of dry mass, spatial coordinates, the biological level of organization (e.g., organ- or individual-level measurement, mean per species or population), taxonomic information (e.g., from species or morphospecies to kingdom), realm identity, the ontogenetic stage when possible (e.g., larvae, juvenile, adult, seed, sprout). Additionally, the dataset incorporates any other relevant information provided by contributors. A metadata file ‘StoichLife_metadata.xlsx’ accompanies the dataset, detailing column headings, measurement units, and descriptions of numerical variables. This file also includes a comprehensive list of data sources used in compiling the database, ensuring full transparency and reproducibility.
Technical Validation
Each dataset underwent thorough inspection using R (version 4.0.0 2020–04–2493) to ensure data integrity and consistency. We generated histograms and estimated value ranges for each element and elemental ratio to detect extreme values, potential outliers, or measurement errors. In most cases, extreme values were retained unless they were deemed implausible based on clear indications of measurement errors. Only four values were excluded due to implausibility: one C value exceeding 80%, one N value below 0.01%, and two P values exceeding 20%. Importantly, original datasets with missing information were not excluded from the StoichLife dataset. Some datasets contain missing data for specific variables, such as sampling location, specifically longitude, body mass, and trophic group (Supplementary Table 1). These gaps are retained to preserve data availability and allow future users to apply imputation or additional validation as needed.
References
da Silva, J. J. R. F. & Williams, R. J. P. The Biological Chemistry of the Elements: The Inorganic Chemistry of Life. (OUP Oxford, 2001).
Kaspari, M. & Powers, J. S. Biogeochemistry and Geographical Ecology: Embracing All Twenty-Five Elements Required to Build Organisms. Am Nat 188 (Suppl 1), S62–73 (2016).
Sterner, R. W. & Elser, J. J. Ecological Stoichiometry: The Biology of Elements from Molecules to the Biosphere. (Princeton University Press, 2002).
Kay, A. D. et al. Toward a stoichiometric framework for evolutionary biology. Oikos 109, 6–17 (2005).
Jeyasingh, P. D., Cothran, R. D. & Tobler, M. Testing the ecological consequences of evolutionary change using elements. Ecology and Evolution 4, 528–538 (2014).
González, A. L., Dézerald, O., Marquet, P. A., Romero, G. Q. & Srivastava, D. S. The Multidimensional Stoichiometric Niche. Frontiers in Ecology and Evolution 5 (2017).
Leal, M. C., Seehausen, O. & Matthews, B. The Ecology and Evolution of Stoichiometric Phenotypes. Trends in Ecology & Evolution 32, 108–117 (2017).
González, A. L. et al. Ecological mechanisms and phylogeny shape invertebrate stoichiometry: A test using detritus-based communities across Central and South America. Functional Ecology 32, 2448–2463 (2018).
Meunier, C. L. et al. From Elements to Function: Toward Unifying Ecological Stoichiometry and Trait-Based Ecology. Front. Environ. Sci. 5 (2017).
Elser, J. J. et al. Nutritional constraints in terrestrial and freshwater food webs. Nature 408, 578–580 (2000).
Kay, A. D. & Vrede, T. Ecological Stoichiometry: Evolutionary and biochemical aspects. https://researchonline.stthomas.edu/esploro/outputs/bookChapter/Ecological-Stoichiometry-Evolutionary-and-biochemical-aspects/991015132038903691 (2008).
Fagan, W. F. et al. Nitrogen in Insects: Implications for Trophic Complexity and Species Diversification. The American Naturalist 160, 19 (2002).
González, A. L. et al. StoichLife: A global database of plant and animal elemental content [Dataset]. Dryad https://doi.org/10.5061/dryad.3tx95x6r2 (2025).
Korotkevich, A. Y., Potapov, A. M., Tiunov, A. V. & Kuznetsova, N. A. Collapse of trophic-niche structure in belowground communities under anthropogenic disturbance. Ecosphere 9, e02528 (2018).
Scheunemann, N., Digel, C., Scheu, S. & Butenschoen, O. Roots rather than shoot residues drive soil arthropod communities of arable fields. Oecologia 179, 1135–1145 (2015).
