Abstract
Tackling the current global biodiversity crisis requires large-scale spatially accurate biodiversity data to rapidly assess knowledge gaps and set conservation priorities. Obtaining such data is often challenging because surveying biodiversity across broad spatial scales requires massive logistical and economic efforts. Here, we provide high-resolution (0.81 to 81 km2, depending on species ecology) habitat suitability raster maps for all 225 widespread breeding bird species in Italy. Maps were generated by means of species distribution models based on ~2.5 million spatially accurate (≤1 km-scale) and expert-validated occurrence records. Occurrence data were collected during the breeding seasons 2010–2016 by over 3000 skilled observers, mostly through the Ornitho.it web platform, with the aim of realizing the second Atlas of Breeding Birds in Italy, released in 2022. These raster maps will be useful to ecologists, conservation scientists and practitioners for investigating broad spatial patterns in avian diversity and identifying conservation priorities. We discuss potential applications of this dataset for inferring the composition of ecological communities and species distributions at the Italian scale.
Similar content being viewed by others
Background & Summary
The ongoing biodiversity crisis is leading to alarming rates of population decline and species extinction1,2. Actions and policies are needed to address biodiversity losses through mitigating impacts, and where possible reversing population declines by restoring degraded natural habitats or improving habitat quality. However, their effectiveness is often hampered by deficient data and knowledge gaps regarding basic aspects of the ecology of declining species (e.g. demography and distribution)3. Indeed, obtaining accurate spatial data on biodiversity is challenging because of the relatively limited economic resources allocated to conservation science4 and the high costs and massive logistical effort that biodiversity data collection at broad spatial scales requires5.
Hence, large-scale biodiversity assessments are often difficult to perform, especially for environmentally heterogeneous areas with variable levels of accessibility6. This is often the case even for well-known taxa, such as birds, for which most information is concentrated in densely sampled areas, while others lack adequate coverage7,8,9. In those contexts, it is possible to capitalise on data available from highly investigated areas to derive information about the possible occurrence of species also in less densely monitored areas through adequate quantitative approaches.
The development of Species Distribution Models (SDMs) in the last few decades has indeed promoted quantitative estimations of species range based on presence probabilities or environmental suitability. SDMs (also called Environmental Niche Models, Environmental or Habitat Suitability Models) link the presence (geographical locations) of a species to environmental variables within a study area for which such variables are spatially explicit and provide an effective measure of the environmental suitability of the reference area for the investigated species10. This enables not only improved estimation of current distributions, but also potential predictions of temporal variations in distributions due to climate change11,12,13 changes in land use, disturbance14,15 or other anthropogenic pressures.
The use of SDMs has increasingly become standard practice in many conservation applications. Examples include defining areas of major importance for conservation16, inferring ecological networks17 and future variations in connectivity18, and improving the interpretation of demographic trends19. In some cases, relevant demographic parameters, such as the abundance or reproductive success of a species, strongly correlate with environmental suitability calculated from SDMs20,21,22.
SDMs allow inferring species distributions over large areas, using standardised survey data23 or opportunistic datasets24, drastically reducing the costs and efforts required to obtain comparable information through dedicated large-scale surveys. Hence, SDMs have been increasingly adopted for biodiversity mapping by inferring the potential distribution of species9,25,26, at least partly ‘filling’ knowledge gaps by estimating environmental suitability (or the probability of presence) in unsurveyed/poorly surveyed areas. SDMs are highly flexible and can be based both on presence-absence data27 or presence-only data28.
Here, we provide habitat suitability maps for 225 breeding bird species in Italy, i.e. all widespread bird species breeding in the country, corresponding to ~83% of the total number of breeding species (Fig. 1). These suitability maps have a species-specific spatial resolution, with three different scales: 0.81 km2 (0.9 × 0.9 km; 176 species showing small-size breeding homeranges), 9 km2 (3 × 3 km; 45 species with larger home ranges), and 81 km2 (9 × 9 km; four species regularly exploiting very large areas during the breeding period). The models used for producing these maps were based on data collected for the second Italian breeding bird atlas by over 3,000 skilled observers, mainly through the Ornitho.it web portal (www.ornitho.it)9.
Synthetic workflow for generating habitat suitability maps for 225 widespread breeding bird species in Italy with data collected for realizing the second Italian breeding bird atlas9.
We encourage researchers and policymakers to use these high-resolution raster files for e.g. improving biodiversity monitoring schemes, evaluating the effectiveness of the protected areas network, identifying priority areas for conservation, landscape planning, and characterising threats to Italian biodiversity, including the assessment of current and future interactions with human infrastructures (e.g. roads, powerlines, wind power generation infrastructures29,30,31). In Usage Notes, we discuss potential applications of the dataset and call for careful use of habitat suitability values, while in the Supplementary Information 1 we provide an example of the use of the dataset for investigating the macroecological drivers of avian species richness in Italian urban areas.
Methods
Italian breeding bird atlas and occurrence data
Bird taxonomy followed HBW-BirdLife International32, adopted also for the checklist of Italian birds33 and the European Atlas EBBA225. Most occurrence data for the second Italian breeding bird atlas were bird observations/counts collected through the Ornitho.it web portal by over 3,000 skilled observers (2,360,284 records), but included also data collected through other methods (e.g., ringing), standardised monitoring or regional survey projects (630,312 records; see Lardelli et al.9 for further details). The data were used to generate distribution maps at the 10 km scale9 [Universal Transverse Mercator (UTM) projection system, 10 × 10 km grid cells].
The vast majority of Ornitho.it occurrence data were occasional observations, followed by complete lists of species recorded over variable (and generally unknown) spatial extents. Such heterogeneous data had been previously used to develop robust SDMs34. Among data derived from sources different than Ornitho.it, 80% originated from the Farmland Bird Index project35, a large-scale and country-wide monitoring initiative based on point counts and contributing to the PanEuropean Common Bird Monitoring Scheme (www.pecbms.info).
