Abstract
Hyacinth (Hyacinthus orientalis L.) is a popular floricultural crop. Its cultivars exhibit a wide range of phenotypic variations, especially flower colours. Yet, the cultivar pedigree was poorly recorded, impeding efficient breeding in producing novel cultivars. In addition, scarce genomic resource of the species hinders the exploration on the molecular mechanism in controlling the diversification of floral colour. In this study, transcriptome profiling was conducted on seven hyacinth cultivars, each representing a major flower colour. RNA-Seq libraries were prepared from 189 samples that were collected in three perianth partitions at three consecutive developmental stages in biological triplicates. A total of 1,256.8 gigabytes data were generated. The reproducibility and variability of our dataset were assessed through correlation analysis and principal component analysis, respectively. In addition, the usability of the dataset was demonstrated by the identification of differentially expressed genes, functional annotation and functional enrichment analysis. This study provides the first spatiotemporal profiling of the gene expression of hyacinths, contributing to molecular breeding of hyacinth cultivars with novel flower colours.
Similar content being viewed by others
Background & Summary
Hyacinthus orientalis L., commonly known as hyacinth or garden hyacinth, is one of the most popular floricultural crops. In Angiosperm Phylogenetic Group (APG) IV system1, the species is classified under the family Asparagaecae subfamily Scilloideae, which is signatured by its storage organ as bulb, basal leave arrangement and raceme inflorescence2. With strong fragrance and brilliant flower colours, hyacinth is always a frequent candidate for potted ornamentals, cut flowers, floral arrangement and flowerbeds. Hyacinth is native to the eastern Mediterranean including countries like Turkey3, Israel4, Lebanon5, Cyprus4,6 and Syria7,8. Since its introduction to Europe in 15629, the long-lasting domestication for over 460 years has shaped this plant into diversified phenotypes, including cultivars with distinct flower colours and forms. Double-flower cultivars were once abundant in hyacinth in the 18th century10, yet most of them were unable to survive9. Nowadays, most of the existing cultivars of hyacinths are single flowered, with flower colours almost covered the spectrum of visible light which can be categorised into 8 classes - red, pink, orange, yellow, white, blue, purple and nearly black. More importantly, flower colour is still one of the major targets for breeding novel cultivar to obtain considerable revenue, as demonstrated by the case of the black-flower cultivar ‘Midnight Mystique’11.
The ancestor of modern hyacinth cultivars is far less attractive, as shown in its specimens (lectotype - G. Clifford, BM00055852712; syntype - van Royen, L005277913; specimen of wild type – BATMAN 014, Supplementary Figure S1) with only a few bell-shaped flowers in pale blue sparsely arranged on a slender spike. Spontaneous mutation and hybridisation were the major traditional breeding methods for producing new hyacinth cultivars with novel flower colours14. The order of new flower colour appearance in hyacinth cultivar was loosely recorded in the domestication history15: white (in 1582), purple (in 1596), red (in 1614) and yellow (in 1767). The power of domestication has miraculously turned this monotonous plant into a colourful floricultural crop with economic importance. Particularly, hyacinth hold the balance of power to the floricultural industry of the Netherlands, where contributes 95% worldwide production16. Noteworthily, hyacinth is the fifth best-selling bulbous plant in the global flower bulb trade14, contributing enormous economic income to the society. Additionally, hyacinth is the only bulbous floricultural crop with cultivars in diversified flower colours sharing a single protospecies as Hyacinthus orientalis L. It is contrasting to other famous bulbous ornamentals like tulips17, lilies18 and daffodils19, of which the cultivars were crossbred from a number of species. Unlocking the mechanism underlying the diversification of floral pigments is essentially critical to molecular breeding of bulbous ornamentals, which take years for the first flowering from seeds.
The wide range of flower colours in hyacinth cultivars is contributed by the flavonoids, including anthocyanins and anthoxanthins. The three anthocyanins primarily synthesised in the anthocyanin biosynthetic pathway – cyanidin, pelargonidin and delphinidin20 – are known to give magenta-red, orange-red and purplish-blue colour, respectively21. Further structural modification results in other existing forms. Peonidin in magenta is derived from cyanidin, while malvidin in purplish-blue and petunidin in dark purple or dark red are derived from delphinidin20,21. Anthoxanthins, the collective name of flavones and flavonols, give white to yellow colour or even colourlessness20.
The composition of flavonoids varies across hyacinth cultivars. In the red-flower cultivar ‘Hollyhock’, derivatives of pelargonidin and cyanidin were identified22,23,24, while in the blue-flower cultivar ‘Delft Blue’, derivatives of delphinidin, petunidin, cyanidin and pelargonidin were found25. A systematic profiling on the floral pigments of twelve hyacinth cultivars was done by Tao et al. in 201526. Pelargonidin derivatives were found in the cultivars with pink and orange flowers, whereas both pelargonidin derivatives and cyanidin derivatives were identified in cultivars with red, purple, violet and blue flowers. Flavones exist in the perianths of all studied cultivars, as reflected by the yellowish colouration in 30% ammonia. In 2019, Su et al. determined the floral pigments in 27 hyacinth cultivars by observing colour reaction in different reagents27. It was found that flavonols exist in the red flowers of ‘Jan Bos’ as 25% ammonia water turned dark yellow. Both Tao’s and Su’s studies revealed that perianths of hyacinth cultivars are free of coloured carotenoids, since no colour change was observed in petroleum ether, regardless of flower colour26,27. In addition, no anthocyanin was found in the flowers of the cultivars blossoming in yellow and white.