Scheunemann, N., Scheu, S. & Butenschoen, O. Incorporation of decade old soil carbon into the soil animal food web of an arable system. Applied Soil Ecology 46, 59–63 (2010).
Scheunemann, N. et al. Incorporation of root C and fertilizer N into the food web of an arable field: Variations with functional group and energy channel. Food Webs 9, 39–45 (2016).
Pollierer, M. M., Langel, R., Scheu, S. & Maraun, M. Compartmentalization of the soil animal food web as indicated by dual analysis of stable isotope ratios (15N/14N and 13C/12C). Soil Biology and Biochemistry 41, 1221–1226 (2009).
Potapov, A. A., Semenina, E. E., Korotkevich, A. Y., Kuznetsova, N. A. & Tiunov, A. V. Connecting taxonomy and ecology: Trophic niches of collembolans as related to taxonomic identity and life forms. Soil Biology and Biochemistry 101, 20–31 (2016).
Klarner, B. et al. Trophic shift of soil animal species with forest type as indicated by stable isotope analysis. Oikos 123, 1173–1181 (2014).
Klarner, B. et al. Trophic niches, diversity and community composition of invertebrate top predators (Chilopoda) as affected by conversion of tropical lowland rainforest in Sumatra (Indonesia). PLOS ONE 12, e0180915 (2017).
Seeber, J. et al. Abundance and trophic structure of macro-decomposers on alpine pastureland (Central Alps, Tyrol): effects of abandonment of pasturing. Pedobiologia 49, 221–228 (2005).
Seeber, J., Langel, R., Meyer, E. & Traugott, M. Dwarf shrub litter as a food source for macro-decomposers in alpine pastureland. Applied Soil Ecology 41, 178–184 (2009).
Goncharov, A. A., Tsurikov, S. M., Potapov, A. M. & Tiunov, A. V. Short-term incorporation of freshly fixed plant carbon into the soil animal food web: field study in a spruce forest. Ecological Research 31, 923–933 (2016).
González, A. L. et al. Bromeliad growth and stoichiometry: responses to atmospheric nutrient supply in fog-dependent ecosystems of the hyper-arid Atacama Desert, Chile. Oecologia 167, 835–845 (2011).
Rizzuto, M. et al. Patterns and potential drivers of intraspecific variability in the body C, N, and P composition of a terrestrial consumer, the snowshoe hare (Lepus americanus). Ecology and Evolution 9, 14453–14464 (2019).
Ludwig, L., Barbour, M. A., Guevara, J., Avilés, L. & González, A. L. Caught in the web: Spider web architecture affects prey specialization and spider–prey stoichiometric relationships. Ecology and Evolution 8, 6449–6462 (2018).
Mozsár, A., Sály, P., Antal, L., Nagy, S. A. & Boros, G. Organismal stoichiometry at the temporal scale: Seasonal variability shapes interspecific differences in fish. Freshwater Biology 64, 244–254 (2019).
Jochum, M. et al. Decreasing Stoichiometric Resource Quality Drives Compensatory Feeding across Trophic Levels in Tropical Litter Invertebrate Communities. The American Naturalist 190, 131–143 (2017).
Drescher, J. et al. Ecological and socio-economic functions across tropical land use systems after rainforest conversion. Philosophical Transactions of the Royal Society B: Biological Sciences 371, 20150275 (2016).
Chemical Analysis of Ecological Materials: With Special Contrib. on Organic Pesticides by John A. Parkinson, X-Ray Fluorescence Spectrometry by Colin C. Evans,Data Processing and Statistical Analysis by H. Max Grimshaw David K. Lindley. (Blackwell, Oxford, 1974).
Fassbender, H. W. Simulate P-Bestinmung in N-Kjeldahl Ausfschlub von Bodenproben. Die phosphörsäure 30, 44–53 (1973).
Filipiak, M., Woyciechowski, M. & Czarnoleski, M. Stoichiometric niche, nutrient partitioning and resource allocation in a solitary bee are sex-specific and phosphorous is allocated mainly to the cocoon. Sci Rep 11, 652 (2021).