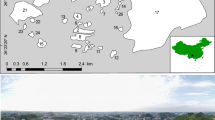
The database contained records of avian species with reproductive evidence (possible, probable or confirmed breeding; behavioural or physiological, including territorial behaviour, nest building, observations of fledged nestlings, etc.; reproductive evidence was assigned by observers in the field according to the standard European Bird Census Council methodology, see https://ebba2.info/about/methodology/) collected during the breeding seasons spanning from 2010 to 2016. Data archived in Ornitho.it are subjected to regular expert validation on a regional/local basis, and were subjected to additional quality controls before generating both distribution maps and SDMs. We excluded incomplete or incorrect data36, including those supported by weak breeding evidence for colonial and rare species. Only spatially accurate occurrence records (see Data selection, spatial scales and background points) were used for building SDMs and realising habitat suitability maps for all widespread species (see Lardelli et al.9). Figure 2 shows the distribution of five sample species at the 10 km scale9.
Original bird occurrence data used for generating distribution maps and building SDMs could not be made publicly available due to restrictions on their public release by data owners (i.e. individual observers or entities contributing data for realizing the atlas), especially for conservation-reliant taxa or those highly sensitive to human disturbance (see Lardelli et al.9).
Modelling habitat suitability
Due to the nature of the occurrence data (i.e., presence-only9), SDMs were built using MaxEnt37,38, the most effective method for modelling this kind of data39. Known drawbacks of using MaxEnt40,41 have been dealt with through careful selection of background points and parametrization and tuning of models (see below). MaxEnt relies on background data to define available environmental conditions for the target species within the study region. We parametrized MaxEnt models accounting for non-uniform sampling and optimized model complexity by carefully selecting environmental variables, in order to obtain robust and generalizable results20,42,43,44,45. The following paragraphs summarise the approach adopted for model development, from the preparation of input data to model validation (Fig. 1).
Cumulative distribution (grey dots) at the 10 km scale of a sample of five common Italian breeding species (Parus major, Sturnus vulgaris, Anthus spinoletta, Corvus cornix, Sylvia melanocephala) representative of different habitats, species groups or biogeographical regions (respectively, forest, farmland, mountain, generalist and Mediterranean species), accounting for 10.7% of records (UTM grid), highlighting the relatively homogeneous spatial coverage of the dataset used for producing the second Italian breeding bird atlas9 and species distribution models.
Data selection, spatial scales and background points
To build SDMs, we used occurrence data with high spatial accuracy (<1 km; i.e., excluding records associated with broader areas, such as municipalities or protected areas, depending on how data were uploaded on Ornitho.it). These were assigned to 1 × 1 km UTM grid cells. Eventually, the final database consisted of 2,577,222 unique occurrence records (i.e., including only a single breeding record per species per cell).
Considering the substantial differences in species’ ecology, spatial scales for SDM development were appropriately differentiated in order to assess the relationships between species and the environment at the scale at which they are most ecologically relevant46. For instance, small passerines are influenced by environmental variables at a very local scale (i.e., thousands of square metres or a few hectares47). By contrast, large raptors are influenced by environmental characteristics of much larger territories48. Given the resolution of both bird occurrence (1 × 1 km UTM grid) and environmental data (see ‘Environmental variables’), we set the lowest spatial scale at 0.81 km². Additionally, two broader spatial scales were identified, taking into account the average home range sizes of the analysed species. The exact scale at which models were implemented depended on the exact number of cells that, under a focal feature approach (see ‘Environmental variables’), provided the closest match to the desired scale. The three spatial scales considered for environmental variables were: ‘micro’ (variables computed at 0.81 km² resolution), ‘meso’ (variables computed at 9 km² resolution), and ‘mega’ (variables computed at 81 km² resolution). Additional details on generated maps are provided in Supplementary Table 1.
The generation of background is a fundamental aspect for developing realistic and robust SDMs, and it could be even more important in the case of modelling citizen science data unevenly collected over broad areas49. For instance, setting background points in unsurveyed areas could result in the environmental conditions observed at those points being considered as unsuitable for a species due to the lack of observations, rather than representing actual counter-selection for those conditions. We managed to limit as much as possible the effects of undersampling in the definition of background. To avoid including unsurveyed areas, we used the centroids of effectively sampled 1 × 1 km UTM cells as background points. For diurnal species, given the significantly larger number of observations (2,542,772 data points) compared to nocturnal species (34,450 data points), we chose as background points the centroids of the most intensively sampled cells (considering only those with at least 20 occurrence records), whereas for nocturnal species we selected all sampled cells. This resulted in 30,180 background points for diurnal species and 14,672 for nocturnal ones.
Environmental variables
The environmental variables used in the models belonged to three distinct groups: climatic, topographic, and land use. Climatic variables (1981–2010) were retrieved from the CHELSA (https://chelsa-climate.org) database at a 30 arc sec resolution (less than 1 km at the study latitude)50. Even if the period to which climatic variables refer does not perfectly overlap with the bird occurrence data, it is likely still highly representative of the relative effect of predictors, and definitely relevant to understand the indirect effect of climate on bird distribution, through e.g. effects on vegetation characteristics51, which are more dependent on long-term climate rather than on year to year weather variations. Several bioclimatic variables, summarising monthly values to obtain biologically meaningful predictors commonly used in SDMs52, were calculated. As bioclimatic variables from the CHELSA database were not available for open water bodies (coastal lagoons), the values of bioclimatic variables for cells with these habitats were recalculated from the original climate data using CHELSA database algorithms53.
Topographic variables were calculated through GRASS54 from a digital elevation model (DEM; https://www.eea.europa.eu/en/datahub/datahubitem-view/d08852bc-7b5f-4835-a776-08362e2fbf4b) with a resolution of 25 m. These were slope (in degrees) and solar radiation (total daily value, in kWh/m2) for summer solstice, considering the shading effect of relief. Land use/land cover variables were obtained from the CORINE Land Cover database55. The 2012 edition was chosen as it best overlapped with the occurrence data collection period (i.e. 2010–2016).