Application of multi-omics and advance sequencing technologies, including next-generation sequencing (NGS) and the third-generation sequencing (TGS), in studying model plants28 and other economically valuable plants29 are common. Unfortunately, hyacinth was always out of the list. Till now there is no reference genome at chromosome level assembled for hyacinth, but only an unpublished genome assembled at scaffold-level deposited in GenBank (GCA_031762755.1). Limited genomic resources were available for this understudied plant. Our group contributed the complete plastid genomes of the species in a study exploring phylogenetic relationships of seven cultivars with distinct flower colours30. However, these data cannot unlock the molecular mechanism for the diversification of floral pigmentation. More specifically speaking, which genes can affect the biosynthesis of floral pigments? How does the expression (upregulation or downregulation) of these genes contribute to the pigment composition? Would the co-expressions of these differentially expressed genes (DEGs) formulate a particular flower colour? Transcriptomic data could address these research questions and assist the breeders in creating novel hyacinth cultivars with desired and even undiscovered traits.
In 2020, Li and collaborators published the mechanism of flower colour diversification in hyacinth cultivars studied by transcriptomic technologies31. They revealed that HoF3’5’H1 and HoFOMT2 are key genes involved in peonidin synthesis in a black-flower cultivar, while HoDFR2 is related to the biosynthesis of pelargonidin in the cultivars with blue and red flowers. The transcription factor HoMYB5 is a key gene in the suppression of anthocyanin biosynthesis in a white-flower cultivar31. However, their transcriptomic dataset was prepared from a single cultivar. We therefore set forth to explore the DEGs between cultivars, developmental stages and floral partitions.
In this study, we applied top-down approach in exploring the underlying genotypes on the seven selected cultivars with different flower colours. This study was designed to provide transcriptomic data which are referrable and valuable to both scientists and breeders to explore the biosynthesis of floral pigments in hyacinth and its closely related ornamentals, contributing to the new generation of molecular breeding.
Methods
Plant materials and collection of RNA samples
Seven hyacinth cultivars with distinct flower colours were selected, namely ‘Jan Bos’ (JB) in red, ‘Pink Pearl’ (PP) in pink, ‘Gipsy Queen’ (GQ) in orange, ‘City of Haarlem’ (CH) in yellow, ‘China Pink’ (CP) in pinkish white, ‘Delft Blue’ (DB) in blue and ‘Peter Stuyvesant’ (PS) in dark purple. The bulbs were imported from the Netherlands (Simple Pleasures Flowerbulbs & Perennials Inc.) in September 2022. They were firstly immersed in tetrachlorophthalonitrile for 1 hour to prevent fungal infection. Air-dried bulbs were stored at 4 °C for three months. The vernalised bulbs were planted on 3rd January 2023, in an outdoor environment outside Li Dak Sum Yip Yio Chin R&D Centre for Chinese Medicine (GPS: 22.419292, 114.210200) with natural sunlight. Perianths were collected during 16th January to 4th February 2023.
Three consecutive developmental stages were targeted, namely the first stage with green flower buds (B), the second stage with coloured flower buds (C) and the third stage with flowers in full anthesis (A; Fig. 1a). Three perianth partitions of each flower were collected separately, namely the outer perianth lobes (o), the inner perianth lobes (n), and the perianth tube (t; Fig. 1b). The perianth partitions of a single flower were dissected with sterile blade, with removal of androecium and gynoecium using autoclaved forceps. Divided floral partitions were fragmented with sterile blade, then immediately submerged in 1.2 mL RNAlater™ Stabilization Solution (Invitrogen, MA, USA) and stored overnight at 4 °C allowing full penetration before long-term storage at −20 °C. Three biological replicates were collected for each perianth partitions at different timepoints. A total of 189 samples were collected for RNA sequencing. The voucher specimens of the seven studied cultivars (K. H. Wong 327, 328, 330, 332, 333, 335 & 336; Supplementary Figures S2–S8) were deposited in Shiu-Ying Hu Herbarium (herbarium code: CUHK), School of Life Sciences, the Chinese University of Hong Kong. Absorbent boards (catalogue number PW104) manufactured by Kunming Plantwise Biotech Co., Ltd. (Yunnan, China) were adopted for specimen pressing to retain flower pigments from degradation.