Hood II JM. Consumer nutrient stoichiometry: patterns, homeostasis, and links with fitness. (University of Minnesota, Minnesotta, USA, 2010).
Jackson, M. C. et al. Food web properties vary with climate and land use in South African streams. Functional Ecology 34, 1653–1665 (2020).
Jackson, M. C. et al. Trophic overlap between fish and riparian spiders: potential impacts of an invasive fish on terrestrial consumers. Ecology and Evolution 6, 1745–1752 (2016).
Harrower WL. Changes in trophic structure along a gradient of water availability in temperate montane grasslands. (University of British Columbia, Vancouver, Canada, 2017).
Nash, L. et al. Tropical and temperate differences in the trophic structure and aquatic prey use of riparian predators. 1423245 bytes Dryad https://doi.org/10.5061/DRYAD.HT76HDRNB (2023).
Nash, L. N. et al. Tropical and temperate differences in the trophic structure and aquatic prey use of riparian predators. Ecology Letters 26, 2122–2134 (2023).
Ebel, J. D., Leroux, S. J., Robertson, M. J. & Dempson, J. B. Ontogenetic differences in Atlantic salmon phosphorus concentration and its implications for cross ecosystem fluxes. Ecosphere 6, 1–18 (2015).
Ebel, J. D., Leroux, S. J., Robertson, M. J. & Dempson, J. B. Whole body-element composition of Atlantic salmon Salmo salar influenced by migration direction and life stage in three distinct populations. Journal of Fish Biology 89, 2365–2374 (2016).
Ebel, J., Leroux, S., Robertson, M. & J. Brian Dempson. Atlantic salmon %P and ecosystem flux. 205866 Bytes figshare https://doi.org/10.6084/M9.FIGSHARE.1266298 (2014).
Moody, E. K. et al. Data from: Consumption explains intraspecific variation in nutrient recycling stoichiometry in a desert fish. 57972 bytes Dryad https://doi.org/10.5061/DRYAD.7F753F0 (2019).
Moody, E. K. et al. Consumption explains intraspecific variation in nutrient recycling stoichiometry in a desert fish. Ecology 99, 1552–1561 (2018).
Moody, E. K., Lujan, N. K., Roach, K. A. & Winemiller, K. O. Data from: Threshold elemental ratios and the temperature dependence of herbivory in fishes. 56629 bytes Dryad https://doi.org/10.5061/DRYAD.D5F6H00 (2019).
Benke, A. C., Huryn, A. D., Smock, L. A. & Wallace, J. B. Length-Mass Relationships for Freshwater Macroinvertebrates in North America with Particular Reference to the Southeastern United States. Journal of the North American Benthological Society 18, 308–343 (1999).
American Water Works Association (AWWA) and Water Pollution Control Federation (WPCF). Washington DC. APHA Standard Methods for the Examination of Water and Wastewater. 18th Edition, American Public Health Association. (1992).
Alfaro Ayllon F. Causes and consequences of nitrogen limited chronosequences: evidence from the andean dry puna. (Pontificia Universidad de Chile, Santiago, Chile, 2014).
Verardo, D. J., Froelich, P. N. & McIntyre, A. Determination of organic carbon and nitrogen in marine sediments using the Carlo Erba NA-1500 analyzer. Deep Sea Research Part A. Oceanographic Research Papers 37, 157–165 (1990).
Paseka R. Linking host-parasite interactions and ecosystem processes with energy and elements. (Rutgers University, NJ, USA, 2018).
Langel, R. & Dyckmans, J. Combined 13C and 15N isotope analysis on small samples using a near‐conventional elemental analyzer/isotope ratio mass spectrometer setup. Rapid Comm Mass Spectrometry 28, 1019–1022 (2014).
Recalde, F. C., Postali, T. C. & Romero, G. Q. Unravelling the role of allochthonous aquatic resources to food web structure in a tropical riparian forest. Journal of Animal Ecology 85, 525–536 (2016).
Thomisch K. Studiengang Biologische Diversität und Ökologie Struktur des Bodennahrungsnetzes in einem Pflanzendiversitätsgradienten vorgelegt von Matrikelnummer. (Georg-August-Universität Göttingen, Göttingen, Germany, German).