Environmental variables were calculated for raster grids of 100 × 100 m and 1 × 1 km. Environmental variables were attributed to cells using a ‘focal features’ (or ‘moving window’) approach, which aggregates information from neighbouring cells at the pixel level56. For each focal cell (i.e. the raster cell where the centroid of the UTM cell associated with observations or the background point fell), we calculated a summary value of each environmental variable within a neighbourhood of surrounding cells (Fig. 3). At each spatial scale, environmental variables were measured in a neighbourhood of focal cells established to approximate as much as possible the spatial scale of bird data. At the micro-scale, 9 × 9 cells of 100 m each were considered, covering a total area of 0.81 km² within which to calculate the values of environmental variables associated with the focal cell. At the meso-scale, 3 × 3 cells of 1 km each were considered (total 9 km²), while at the mega-scale, predictor values were calculated using 9 × 9 cells of 1 km each (total 81 km²). This approach allowed us to best handle calculations of environmental variables for those UTM grid cells that spanned across different UTM zones, which had slightly irregular and deformed shapes. Furthermore, it improved the ecological realism of bird-habitat associations, especially at the broader spatial scales.
Schematic representation of the focal features (moving window) approach for calculating environmental variables at the ecologically relevant scale for each species: for each cell (focal cell, in darker colour), the values of different environmental variables were computed for the surrounding cells (in light colour). The arrows show the directions over which the number of cells were counted for the computation at the micro-scale (9 × 9 cells of 100 m each; see the text for the other scales).
Training and test datasets
To develop robust and generalizable SDMs, it is crucial to assess their predictive capacity on independent datasets. To this end, a fraction of the data was not used for model building and kept as a test dataset to evaluate model performance. To select spatially independent data for the test dataset, we relied on the ‘checkerboard 2’ function of the ENMeval R package43. Checkerboard 2 was chosen to obtain subgroups of occurrence datasets that were spatially independent but still representative of the entire study region, as well as to obtain a desirable subdivision of occurrence records between training and testing data. Only presence data, not background data, were split between training and test datasets. At each scale, the procedure produced a ‘double checkerboard’, with 10 and 2 as grouping factors at the two levels, at each scale. This resulted in four different subsets of data. Three subsets constituted the training dataset, while the remaining subset was used as the test dataset. This process yielded two spatially independent sets of data, corresponding roughly to ¾ and ¼ of the original data (training and test datasets, respectively).
Model building
To reduce the number of variables included in the models and to avoid collinearity, sets of environmental variables were defined for different species groups based on their ecology. Hence, different environmental variables were considered for forest, farmland, wetland, mountain, and generalist species (Supplementary Table 2).
Although machine learning methods (such as MaxEnt) are much less sensitive to the effects of correlated predictors compared to classical statistical methods, including strongly collinear predictors may lead to unpredictable effects in the extrapolation phase45. Therefore, at each spatial scale and for each subgroup of variables identified according to species’ ecology, correlations among all predictors were assessed and we avoided including collinear variables in models as described in step 4 below.
Model calibration was performed on the training dataset and aimed at identifying the best combination of model parameters for each species. Accurately parameterizing models yields significant improvements in model predictions57, prevents overfitting and increases the ecological relevance of species-habitat relationships, resulting in robust and effective predictions of species distributions.
MaxEnt models for each species were tuned as described below:
-
1.
functions for species-habitat relationships: to avoid overfitting, we considered only linear or quadratic effects of environmental predictors, i.e. simple fitting functions that can be easily evaluated in terms of ecological realism. This prudential approach may slightly reduce the model’s accuracy on training data, but it reduces the risk of considering unlikely species-habitat relationships, inconsistent with real ecological effects.
-
2.
number of iterations: the number of iterations was set to 1,000; if the model converges earlier, the actual number will be smaller, otherwise it will continue to seek convergence until that value. In fact, for all species, the number of iterations in the final model was smaller.
-
3.
value of regularization multiplier: the regularization multiplier is a crucial parameter for SDMs, as it determines whether distributions are more fragmented or more homogeneous, relaxing or tightening the effect of environmental parameters on suitability. The selection of the most suitable value was performed by testing values from 0.5 to 4, in 0.5 steps (i.e. 8 values)58. The AICc value (Akaike’s Information Criterion, corrected for small samples59,60) was then calculated for each model, and the value producing the most parsimonious model was chosen.
-
4.
selection of environmental variables: after step 3), we first calculated, for all cells with at least one occurrence record at the relevant spatial scale, the correlation among all possible pairs of environmental predictors, in order to consider a large and representative set of environmental conditions. We then identified pairs of variables that were highly correlated (|r| ≥ 0.8) (a threshold similar to that recommended by Dormann et al.61 and Feng et al.45) (three pairs at the micro scale, and two at the broader scales). For each pair of highly correlated variables, we built SDMs by including each predictor singly, keeping for subsequent analyses only the predictor leading to the most supported model (lower AICc). We then built a model including all non-collinear predictors. We simplified this model by first removing any variable with a lambda coefficient (i.e. index for variables’ contribution in predicting distribution) equal to zero, and hence irrelevant. Finally, a variable selection procedure based on AICc was performed: for each remaining predictor, its permutation importance (importance of the specific factor in explaining the species’ distribution according to MaxEnt) was calculated. The variable with the lowest permutation importance value was removed from the model, and the AICc was calculated; if the model improved (i.e., the AICc decreased), we continued with removing the least important variable until the model showed no further improvements in AICc. The resulting model was considered as the final model for each species. For the calculation of AICc, a recently developed ad hoc method was used34.