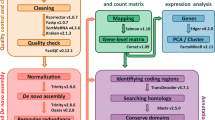
Summary of experiment design, sampling and workflow of the study. (a) Three consecutive developmental stages were targeted for the seven studied hyacinth cultivars. The flower buds of all cultivars were initially green at the first stage (B). Changes and differentiation of colour on flower buds were observed at the second stage (C). Flowers were in full anthesis at the third stage (A). The side view and top view of the flower at the third stage were documented for each cultivar, showing the colour differences of each partition. The white scale bar in each photo represents the length of 1 cm. (b) For each biological sample at each developmental stage, the perianth was further divided into three partitions, namely the outer perianth lobes (o), the inner perianth lobes (n) and the perianth tube (t). Perianth partitions are numbered as in the key. 1,2,3 - outer perianth lobes; 4,5,6 - inner perianth lobes; 7,8 - perianth tube; 2,5,8 - abaxial side; 1,3,4,6,7 - adaxial side. The white scale bar in each photo represents the length of 1 cm. A total of 189 RNA samples were collected, extracted and sequenced. The RNA-Seq libraries were sequenced on NovaSeq X Plus Platform, with a paired-end read size of 150 nucleotides. Before de novo assembly using Trinity, quality control was carried on the raw reads, during which the adaptors and low-quality reads were removed to generate clean reads. After assembly, gene expression levels were first quantified and then normalised into FKPM. Analyses including differential expression analysis, functional annotations and functional enrichment analysis were conducted.
RNA extraction, sequencing and de novo assembly
Total RNA was extracted using Qiagen RNeasy Plant Mini Kit (Qiagen Co., Hilden, Germany) following the instructions of manufacturer. The quality and quantity of extracted RNA were assessed by 1.5% agarose gel electrophoresis and Nanodrop Lite Spectrophotometer (Thermo Fisher Scientific, MA, USA), respectively. The gel records (Supplementary Figures S9–S19) and readings of spectrophotometer (Supplementary Table S1) were included in the additional information. Sequencing, de novo assembly and standard analyses of RNA-Seq libraries were done by Novogene Co., Ltd. (http://en.novogene.com/, Beijing, China). Libraries of mRNA with poly-A enrichment were sequenced on Illumnia NovaSeq X Plus platform, with paired-end sequencing of 150 nucleotides. About 6 GB data were targeted for each sample. A total of 189 RNA-Seq datasets in 1,256.8 GB were generated. Each biological replicate was given a unique sample code. For example, GQ1oA, GQ2oA, GQ3oA are the three biological replicates of the outer perianth lobes of the cultivar ‘Gipsy Queen’ collected at the third stage (flowers in full anthesis). The sample code is equivalent to the library ID of that sample (Table 1).
The raw reads were cleaned by removing the adaptor sequences, the reads with over 0.1% undetermined nucleotides and the reads with over 50 bases showing quality value less than or equal to 5. The Q20, Q30 and GC content of each RNA library are visualised in Fig. 2a. The Q20 of all libraries were above 97%, with only a few outliers showing Q30 lower than 95%. The GC contents of all libraries ranged from 48.85% to 51.55%, with an average value of 50.47%. Meanwhile, the error rate of all libraries was mostly controlled at 0.02% but not exceeding 0.03%. The quality control data of the RNA sequencing is listed in Supplementary Table S2. In addition, the quality of the bidirectional reads for each library were assessed using the software FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and the software multiQC32 were adopted to summarise the FastQC reports. The mean quality scores per nucleotide position of all sequencing reads are above the Phred score of 34.00 (Fig. 2b), and most of the sequences are in high quality reflected by the majority scored above 35.00 in Phred score (Fig. 2c). These figures support the high accuracy of the sequencing data.
Quality control of sequencing reads and de novo assembly. Quality assessment of RNA sequencing and de novo assembly. (a) The qualities of the 189 transcriptome libraries reflected by the values of Q20, Q30 and GC content. (b) Mean quality score per position of the bidirectional reads of the 189 libraries. (c) Quality score distribution over the bidirectional reads of all libraries. (d) The frequency of transcripts at different lengths. (e) The frequency of unigenes at different lengths. (f) The length distribution of assembled transcripts and unigenes categorised into different length intervals. (g) The results of BUSCO assessment on the completeness of assembled transcripts in a whole dataset (Trinity.fasta), non-redundant transcripts (cluster_all.fasta) and also the longest sequence of each cluster (unigene.fasta).