Murphy, J. & Riley, J. P. A modified single solution method for the determination of phosphate in natural waters. Analytica Chimica Acta 27, 31–36 (1962).
Stainton MP, Capel MJ, & Armstrong FAJ. The Chemical Analysis of Fresh Water. (1974).
Ikeda, T. & Skjoldal, H. R. Metabolism and elemental composition of zooplankton from the Barents Sea during early Arctic summer. Marine Biology 100, 173–183 (1989).
Vanni, M. J. et al. A global database of nitrogen and phosphorus excretion rates of aquatic animals. Ecology 98, 1475–1475 (2017).
Lovelock, C. E., Reef, R. & Pandolfi, J. M. Regional variation in δ13C of coral reef macroalgae on the Great Barrier Reef. Supplement to: Lovelock, CE et al. (accepted): Regional variation in δ13C of coral reef macroalgae. Global Ecology and Biogeography PANGAEA https://doi.org/10.1594/PANGAEA.900711 (2019).
Vergutz, L., Manzoni, S., Porporato, A., Novais, R. F. & Jackson, R. B. A Global Database of Carbon and Nutrient Concentrations of Green and Senesced Leaves. ORNL DAAC https://doi.org/10.3334/ORNLDAAC/1106 (2012).
Borer, E. T. et al. Global biogeography of autotroph chemistry: is insolation a driving force? Oikos 122, 1121–1130 (2013).
Benstead, J. P. et al. Biotic and abiotic controls on the ecosystem significance of consumer excretion in two contrasting tropical streams. Freshwater Biology 55, 2047–2061 (2010).
Bertram, S. M., Bowen, M., Kyle, M. & Schade, J. D. Extensive natural intraspecific variation in stoichiometric (C:N:P) composition in two terrestrial insect species. Journal of Insect Science 8, 26 (2008).
Christian, A. D., Crump, B. G. & Berg, D. J. Nutrient release and ecological stoichiometry of freshwater mussels (Mollusca:Unionidae) in 2 small, regionally distinct streams. Journal of the North American Benthological Society 27, 440–450 (2008).
Cross, W. F., Benstead, J. P., Rosemond, A. D. & Bruce Wallace, J. Consumer-resource stoichiometry in detritus-based streams. Ecology Letters 6, 721–732 (2003).
Dierenfeld, E. S., Norton, T. M., Hyslop, N. L. & Stevenson, D. J. Nutrient Composition of Prey Items Consumed by Free-Ranging Drymarchon couperi (Eastern Indigo Snakes). Southeastern Naturalist 14, 551–560 (2015).
Durston, D. J. & El-Sabaawi, R. W. Bony traits and genetics drive intraspecific variation in vertebrate elemental composition. Functional Ecology 31, 2128–2137 (2017).
El-Sabaawi, R. W. et al. Widespread intraspecific organismal stoichiometry among populations of the Trinidadian guppy. Functional Ecology 26, 666–676 (2012).
El-Sabaawi, R. W., Warbanski, M. L., Rudman, S. M., Hovel, R. & Matthews, B. Investment in boney defensive traits alters organismal stoichiometry and excretion in fish. Oecologia 181, 1209–1220 (2016).
Filipiak, M. et al. Ecological stoichiometry of the honeybee: Pollen diversity and adequate species composition are needed to mitigate limitations imposed on the growth and development of bees by pollen quality. PLOS ONE 12, e0183236 (2017).
Filipiak, M., Sobczyk, Ł. & Weiner, J. Fungal Transformation of Tree Stumps into a Suitable Resource for Xylophagous Beetles via Changes in Elemental Ratios. Insects 7, 13 (2016).
Filipiak, M. & Weiner, J. How to Make a Beetle Out of Wood: Multi-Elemental Stoichiometry of Wood Decay, Xylophagy and Fungivory. PLOS ONE 9, e115104 (2014).
Filipiak, M. & Weiner, J. Nutritional dynamics during the development of xylophagous beetles related to changes in the stoichiometry of 11 elements. Physiological Entomology 42, 73–84 (2017).