Data Records
The habitat suitability maps for 225 widespread breeding bird species in Italy during 2010-2016, including SDM statistical outputs, is publicly available at https://doi.org/10.13130/RD_UNIMI/LUC3K662.
Suitability maps are provided as raster (.tif) files. The raster file names include the EURING species codes (see https://euring.org/data-and-codes/euring-codes) and the first three letters of genus and species (e.g. “E00070_Tac.ruf.tif” for the little grebe Tachybaptus ruficollis).
The statistical outputs corresponding to each species’ SDM are available as folders, named with the scientific name of the species. Folders contain: (1) model evaluation (.csv file); (2) model results (.csv file); (3) permutation importance of used environmental predictors (.csv file); (4) barplot of the five most important predictors (.jpg); and (5) all the response plots to environmental predictors (.jpg). For ease of visualisation, we provide a single PDF file with the response plots for the five most important predictors for each species, as well as the barplot with their permutation importance (Supplementary Information 2).
Species-specific threshold values, Maximum Training Sensitivity plus Specificity (MTSS) and 10th percentile, useful for deriving binary predictions of species occurrence63 are reported in Supplementary Table 3.
Technical Validation
Model evaluation and validation
We tested the robustness of models for each species using the test dataset. Statistics used for the evaluation and validation were calculated using the final models based on both the training and the test datasets. The most important aspect of validation was the consistency of predictive ability on both the training and test datasets64. Comparable values indicate a generalizable model unconstrained from overfitting issues and able to predict environmental suitability in sites not used for its construction, as well as in those used for its development.
To evaluate model performance, four reference statistics were considered:
-
1.
AUC (Area Under the Curve of the Receiver Operating Characteristic plot), which assesses the discriminatory ability of a model65. Values equal to 0.5 represent a chance-level performance, while 1 indicates a perfect ability to distinguish between presences and background points. In absolute terms, AUC is not a good measure of model accuracy, as rare species tend to have higher AUC values than common ones66; therefore, we mainly used AUC to compare values calculated on training and test datasets for the same species. A model can be regarded as valid and generalizable when similar AUC values for the training and test datasets are obtained (difference < 0.05).
-
2.
TSS (True Skill Statistic), which compares the number of correct predictions, minus those attributable to chance, to those of a hypothetical set of perfect predictions (defined as sensitivity + specificity - 1). TSS ranges from −1 to +1, with the maximum value indicating a perfect match and zero indicating performance equal to chance67. Differences in the values of TSS between test and training dataset models >0.05 suggest possible overfitting issues.
-
3.
Minimum training presence omission rate on test dataset, which evaluates the proportion of occurrences included in the test dataset falling below the lower suitability value at which the species occurs, based on the locations used to develop the model (training dataset). Ideally, it should be zero or close to zero (no or a very few test locations occurring at suitability values lower than the minimum values recorded at training locations).
-
4.
10th percentile omission rate on the test dataset, which evaluates the proportion of occurrences included in the test dataset falling below the threshold value of the 10th percentile from occurrences of the training dataset. Ideally, it should be close to 10% of records. Values higher than 0.1 (e.g. >0.2–0.3) indicate likely overfitting issues.
Validation statistics for all SDMs are reported in Supplementary Table 4. The minimum sample size used for implementing SDMs was set at 50 occurrence records for non-colonial species and 20 for colonial ones (for which occurrence records are necessarily scarcer because of the aggregated distribution). In the case of a few species for which models were based on a very low sample of occurrence records [i.e. Mediterranean gull (Larus melanocephalus), n = 20; Eurasian spoonbill (Platalea leucorodia), n = 30; Savi’s warbler (Locustella luscinioides), n = 67; great spotted cuckoo (Clamator glandarius), n = 78], the evaluation statistics were unreliable and model validity was instead visually assessed on the basis of the concordance between predicted and observed distributions. Overall, the average difference between training and testing dataset was 0.00 ± 0.03 for TSS (mean ± SD; only 10 species showed a difference >0.05 but always <0.09) and 0.00 ± 0.01 for AUC (only one species showed a difference >0.05, being 0.06), indicating very good performances for this indicator. Also, omission rates at minimum training presence and 10th percentile generally showed optimal values, with a few exceptions mostly related to species with a low sample of occurrence records. Furthermore, the reliability of each final model was verified by means of visual check of the resulting habitat suitability map, conducted by species’ experts (see Lardelli et al.9).
Restriction of environmental suitability predictions to presence-only areas
Due to biogeographical reasons, not all species may actually occur in all areas classified as suitable by distribution models68. Given the main purpose of these models (i.e. assisting in defining species distribution, even in poorly investigated areas), we excluded these regions from environmental suitability maps, setting environmental suitability to zero in areas where a given species has never been recorded. To this end, environmental suitability maps were intersected with actual distribution maps in order to exclude regions located outside a given species’ distribution range from potentially suitable sites. For instance, the Eurasian nuthatch (Sitta europaea) does not breed in Sardinia due to biogeographical/historical reasons, hence we set suitability to zero there despite suitable woodland habitats being identified by SDMs. The types of correction applied to environmental suitability maps based on species distribution were as follows (see Supplementary Table 1 for details and species concerned):
-
1.
Exclusion of mainland Italy from suitable areas for species limited to the islands.
-
2.
Exclusion of Sardinia and/or Sicily and/or adjacent smaller islands from suitable areas for species breeding only on the Italian mainland.
-
3.
Restriction to a buffer surrounding actual occurrence sites: for species with a concentrated distribution but not falling into any of the previous cases, an informative layer representing the actual range was generated. For most species, a distance of 200 km (empirically set based on previous experience) around the presence sites was used; for some species with a particularly restricted or concentrated distribution (e.g. some grouse and owl species occurring only in the Alps), such distance was set to 50 km. For species distributed throughout Italy, even if scattered, no range restriction was applied.