The cleaned reads were de novo assembled into transcripts using the software Trinity33. The software CORSET34 was adopted to remove the redundant transcripts, through the method of hierarchical clustering based on the proportion of shared reads and expressions. The longest transcript of each cluster was selected as the unigene. The results and quality of de novo assembly are visualised in Fig. 2d–f. The maximum length of both transcript and unigene were in 26,237 bp, and the minimum length of both were in 301 bp (Fig. 2d,e). The mean lengths of transcript and unigene were 873 bp and 809 bp, respectively. The N50s of transcript and unigene were 1,098 bp and 1,012 bp, respectively. The largest portion of transcript (36.62%) was in the range of 501–1,000 bp, while the largest portion of unigene (42.74%) fell in the interval of 301–500 bp (Fig. 2f). To evaluate the completeness of the transcript, BUSCO assessment35 was done on the whole transcriptome dataset, transcript clusters and unigenes. Before hierarchical clustering, the whole dataset has 70.7% complete transcripts, including 15.1% single and 55.6% duplicated transcripts (Fig. 2g). After clustering, both the cluster and unigene datasets show the same BUSCO values, i.e. 59.5% complete transcripts consisting of 55.3% single and 4.2% duplicated transcripts. There remained 19.7% and 20.8% transcripts that were fragmented and missing, respectively, in the cluster and unigene datasets.
Quantification and normalisation of gene expression
To quantify the expression level, the software RNA-Seq by Expectation Maximization (RSEM)36 was adopted to map the reads to a set of reference transcript sequences filtered by CORSET. The read count as the relative abundance of unigene was calculated based on the alignment of reads against the reference transcript using Bowtie37. Then, the read counts were normalised into Fragments Per Kilobase of transcript sequence per Millions (FPKM) value, which considers the effects of both sequencing depth and gene length38.
Specifying colour using RHS Colour Chart
The flower colours of each hyacinth cultivar at Stage A were specified using the Royal Horticultural Society (RHS) Colour Chart (Sixth Edition 2019 reprint). The colour of five divisions, namely the midrib on adaxial side of perianth lobes (I), the periphery on adaxial side of perianth lobes (II), the midrib on abaxial side of perianth lobes (III), the periphery on abaxial side of perianth lobes (IV), and the surface of perianth tube (V), were documented. For each cultivar, 10 flowers in full anthesis (Stage A) were collected for measuring colour, and the most frequent colour was adopted26. The results are displayed in Table 2, and the measurement records are attached as Supplementary Table S3.
Extraction of anthocyanins and flavonoids
The flowers in full anthesis (Stage A) of each cultivar were frozen in liquid nitrogen and stored at −80 °C until being used. About 1 g of perianth were ground into fine powder with liquid nitrogen, then extracted with 2 mL anthocyanin extraction buffer (AEB, Methanol:Water:Formic acid:Trifluoroacetic acid in 70∶27∶2∶1, by volume) for 24 hours under dark39. The lysates were centrifuged at maximum speed (15,000 rpm) for phase separation. The supernatants were collected and passed through a 0.45 μm filter membrane. Three biological replicates of each cultivar were extracted. Photo documentation and summary of the extracts are recorded in Supplementary Figure S21 and Supplementary Table S4, respectively. The HPLC-graded standards of anthocyanins (pelargonidin 3-O-glucoside chloride, Pg3O, cyanidin-3-O-glucoside chloride, Cy3O and delphinidin-3-O-glucoside chloride, Dp3O) and flavonoids (kaempferol, quercetin and myricetin) were purchased from Shanghai Yuanye Bio-Technology Co., Ltd (Shanghai, China).
Data Records
The data underlying this article are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database under the accession number SRP48179440. Each independent RNA-Seq library are under the accession number of SRR27431542 to SRR27431691 and SRR27433589 to SRR27433627 (Table 3). The gene expression data derived from our original transcriptomic datasets are now deposited in NCBI Gene Expression Omnibus (GEO) under the accession number GSE28640641. The results of de novo assembly, quantification of gene expression level, differential expression analysis, functional annotation, functional enrichment analysis, CDS prediction, detection of SSR, SNP and InDels, together with the supplementary files, were deposited in CUHK Research Data Repository (https://researchdata.cuhk.edu.hk/dataverse/hyacinth_transcriptome)42 and available to the public.
Technical Validation
Correlation analysis was conducted to assess the reproducibility of dataset. As shown in Fig. 3a, the biological replicates of each cultivar at different developmental stages and perianth partitions were closely related to each other (coefficient of determination R2 close to 1), reflecting high similarities between the replicates. In addition, similarities were also observed between developmental stages and cultivars. Except ‘Peter Stuyvesant’, the replicates of Stage C and Stage A for all other cultivars showed a stronger correlation. For ‘Peter Stuyvesant’, the correlation between the replicates of Stage C and Stage B and was much stronger than that between the replicates of Stage C and Stage A. Meanwhile, the replicates of ‘China Pink’ and ‘Delft Blue’ also showed similar pattern in the correlation plot, which may imply their similarity in gene expression, and further, their phylogenetic relationship.
Results of correlation analysis and principal component analysis. (a) Correlation plot of the 189 RNA-Seq libraries. The columns and rows are equivalent, under the sequential order of cultivars, developmental stages, perianth partitions and biological replicates. (b–d) PCA plots of the 189 transcriptome libraries in 2 dimensions, grouped in (b) cultivars, (c) developmental stages and (d) partitions of perianth.