Follum, O. A. & Gray, J. S. Nitrogenous excretion by the sediment-living bivalve Nucula tenuis from the Oslofjord, Norway. Mar. Biol. 96, 355–358 (1987).
Frost, P. C., Tank, S. E., Turner, M. A. & Elser, J. J. Elemental Composition of Littoral Invertebrates from Oligotrophic and Eutrophic Canadian Lakes. Journal of the North American Benthological Society 22, 51–62 (2003).
Gong, X. et al. Spatial Patterns of Leaf Carbon, Nitrogen, and Phosphorus Stoichiometry of Aquatic Macrophytes in the Arid Zone of Northwestern China. Front. Plant Sci. 9 (2018).
Hambäck, P. A. et al. Effects of body size, trophic mode and larval habitat on Diptera stoichiometry: a regional comparison. Oikos 118, 615–623 (2009).
Horn, S. & de la Vega, C. Relationships between fresh weight, dry weight, ash free dry weight, carbon and nitrogen content for selected vertebrates. Journal of Experimental Marine Biology and Ecology 481, 41–48 (2016).
Huang, L., Wu, Y., Wan, R. & Zhang, J. Carbon, nitrogen and phosphorus stoichiometry in Japanese anchovy (Engraulis japonicus) from the Huanghai Sea, China. Acta Oceanol. Sin. 31, 154–161 (2012).
Jankowski, T. Chemical composition and biomass parameters of a population of Craspedacusta sowerbii Lank1880 (Cnidaria: Limnomedusa). Journal of Plankton Research 22, 1329–1340 (2000).
Jansen, H. M., Verdegem, M. C. J., Strand, O. & Smaal, A. C. Seasonal variation in mineralization rates (C-N-P-Si) of mussel Mytilus edulis biodeposits. Mar Biol 159, 1567–1580 (2012).
Leal, M. C., Best, R. J., Durston, D., El-Sabaawi, R. W. & Matthews, B. Stoichiometric traits of stickleback: Effects of genetic background, rearing environment, and ontogeny. Ecology and Evolution 7, 2617–2625 (2017).
Li, Z. et al. Incorporation of root-derived carbon into soil microarthropods varies between cropping systems. Biol Fertil Soils 56, 839–851 (2020).
Moody, E. K., Lujan, N. K., Roach, K. A. & Winemiller, K. O. Threshold elemental ratios and the temperature dependence of herbivory in fishes. Functional Ecology 33, 913–923 (2019).
Moody, E. K., Rugenski, A. T., Sabo, J. L., Turner, B. L. & Elser, J. J. Does the Growth Rate Hypothesis Apply across Temperatures? Variation in the Growth Rate and Body Phosphorus of Neotropical Benthic Grazers. Front. Environ. Sci. 5 (2017).
Paseka, R. E. & Grunberg, R. L. Allometric and trait-based patterns in parasite stoichiometry. Oikos 128, 102–112 (2019).
Prater, C. et al. Understanding variation in salamander ionomes: A nutrient balance approach. Freshwater Biology 64, 294–305 (2019).
Small, G. E. & Pringle, C. M. Deviation from strict homeostasis across multiple trophic levels in an invertebrate consumer assemblage exposed to high chronic phosphorus enrichment in a Neotropical stream. Oecologia 162, 581–590 (2010).
Sullivan, M. L., Zhang, Y. & Bonner, T. H. Carbon and nitrogen ratios of aquatic and terrestrial prey for freshwater fishes. Journal of Freshwater Ecology 29, 259–266 (2014).
Tande, K. S. Ecological investigations on the zooplankton community of Balsfjorden, northern Norway: Generation cycles, and variations in body weight and body content of carbon and nitrogen related to overwintering and reproduction in the copepod Calanus finmarchicus (Gunnerus). Journal of Experimental Marine Biology and Ecology 62, 129–142 (1982).
Woods, H. A., Fagan, W. F., Elser, J. J. & Harrison, J. F. Allometric and phylogenetic variation in insect phosphorus content. Functional Ecology 18, 103–109 (2004).
Zhang, G. & Han, X. N:P stoichiometry in Ficus racemosa and its mutualistic pollinator. Journal of Plant Ecology 3, 123–130 (2010).