Interpretation of environmental suitability maps
The raster maps indicated the environmental suitability for each species based on the environmental suitability model. The reported values in each raster’s cell (obtained through the cloglog transformation of raw MaxEnt output) range from zero (i.e. unsuitable) to one (i.e. maximum suitability). For a correct interpretation of the habitat suitability maps, it should be kept in mind that: (1) they represent habitat suitability, not a true species’ probability of presence, given that absence sites are unavailable and prevalence is unknown69; although these two variables are highly correlated and the cloglog transformation is meant to approximate the occurrence probability, an environmental suitability of 0.5 does not (necessarily) correspond to a 50% probability of species presence; (2) they do not represent abundance, even though there is published evidence of positive correlations between environmental suitability and local density20,21.
Usage Notes
The habitat suitability raster maps we have generated may be used for a variety of applied or theoretical purposes. Suitability rasters can allow a rapid evaluation of potential occurrence of species that are of conservation relevance, sensitive to disturbance or alteration, for management and planning purposes. Similarly, they can be used to identify species-rich and priority areas for different groups of species70, or to model community patterns at varying spatial scales, up to the national level. For instance, in the Supplementary Information 1, we provide and thoroughly discuss a study case focusing on patterns and drivers of bird species richness in Italian urban areas.
When using SDMs to predict species occurrence or community traits, it is fundamental to bear in mind some practical aspects related to both the intrinsic characteristics of the models and the data used to generate them. First, even if the sampling coverage was generally satisfactory at the national level (see Fig. 2), at smaller scales survey intensity was much more heterogeneous. Hence, we cannot exclude that some extrapolation over unsampled combinations of environmental predictors may have occurred: if any, such cases are likely to be extremely limited, but care should be applied when interpreting habitat suitability values at local scales. At these scales, other factors not taken into account in the modelling procedure (e.g., interspecific interactions, local disturbance, availability of key specific resources, etc.) may also become increasingly relevant in determining the actual occurrence or absence. Second, we did not explicitly model spatial autocorrelation. Therefore, we cannot exclude that for some species models might not be the best performing ones (and resulting spatial suitability patterns the most accurate) because of unmodelled spatial patterns in the data. This is somehow exemplified by the latitudinal effect in inferred species richness that was detected among Italian urban bird communities (see Supplementary Information 1). Third, the selection of thresholds for binary classification and generating predicted distributions is somehow species- and context-dependent. In our worked example, we found that MTSS led to more reliable outcomes than 10th percentile, but different thresholds may be more suited depending on model, species and also purposes of the reclassification. Fourth, the buffer distances we used to exclude unoccupied areas from models’ predictions (200 km and 50 km buffers) might potentially exclude some occupied areas, or conversely include unoccupied ones. A careful investigation of local contexts and of actual, updated information on regional species distribution are required to properly evaluate models’ outcomes towards the margins of ‘cropped’ suitability. Fifth, habitat selection often occurs at multiple spatial scales71. Unfortunately, testing environmental drivers of habitat suitability at multiple scales that are ecologically relevant with available data (presence-only citizen science data at a 1 km-resolution) was unfeasible, given that for most species habitat selection works at finer spatial scales. However, for some species models would have been even more informative and representative with the integration of multiple spatial scales, and the latter could be something to pursue in other applications where it is possible to collect records at very fine scales. Finally, we point out that we built SDMs with different spatial scales and specific combinations of environmental predictors according to broad ecological groups of species (Supplementary Table 2). Although we calibrated and tuned models according to species-specific results, we adopted the same modelling framework for all species assigned to a given ecological group. It could be argued that for some species different approaches might lead to better results, both for model construction and for subsequent cropping of suitable areas using the predefined buffers.
References
Butchart, S. H. et al. Global biodiversity: indicators of recent declines. Science 328(5982), 1164–1168, https://doi.org/10.1126/science.1187512 (2010).
Cowie, R. H., Bouchet, P. & Fontaine, B. The Sixth Mass Extinction: fact, fiction or speculation. Biol. Rev. 97(2), 640–66, https://doi.org/10.1111/brv.12816 (2022).
Kindsvater, H. K. et al. Overcoming the data crisis in biodiversity conservation. Trends Ecol. Evol. 33(9), 676–688, https://doi.org/10.1016/j.tree.2018.06.004 (2018).
Gardner, T. A. et al. The cost‐effectiveness of biodiversity surveys in tropical forests. Ecol. Lett. 11(2), 139–150, https://doi.org/10.1111/j.1461-0248.2007.01133.x (2008).
Marta, S., Lacasella, F., Romano, A. & Ficetola, G. F. Cost-effective spatial sampling designs for field surveys of species distribution. Biodivers. Conserv. 28(11), 2891–2908, https://doi.org/10.1007/s10531-019-01803-x (2019).
Callaghan, C. T., Poore, A. G., Hofmann, M., Roberts, C. J. & Pereira, H. M. Large-bodied birds are over-represented in unstructured citizen science data. Sci. Rep. 11(1), 19073, https://doi.org/10.1038/s41598-021-98584-7 (2021).
Leitão, P. J., Moreira, F. & Osborne, P. E. Effects of geographical data sampling bias on habitat models of species distributions: a case study with steppe birds in southern Portugal. Int. J. Geogr. Inf. Syst. 25(3), 439–454, https://doi.org/10.1080/13658816.2010.531020 (2011).
Meschini, E. & Frugis. S. Atlante degli uccelli nidificanti in Italia. Supplemento Ricerche Biologia della Selvaggina, vol. XX, (1993).
Lardelli, R. et al. Atlante degli uccelli nidificanti in Italia. Edizioni Belvedere. (2022).
Engler, J. O. et al. Avian SDMs: current state, challenges, and opportunities. J. Avian Biol. 48(12), 1483–1504, https://doi.org/10.1111/jav.01248 (2017).
Huntley, B., Collingham, Y. C., Willis, S. G. & Green, R. E. Potential impacts of climatic change on European breeding birds. PloS one 3(1), e1439, https://doi.org/10.1371/journal.pone.0001439 (2008).