Principal component analysis (PCA) was conducted to evaluate the variability of sequencing libraries influenced by different factors. The influences from cultivars and developmental stages were observed. As shown in Fig. 3b, clustering of libraries from different cultivars was particularly obviously on the y-axis (PC2), forming distinct layers for almost all cultivars, except ‘China Pink’ and ‘Delft Blue’ which were clustered together. It could be explained by the smaller genetical difference between the two than that between other cultivars, since ‘China Pink’ is the sport (mutant) of ‘Delft Blue’43. Developmental stages also influenced the grouping of sequencing libraries on the x-axis (PC1), forming two distinct pillars (Fig. 3c). Some libraries of Stages C and A were clustered together, yet their differences were still observed by their condensation of majorities. Perianth partitions induced no influence on the grouping, as the sequencing libraries of all three partitions clustered together (Fig. 3d).
Transcriptomic analyses, including differential expression analysis, functional annotation and functional enrichment analysis, were performed to demonstrate the usability of our dataset while investigating gene expression profiles of hyacinth. The read counts were normalised using DESeq 244 with negative binomial distribution p-value estimation model. The adjusted p-values (padj) were estimated using BH procedure45. The DEGs were firstly screened through the standard |log2(FoldChange)| > 1 and padj <0.05. To further filter DEGs across different comparison groups, the threshold DESeq 2 p-value ≤ 0.05 & |log2FoldChange| ≥ 1.0 were adopted. The DEGs identified across different factors, i.e. cultivars, developmental stages and perianth partitions, are visualised in Fig. 4.
Results of differential expression analysis. Visualisation of DEGs across different parameters. (a–c) Heatmaps showing the DEGs across different comparisons. Boxes filled in red represent the number of upregulated genes and those filled in blue represent the number of downregulated genes. The numbers in the boxes represent the number of DEGs across the comparison of (a) different cultivars, (b) different developmental stages and (c) different partitions of perianth. (d) Flower diagram showing the unique and shared DEGs (as core genes) of the seven studied cultivars. The numbers in the brackets indicate the total number of the transcriptome datasets of that cultivar. (e,f) Venn diagrams showing the number of unique (non-overlapped regions) and shared DEGs (overlapped regions) between (e) developmental stages and (f) perianth partitions. (g–i) Heatmap visualising the FPKM cluster analysis of DEGs across different parameters, using the log10(FPKM + 1) value. Boxes in orange represent upregulated DEGs and those in blue represent downregulaed DEGs. The expression patterns of the selected top 50 DEGs are visualised in (g) cultivars, (h) developmental stages and (i) perianth partitions.
The numbers of upregulated and downregulated DEGs identified in each comparison group are visualised in the heatmaps (Fig. 4a–c). Comparing the cultivars, the smallest number of upregulated (4,126) and downregulated genes (2,796) were found in the same comparison group as ‘China Pink’ versus ‘Delft Blue’ (Fig. 4a). The greatest numbers of upregulated (41,612) and downregulated genes (53,395) were observed in two distinct comparison groups, as ‘China Pink’ versus ‘Peter Stuyvesant’ and ‘Gipsy Queen’ versus ‘Delft Blue’, respectively. Comparing the developmental stages, the greatest numbers of upregulated (76,986) and downregulated (34,791) genes were identified in the comparison group Stage B versus Stage A (Fig. 4b). Comparing the perianth partitions, the differential expression between the outer and inner perianth lobes was the smallest (1,317 upregulated and 373 downregulated genes; Fig. 4c).
The number of shared (core) and unique DEGs across the cultivars are visualised in the flower diagram (Fig. 4d). The seven cultivars shared a total of 50,344 DEGs. ‘City of Haarlem’ holds the greatest number of unique DEGs (5,570), while ‘China Pink’ has the smallest number of unique DEGs (1,529). The Venn diagrams (Fig. 4e,f) show the number of shared and unique DEGs identified between different developmental stages and perianth partitions. A total of 100,341 DEGs were found common across all three developmental stages (Fig. 4e). Stage B showed the greatest number of unique DEGs (40,307), while Stage C had the least unique DEGs (2,075). These two stages shared 12,441 DEGs. The commonly shared DEGs in all three perianth partitions accounted for 123,498, and the greatest number of unique DEGs was found in perianth tube (10,125; Fig. 4f).
The expression patterns of the top 50 DEGs across different factors are visualised in the heatmaps (Fig. 4g–i) generated by SRplot46. The read counts in FPKM were firstly normalised into the value of log10(FPKM + 1). The 50 candidate DEGs in which had significant expression levels were then selected based on the thresholds, i.e. log10(FPKM + 1) ≥3.10 for cultivars, ≥3.00 for developmental stages and ≥3.28 for perianth partitions. The DEGs identified across the cultivars at different developmental stages are visualised in the heatmaps in Supplementary Figure S20. These findings facilitate the identification of candidate genes strongly associated with each factor for future validation.