Andrieux, B., Signor, J., Guillou, V., Danger, M. & Jabot, F. Body stoichiometry of heterotrophs: Assessing drivers of interspecific variations in elemental composition. Global Ecology and Biogeography 30, 883–895 (2021).
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. (2020).
Norman, K. E. A., Chamberlain, S. & Boettiger, C. taxadb: A high-performance local taxonomic database interface. Methods in Ecology and Evolution 11, 1153–1159 (2020).
Ackerman, D., Millet, D. B. & Chen, X. Global Estimates of Inorganic Nitrogen Deposition Across Four Decades. Global Biogeochemical Cycles 33, 100–107 (2019).
González, A. L. et al. StoichLife: A global dataset of plant and animal elemental content. Zenodo https://doi.org/10.5281/ZENODO.13384716 (2025).
Acknowledgements
This paper is a joint effort of the working group sBiomaps kindly supported by sDiv, the Synthesis Centre of the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig, funded by the German Research Foundation (FZT 118, 202548816). Members of sBiomaps include ALG, OD, JM, KA, UB, MF, MJa, Mjo, WSH, HH, SJL, MPN, REO, GLWP, RP, AR, JS, ES, MS, and EZ. This research was partially supported by a grant to ALG from the National Science Foundation (NSF: DEB-1754326). MF was supported by the Jagiellonian University, Faculty of Biology, Institute of Environmental Sciences (N18/DBS/000022). HH was supported by the German Research Foundation (DFG HI848/26–2), SJL by the Natural Sciences and Engineering Research Council of Canada, Discovery Grant (RGPIN-2020-04132), JSs by the Research Foundation Flanders (FWO), EZ by the FAPERJ JCNE (E-26/201.286/2021, E-26/203.213/2017), NE by the DFG FOR 5000, VFF was partially supported by the CNPq-Brazil productivity grant (310119/2018-9), JH by the Federal Aid in Sport Fish Restoration Program (F-69-P, Fish Management in Ohio) administered jointly by the U.S. Fish and Wildlife Service and the ODNR-DOW (project FADX09), PK by the Royal Society, Newton Advanced Fellowship (NAF\R2\180791), CL by the Australian Research Council awards (DP0986179 and DP160103669), AMP by the German Research Foundation (DFG, German Research Foundation) –(DFG 493345801 and FZT 118, 202548816, GQR was supported by the FAPESP grant (2019/08474-8) and by the CNPq-Brazil productivity grant (310119/2018-9), AT was supported by the Russian Science Foundation (22-14-00363).
Author information
Authors and Affiliations
Contributions
A.L.G. and O.D. wrote the first draft of the manuscript with inputs from the sBiomaps working group https://www.idiv.de/en/sbiomaps.html. O.D. and A.L.G. generated all analyses and graphs. A.L.G. and O.D. compiled the StoichLife database and obtained the iDiv funding that supported sBiomaps. A.L.G., O.D., K.A., U.B., M.F., H.H., W.S.H., M.Ja, M.Jo, M.P.N., R.E.O., G.L.W.P., R.P., M.S., A.R., E.S., J.S., S.J.L. and E.Z. cleaned the database and compiled environmental data. A.L.G., O.D., J.M., K.A., U.B., M.F., H.H., W.S.H., M.Ja, M.Jo, M.P.N., R.E.O., G.L.W.P., R.P., M.S., A.R., E.S., J.S., S.J.L. and E.Z. participated in one or more of the three sBiomaps workshops at iDiv where the sBiomaps initiative was organized, and the evaluation of the data and first drafts were discussed. All other authors contributed data. All authors contributed to writing and editing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
González, A.L., Merder, J., Andraczek, K. et al. StoichLife: A Global Dataset of Plant and Animal Elemental Content. Sci Data 12, 569 (2025). https://doi.org/10.1038/s41597-025-04852-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-04852-w
This article is cited by
-
Predicting carbon storage in North American maritime boreal forests under combined disturbances
Landscape Ecology (2025)
-
Nitrogen deposition reveals global patterns in plant and animal stoichiometry
Nature Communications (2025)