Thuiller, W., Guéguen, M., Renaud, J., Karger, D. N. & Zimmermann, N. E. Uncertainty in ensembles of global biodiversity scenarios. Nat. Commun. 10, 1–9, https://doi.org/10.1038/s41467-019-09519-w (2019).
Ferrer Obiol, J. et al. Evolutionarily distinct lineages of a migratory bird of prey show divergent responses to climate change. Nat Commun 16, 3503, https://doi.org/10.1038/s41467-025-58617-5 (2025).
Regos, A. et al. Predicting the future effectiveness of protected areas for bird conservation in Mediterranean ecosystems under climate change and novel fire regime scenarios. Divers. distrib. 22(1), 83–96, https://doi.org/10.1111/ddi.12375 (2016).
Titeux, N. et al. Biodiversity scenarios neglect future land‐use changes. Glob. Chang. Biol. 22(7), 2505–2515, https://doi.org/10.1111/gcb.13272 (2016).
Guisan, A. et al. Predicting species distributions for conservation decisions. Ecol. Lett. 16(12), 1424–1435, https://doi.org/10.1111/ele.12189 (2013).
Rödder, D., Nekum, S., Cord, A. F. & Engler, J. O. Coupling satellite data with species distribution and connectivity models as a tool for environmental management and planning in matrix-sensitive species. Environ. Manage. 58, 130–143, https://doi.org/10.1007/s00267-016-0698-y (2016).
Razgour, O. Beyond species distribution modeling: a landscape genetics approach to investigating range shifts under future climate change. Ecol. Inform. 30, 250–256, https://doi.org/10.1016/j.ecoinf.2015.05.007 (2015).
Giné, G. A. F. & Faria, D. Combining species distribution modeling and field surveys to reappraise the geographic distribution and conservation status of the threatened thin-spined porcupine (Chaetomys subspinosus). PLoS One 13(11), e0207914, https://doi.org/10.1371/journal.pone.0207914 (2018).
Brambilla, M., Bazzi, G. & Ilahiane, L. The effectiveness of species distribution models in predicting local abundance depends on model grain size. Ecology 105(2), e4224, https://doi.org/10.1002/ecy.4224 (2024).
VanDerWal, J., Shoo, L. P., Johnson, C. N. & Williams, S. E. Abundance and the environmental niche: environmental suitability estimated from niche models predicts the upper limit of local abundance. Am. Nat. 174(2), 282–291, https://doi.org/10.1086/600087 (2009).
Van Couwenberghe, R., Collet, C., Pierrat, J. C., Verheyen, K. & Gégout, J. C. Can species distribution models be used to describe plant abundance patterns? Ecography 36(6), 665–674, https://doi.org/10.1111/j.1600-0587.2012.07362.x (2013).
Brambilla, M., Gustin, M., Cento, M., Ilahiane, L. & Celada, C. Predicted effects of climate factors on mountain species are not uniform over different spatial scales. J. Avian Biol., 50(9), https://doi.org/10.1111/jav.02162 (2019).
Van Strien, A. J., Van Swaay, C. A. & Termaat, T. Opportunistic citizen science data of animal species produce reliable estimates of distribution trends if analysed with occupancy models. J. Appl. Ecol. 50(6), 1450–1458, https://doi.org/10.1111/1365-2664.12158 (2013).
Keller, V. et al. European Breeding Bird Atlas 2. Distribution, Abundance and Change. European Bird Census Council & Lynx Edicions, Barcelona. 967 pp. (2020).
Teufelbauer, N. et al. Österreichischer Brutvogelatlas 2013-2018 (1. Aufl.) - 680 S., Wien (Verlag des Naturhistorischen Museum Wien (2023).
Knaus, P. et al. Swiss Breeding Bird Atlas 2013–2016. Distribution and population trends of birds in Switzerland and Liechtenstein. Swiss Ornithological Institute, Sempach (2018).
Medrano, F., Barros, R., Norambuena, H., Matus, R. & Schmitt, F. Atlas de las aves nidificantes de Chile. Red de Observadores de Avesy Vida Silvestre de Chile, Santiago (2018).
Kroeger, S. B. et al. Impacts of roads on bird species richness: A meta-analysis considering road types, habitats and feeding guilds. Sci. Total Environ. 812, 151478, https://doi.org/10.1016/j.scitotenv.2021.151478 (2022).
Bernardino, J. et al. Bird collisions with power lines: State of the art and priority areas for research. Biol. Conserv. 222, 1–13, https://doi.org/10.1016/j.biocon.2018.02.029 (2018).
Assandri, G. et al. Assessing exposure to wind turbines of a migratory raptor through its annual life cycle across continents. Biol. Conserv. 293, 110592, https://doi.org/10.1016/j.biocon.2024.110592 (2024).
HBW & BirdLife International. Handbook of the birds of the world and BirdLife International digital checklist of the birds of the world. Ver. 5. http://datazone.birdlife.org/species/taxonomy (2020).
Baccetti, N., Fracasso, G. & COI, Commissione Ornitologica Italiana. CISO-COI Check-list of Italian birds - 2020. Avocetta, 45(1), https://doi.org/10.30456/AVO.2021_checklist_en (2021).
Brambilla, M. et al. Identifying climate refugia for high‐elevation Alpine birds under current climate warming predictions. Glob. Chang. Biol. 28(14), 4276–4291, https://doi.org/10.1111/gcb.16187 (2022).
RETE RURALE NAZIONALE & LIPU/BirdLife Italy. Farmland Bird Index nazionale e andamenti di popolazione delle specie in Italia nel periodo 2000-2023 [On-line document]. https://www.reterurale.it/en (2024).
Van Eupen, C. et al. The impact of data quality filtering of opportunistic citizen science data on species distribution model performance. Ecol. Modell. 444, 109453, https://doi.org/10.1016/j.ecolmodel.2021.109453 (2021).