Seven databases were employed to predict gene function, namely Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genome Orthology (KO), euKaryotic Orthologous Groups (KOG), NCBI non-redundant protein sequences (Nr), NCBI nucleotide sequences (Nt), Protein family (Pfam) and Swiss-Prot (Fig. 5a). The annotated unigenes vary across the database, ranging from 23,809 (KOG) to 158,125 (Nr), representing 4.81% and 31.98% of unigenes, respectively. Under the classification of GO, the top three terms for biological processes were “cellular process”, “metabolic process” and “biological regulation”, for cellular component were “cellular anatomical entity”, “intracellular” and “protein-containing complex”, and for molecular functions were “binding”, “catalytic activity” and “transporter activity” (Fig. 5b). Over half of the genes were related to the protein families under KEGG classification and were involved in genetic information processing (32.08%), signalling and cellular processes (10.78%), and metabolism (10.46%; Fig. 5c).
Results of functional annotation and functional enrichment analysis. (a–c) Functional annotation across different databases. (a) The unigenes of all transcriptome datasets were annotated by the seven databases. The x-axis represents the databases, and the y-axis represents the number of unigenes annotated by the respective databases, with the percentages of all unigenes showing on the bars. (b) The distribution of GO terms for biological processes (orange-red), cellular components (green) and molecular functions (blue). (c) The percentage of genes annotated under KEGG classification. The letters next to the bar lines on the right indicate Level 1 of the KO Pathway. “O” represents Organismal Systems, “N” represents Not Included in Pathway or Brite, “M” represents Metabolism, “G” represents Genetic Information Processing, “E” represents Environmental Information Processing, “C” represents Cellular Processes and “B” represents Brite Hierarchies. (d) The number of transcripts annotated under different classes of transcription factors. (e–g) Through functional enrichment analysis, upregulated expressions were identified in the genes involved in the biosynthesis of (e) flavonoid, (f) flavone and flavonol, and (g) anthocyanin in cultivars across different comparison groups. The gradient in the colour of the bar indicates -log10 (p-value); the bar length represents the count of upregulation.
A total of 4,684 transcription factors (TF) were identified using iTAK47 and hmmscan (www.hmmer.org). They were classified under different TF families (Fig. 5d). The largest family was C2H2 (570), followed by MYB or MYB-related (287) and C3H (246). TFs involved in floral pigments biosynthesis, including MYB or MYB-related31,48,49 (287), bHLH50 (162), bZIP50 (108) and MADS51 (81), were recovered in the TF analysis.
To verify the presence of DEGs involved in floral pigments biosynthesis, functional enrichment analysis was performed with the aid of KEGG database. Upregulation of DEGs were identified between some studied cultivars, and the results are visualised in enrichment bar charts generated by SRplot46 (Fig. 5e–g). The comparison group ‘Jan Bos’ versus ‘City of Haarlem’ has the greatest count of upregulated DEGs in flavonoid biosynthesis (71; Fig. 5e) and anthoxanthin biosynthesis (26; Fig. 5f), and the second greatest count of upregulated DEGs in anthocyanin biosynthesis (15; Fig. 5g).
The above outcome of the analyses indicate that our dataset is highly valuable for the study of floral pigments biosynthesis in hyacinth cultivars and is supported by accurate sequencing, reproducible experimental design and reliable data. The molecular mechanism behind floral pigment diversification in hyacinth cultivars can be revealed by using this dataset, which thereby contributes to the molecular breeding of this floricultural crop with high aesthetic and economical value.
Code availability
The software and their versions are described in Methods. No custom code was used in this study.
References
Angiosperm Phylogeny Group (APG). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 181, 1–20, https://doi.org/10.1111/boj.12385 (2016).
Pfosser, M. & Speta, F. Hyacinthaceae. Hyacinthus, Ornithogalum, Scilla and their relatives. In: The Tree of Life Web Project http://tolweb.org/Hyacinthaceae/21437/2001.01.01 (2001).
Wendelbo, P. Hyacinthus L. In: Flora of Turkey and the East Aegean Islands Vol. 8 (ed. Davis, P., Mill, R. & Tan, K.), pp 263–264 (Edinburgh Univ. Press, 1984).
Horovitz, A. & Danin, A. Relatives of ornamental plants in the flora of Israel. Isr. J. Plant Sci. 32, 75–95, https://doi.org/10.1080/0021213X.1983.10676966 (1983).
Université Saint-Joseph de Beyrouth (USJ). Hyacinthus orientalis. In: Lebanon Flora http://www.lebanon-flora.org/species.php?id_pl=25 (2023).
European Distributed Institute of Taxonomy (EDIT). Hyacinthus orientalis subsp. orientalis. In: Flora of Cyprus — a dynamic checklist https://www.flora-of-cyprus.eu/cdm_dataportal/taxon/0e347036-269f-4729-bd8c-4911bd62d598 (2023).
Bailey, L. H. & Bailey, E. Z. Hyacinthus L. In: Hortus Third: A Concise Dictionary of Plants Cultivated in the United States and Canada, pp 577 (Macmillian Publishing Company, 1976).