Elith, J. et al. A statistical explanation of MaxEnt for ecologists. Divers. Distrib. 17(1), 43–57, https://doi.org/10.1111/j.1472-4642.2010.00725.x (2011).
Phillips, S. J., Anderson, R. P., Dudík, M., Schapire, R. E. & Blair, M. E. Opening the black box: An open‐source release of Maxent. Ecography 40(7), 887–893, https://doi.org/10.1111/ecog.03049 (2017).
Grimmett, L., Whitsed, R. & Horta, A. Presence-only species distribution models are sensitive to sample prevalence: Evaluating models using spatial prediction stability and accuracy metrics. Ecol. Modell. 431, 109194, https://doi.org/10.1016/j.ecolmodel.2020.109194 (2020).
Lissovsky, A. A. & Dudov, S. V. Species-distribution modeling: advantages and limitations of its application. 2. MaxEnt. Biol. Bull. Rev. 11(3), 265–275, https://doi.org/10.1134/S2079086421030087 (2021).
Yackulic, C. B. et al. Presence‐only modelling using MAXENT: when can we trust the inferences? Methods Ecol. Evol. 4(3), 236–243, https://doi.org/10.1111/2041-210x.12004 (2013).
Cobos, M. E., Peterson, A. T., Osorio-Olvera, L. & Jiménez-García, D. An exhaustive analysis of heuristic methods for variable selection in ecological niche modeling and species distribution modeling. Ecol. Inform. 53, 100983, https://doi.org/10.1016/j.ecoinf.2019.100983 (2019).
Muscarella, R. et al. ENM eval: An R package for conducting spatially independent evaluations and estimating optimal model complexity for Maxent ecological niche models. Methods Ecol. Evol. 5(11), 1198–1205, https://doi.org/10.1111/2041-210X.12261 (2014).
Vollering, J., Halvorsen, R., Auestad, I. & Rydgren, K. Bunching up the background betters bias in species distribution models. Ecography 42(10), 1717–1727, https://doi.org/10.1111/ecog.04503 (2019).
Feng, X., Park, D. S., Liang, Y., Pandey, R. & Papeş, M. Collinearity in ecological niche modeling: Confusions and challenges. Ecol. Evol. 9(18), 10365–10376, https://doi.org/10.1002/ece3.5555 (2019).
Holland, J. D., Bert, D. G. & Fahrig, L. Determining the spatial scale of species’ response to habitat. Biosci. 54(3), 227–233, https://doi.org/10.1641/0006-3568(2004)054[0227:DTSSOS]2.0.CO;2 (2004).
Bas, J. M., Pons, P. & Gómez, C. Home range and territory of the Sardinian Warbler Sylvia melanocephala in Mediterranean shrubland. Bird Study 52(2), 137–144, https://doi.org/10.1080/00063650509461383 (2005).
Pfeiffer, T. & Meyburg, B. U. GPS tracking of Red Kites (Milvus milvus) reveals fledgling number is negatively correlated with home range size. J. Ornithol. 156, 963–975, https://doi.org/10.1007/s10336-015-1230-5 (2015).
Fourcade, Y., Engler, J. O., Rödder, D. & Secondi, J. Mapping species distributions with MAXENT using a geographically biased sample of presence data: a performance assessment of methods for correcting sampling bias. PloS one 9(5), e97122, https://doi.org/10.1371/journal.pone.0097122 (2014).
Karger, D. et al. Climatologies at high resolution for the earth’s land surface areas. Sci. Data 4(1), 1–20, https://doi.org/10.1038/sdata.2017.122 (2017).
Ceresa, F., Kranebitter, P., Monrós, J. S., Rizzolli, F. & Brambilla, M. Disentangling direct and indirect effects of local temperature on abundance of mountain birds and implications for understanding global change impacts. PeerJ 9, e12560, https://doi.org/10.7717/peerj.12560 (2021).
Hijmans, R. J., Cameron, S. E., Parra, J. L., Jones, P. G. & Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25(15), 1965–1978, https://doi.org/10.1002/joc.1276 (2005).
Xu, T. & Hutchinson, M. F. New developments and applications in the ANUCLIM spatial climatic and bioclimatic modelling package. Environ. Model. Softw. 40, 267–279, https://doi.org/10.1016/j.envsoft.2012.10.003 (2013).
Neteler, M., Bowman, M. H., Landa, M. & Metz, M. GRASS GIS: A multi-purpose open source GIS. Environ. Model. Softw. 31, 124–130, https://doi.org/10.1016/j.envsoft.2011.11.014 (2012).
European Environment Agency. Corine Land Cover 2012 [On-line document]. https://doi.org/10.2909/a84ae124-c5c5-4577-8e10-511bfe55cc0d (2016).
Valerio, F. et al. GEE_xtract: High-quality remote sensing data preparation and extraction for multiple spatio-temporal ecological scaling. Ecol. Inform. 80, 102502, https://doi.org/10.1016/j.ecoinf.2024.102502 (2024).
Morales, N. S., Fernández, I. C. & Baca-González, V. MaxEnt’s parameter configuration and small samples: are we paying attention to recommendations? A systematic review. PeerJ 5, e3093, https://doi.org/10.7717/peerj.3093 (2017).
Brambilla, M. et al. Potential distribution of a climate sensitive species, the White-winged Snowfinch Montifringilla nivalis in Europe. Bird Conserv. Int. 30(4), 522–532, https://doi.org/10.1017/S0959270920000027 (2020).
Bartoń, K. MuMIn: Multi-model inference (1.43. 17). Vienna, Austria: The R Foundation for Statistical Computing. https://cran.r-project.org/package=MuMIn (2020).
Burnham, K. P. & Anderson, D. R. Model selection and multimodel inference. A practical information-theoretic approach. Second. NY: Springer-Verlag 63(2020), 10, https://doi.org/10.1007/b97636 (2004).