The Royal Horticultural Society. The New Royal Horticultural Society Dictionary of Gardening 1st edn. (ed. Huxley, A., Griffiths, M. & Levy, M.) (The Macmillan Press Limited, 1992).
Darlington, C. D., Hair, J. B. & Hurcombe, R. The history of the garden hyacinths. Heredity 5, 233–252, https://doi.org/10.1038/hdy.1951.22 (1951).
Doorenbos, J. Notes on the history of bulb breeding in the Netherlands. Euphytica 3, 1–18, https://doi.org/10.1007/BF00028123 (1954).
Wainwright, M. After 16 years of seed testing, the new breed of black hyacinth reveals appeal of dark side of the bloom https://www.theguardian.com/uk/2005/mar/22/homesandgardens.shopping (2005).
JSTOR Global Plants. Lectotype of Hyacinthus orientalis L. [family LILIACEAE] https://plants.jstor.org/stable/10.5555/al.ap.specimen.bm000558527 (2011).
JSTOR Global Plants. Syntype of Hyacinthus orientalis L. [family LILIACEAE] https://plants.jstor.org/stable/10.5555/al.ap.specimen.l0052779 (1998).
Benschop, M., Kamenetsky, R., Le Nard, M., Okubo, H. & de Hertogh, A. The Global Flower Bulb Industry: Production, Utilization, Research. In: Horticultural Reviews. (ed. Daunay, M. C., Merwin, I. & Stover, E.), pp 1–115 (John Wiley & Sons, https://doi-org.easyaccess2.lib.cuhk.edu.hk/10.1002/9780470527238.ch1, 2009).
Kersten, J. H. The cultivation of the hyacinth in Holland. J. R. Hortic. Soc. Lond. 11, 54–63, https://www.biodiversitylibrary.org/page/45698008 (1889).
Nowak, J. & Rudnicki, R. M. Hyacinthus. In: The Physiology of Flower Bulbs (ed. de Hertogh, A., Le Nard, M.), pp 335–347. (Elsevier Science Publishers, 1993).
Christenhusz, M. J. et al. Tiptoe through the tulips–cultural history, molecular phylogenetics and classification of Tulipa (Liliaceae). Bot. J. Linn. Soc. 172, 280–328, https://doi.org/10.1111/boj.12061 (2013).
Lim, K. B. & van Tuyl, J. M. Lily. In: Flower Breeding and Genetics (ed. Anderson, N. O.), pp 517–537 (Springer, https://doi.org/10.1007/978-1-4020-4428-1_19, 2007).
Nunez, D. R., de Castro, C. O., Ruiz, S. R. & Ariza, F. A. The origin of cultivation and wild ancestors of daffodils (Narcissus subgenus Ajax) (Amaryllidaceae) from an analysis of early illustrations. Sci. Hortic. 98, 307–330, https://doi.org/10.1016/S0304-4238(03)00055-4 (2003).
Zhao, D. & Tao, J. Recent advances on the development and regulation of flower color in ornamental plants. Front. Plant Sci. 6, 1–13, https://doi.org/10.3389/fpls.2015.00261 (2015).
Khoo, H. E., Azlan, A., Tang, S. T. & Lim, S. M. Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food & Nutrition Research 61, 1–21, https://doi.org/10.1080/16546628.2017.1361779 (2017).
Hosokawa, K., Fukunaga, Y., Fukushi, E. & Kawabata, J. Acylated anthocyanins from red Hyacinthus orientalis. Phytochemistry 39, 1437–1441, https://doi.org/10.1016/0031-9422(95)00140-3 (1995).
Hosokawa, K., Fukunaga, Y., Fukushi, E. & Kawabata, J. Five acylated pelargonidin glucosides in the red flowers of Hyacinthus orientalis. Phytochemistry 40, 567–571, https://doi.org/10.1016/0031-9422(95)00312-U (1995).
Hosokawa, K., Fukunaga, Y., Fukushi, E. & Kawabata, J. Acylated anthocyanins in red flowers of Hyacinthus orientalis regenerated in vitro. Phytochemistry 42, 671–672, https://doi.org/10.1016/0031-9422(95)00949-3 (1996).
Hosokawa, K., Fukunaga, Y., Fukushi, E. & Kawabata, J. Seven acylated anthocyanins in the blue flowers of Hyacinthus orientalis. Phytochemistry 38, 1293–1298, https://doi.org/10.1016/0031-9422(94)00778-R (1995).
Tao, X. H., Yuan, Y., Xu, Y. Q., Shi, Y. M. & Tang, D. Q. Anthocyanin Profiles in Petals of Different Hyacinthus orientalis. Acta Horticulturae Sinica 42, 301–310, https://doi.org/10.16420/j.issn.0513-353x.2014-0635 (2015).
Su, X. Q., Bao, R. L. & Hu, F. R. Flower color determination and anthocyanin component analysis of different hyacinth varieties. Mol. Plant Breed. 17, 5109–5114, https://doi.org/10.13271/j.mpb.017.005109 (2019).