Dormann, C. F. et al. Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography 36(1), 27–46, https://doi.org/10.1111/j.1600-0587.2012.07348.x (2013).
Ilahiane, L. Replication data for: “High-resolution habitat suitability maps for all widespread Italian breeding bird species”. UNIMI Dataverse https://doi.org/10.13130/RD_UNIMI/LUC3K6 (2024).
Liu, C., White, M. & Newell, G. Selecting thresholds for the prediction of species occurrence with presence‐only data. J. Biogeogr. 40(4), 778–789, https://doi.org/10.1111/jbi.12058 (2013).
Vaughan, I. P. & Ormerod, S. J. The continuing challenges of testing species distribution models. J. Appl. Ecol. 42(4), 720–730, https://doi.org/10.1111/j.1365-2664.2005.01052.x (2005).
Fielding, A. H. & Bell, J. F. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ. Conserv. 24(1), 38–49, https://www.jstor.org/stable/44519240 (1997).
Lobo, J. M., Jiménez‐Valverde, A. & Real, R. AUC: a misleading measure of the performance of predictive distribution models. Glob. Ecol. Biogeogr. 17(2), 145–151, https://doi.org/10.1111/j.1466-8238.2007.00358.x (2008).
Allouche, O., Tsoar, A. & Kadmon, R. Assessing the accuracy of species distribution models: prevalence, kappa and the true skill statistic (TSS). J. Appl. Ecol. 43(6), 1223–1232, https://doi.org/10.1111/j.1365-2664.2006.01214.x (2006).
Elith, J. & Leathwick, J. R. Species distribution models: ecological explanation and prediction across space and time. Annu. Rev. Ecol. Evol. Syst. 40(1), 677–697, https://doi.org/10.1146/annurev.ecolsys.110308.120159 (2009).
Guillera‐Arroita, G., Lahoz‐Monfort, J. J. & Elith, J. Maxent is not a presence–absence method: a comment on Thibaud et al. Methods Ecol. Evol. 5(11), 1192–1197, https://doi.org/10.1111/2041-210X.12252 (2014).
Dalpasso, A. et al. High nature value farmlands to identify crucial agroecosystems for multitaxa conservation. Biol. Conserv. https://doi.org/10.1016/j.biocon.2025.111094 (2025).
Jedlikowski, J., Chibowski, P., Karasek, T. & Brambilla, M. Multi-scale habitat selection in highly territorial bird species: exploring the contribution of nest, territory and landscape levels to site choice in breeding rallids (Aves: Rallidae). Acta Oecol. 73, 10–20, https://doi.org/10.1016/j.actao.2016.02.003 (2016).
Brambilla, M. Species data with variables for the paper “Identifying climate refugia for high-elevation Alpine birds under current climate warming predictions”. UNIMI Dataverse https://doi.org/10.13130/RD_UNIMI/ARAI8C (2022).
Acknowledgements
We are extremely grateful to the 3075 volunteers who collected bird occurrence data, to the dozens of local and national experts who helped screening and managing the original occurrence data, to Terna S.p.A. for financially supporting the analyses, and to L. Corsetti (Edizioni Belvedere) for supporting the publication of the atlas. EC and DEC were supported by the National Biodiversity Future Centre - NBFC [funded by the European Union - NextGenerationEU under the National Recovery and Resilience Plan (NRRP) M4C2 Investment Line 1.4: “Strengthening of research facilities and creation of “national R&D champions” on some Key Enabling Technologies”, project CN_00000033]. The study was partly supported by Ecosistema MUSA – Multilayered Urban Sustainability Action (funded by the European Union – NextGenerationEU under the NRRP M4C2 Investment Line 1.5: Strenghtening of research structures and creation of R&D “innovation ecosystems”, set up of “territorial leaders in R&D”, project ECS_00000037).
Author information
Authors and Affiliations
Contributions
Brambilla, M.: Conceptualisation, Resources, Methodology, Data Curation, Formal Analysis, Writing – Original Draft, Writing – Review & Editing. Ilahiane, L.: Methodology, Data Curation, Formal Analysis, Writing – Original Draft, Writing – Review & Editing. Caprio, E.: Conceptualisation, Methodology, Data Curation, Formal Analysis, Writing – Review & Editing, Funding acquisition. Calvi, G.: Methodology, Data Curation, Formal Analysis. Lardelli, R.: Conceptualisation, Resources, Methodology, Data Curation. Bogliani, G. Conceptualisation, Resources, Methodology, Data Curation, Funding acquisition. Brichetti, P.: Conceptualisation, Resources. Celada, C.: Conceptualisation, Resources, Methodology, Data Curation, Funding acquisition. Conca, G.: Resources. Fraticelli, F.: Conceptualisation, Resources, Methodology, Data Curation, Funding acquisition; Gustin, M.: Resources, Funding acquisition. Janni, O: Resources, Data Curation. Pedrini, P.: Conceptualisation, Resources, Methodology, Data Curation, Funding acquisition; Puglisi, L.: Conceptualisation, Resources, Methodology, Data Curation, Funding acquisition, Writing – Review & Editing. Ruggieri, L.: Conceptualisation, Resources, Funding acquisition; Spina, F.: Resources, Data Curation, Writing – Review & Editing. Tinarelli, R.: Conceptualisation, Resources, Methodology, Data Curation, Funding acquisition. Chamberlain, D. E.: Methodology, Formal Analysis, Writing – Review & Editing, Funding acquisition. Rubolini, D.: Conceptualisation, Resources, Methodology, Data Curation, Formal Analysis, Writing – Original Draft, Writing – Review & Editing, Funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Brambilla, M., Ilahiane, L., Caprio, E. et al. High-resolution habitat suitability maps for all widespread Italian breeding bird species. Sci Data 12, 665 (2025). https://doi.org/10.1038/s41597-025-04973-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-04973-2