Naish, M. et al. The genetic and epigenetic landscape of the Arabidopsis centromeres. Science 374, 1–9, https://doi.org/10.1126/science.abi7489 (2021).
Feng, J. et al. The pineapple reference genome: Telomere‐to‐telomere assembly, manually curated annotation, and comparative analysis. J. Integr. Plant. Biol. 66, 2208–2225, https://doi.org/10.1111/jipb.13748 (2024).
Wong, K. H. et al. Characterisation of the complete chloroplast genomes of seven Hyacinthus orientalis L. cultivars: Insights into cultivar phylogeny. Horticulturae 8, 1–23, https://doi.org/10.3390/horticulturae8050453 (2022).
Li, X., Wu, Y. Y. & Feng, Y. Analysis of anthocyanin content and related gene expression in different varieties of Hyacinthus orientalis. Molecular Plant Breeding 18, 4562–4571, https://doi.org/10.13271/j.mpb.018.004562 (2020).
Ewels, P., Magnusson, M., Lundin, S. & Käller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048, https://doi.org/10.1093/bioinformatics/btw354 (2016).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652, https://doi.org/10.1038/nbt.1883 (2011).
Davidson, N. M. & Oshlack, A. Corset: enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biology 15, 1–14, https://doi.org/10.1186/s13059-014-0410-6 (2014).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212, https://doi.org/10.1093/bioinformatics/btv351 (2015).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 1–16, https://doi.org/10.1186/1471-2105-12-323 (2011).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology 10, 1–10, https://doi.org/10.1186/gb-2009-10-3-r25 (2009).
Trapnell, C. et al. Transcript assembly and abundance estimation from RNA-Seq reveals thousands of new transcripts and switching among isoforms. Nat. Biotechnol. 28, 511–515, https://doi.org/10.1038/nbt.1621 (2010).
Sun, W., Li, C., Wang, L. & Dai, S. Analysis of anthocyanins and flavones in different-colored flowers of chrysanthemum. Chinese Bulletin of Botany 45, 327–336, https://www.chinbullbotany.com/EN/10.3969/j.issn.1674-3466.2010.03.004 (2010).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP481794 (2024).
NCBI Gene Expression Omnibus https://identifiers.org/geo/GSE286406 (2025).
Wong, K. H. Spatiotemporal Transcriptomics of Hyacinthus orientalis L. CUHK Research Data Repository https://researchdata.cuhk.edu.hk/dataverse/hyacinth_transcriptome (2024).
van Scheepen, J. Hyacinthus. In International Checklist for Hyacinths and Miscellaneous Bulbs, pp 273–294 (Royal General Bulbgrowers’ Association, https://assets.kavb.nl/docs/286bfc9a-b827-4c44-8168-a7c8fbd28719.pdf, 1991).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq 2. Genome Biology 15, 1–21, https://doi.org/10.1186/s13059-014-0550-8 (2014).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300, https://doi.org/10.1111/j.2517-6161.1995.tb02031.x (1995).
Tang, D. et al. SRplot: A free online platform for data visualization and graphing. PLOS One 18, 1–8, https://doi.org/10.1371/journal.pone.0294236 (2023).
Zheng, Y. et al. iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Molecular Plant 9, 1667–1670, https://doi.org/10.1016/j.molp.2016.09.014 (2016).
Ma, J., Li, Z. & Liu, Y. Integrating multi-omics analysis reveals the regulatory mechanisms of white–violet mutant flowers in grape Hyacinth (Muscari latifolium). Int. J. Mol. Sci. 24, 1–20, https://doi.org/10.3390/ijms24055044 (2023).
Chen, K., Liu, H., Lou, Q. & Liu, Y. Ectopic expression of the grape hyacinth (Muscari armeniacum) R2R3-MYB transcription factor gene, MaAN2, induces anthocyanin accumulation in tobacco. Front. Plant Sci. 8, 1–13, https://doi.org/10.3389/fpls.2017.00965 (2017).
Nakatsuka, T. et al. Floral organ-and temperature-dependent regulation of anthocyanin biosynthesis in Cymbidium hybrid flowers. Plant Science 287, 1–11, https://doi.org/10.1016/j.plantsci.2019.110173 (2019).
Li, B. J. et al. New insight into the molecular mechanism of colour differentiation among floral segments in orchids. Communication Biology 3, 1–13, https://doi.org/10.1038/s42003-020-0821-8 (2020).
Acknowledgements
The research work was financially supported by a donation fund from Wu Jieh Yee Charitable Foundation Limited. The services of mRNA sequencing, de novo assembly and standard analyses were provided by Novogene Co., Ltd. (Beijing, China). UBC Botanical Garden and Gothenburg Botanical Garden provided research materials of the wild individuals of Hyacinthus orientalis L. (BATMAN 014) and Hyacinthus orientalis L. subsp. chinophilus (BATMAN 407), respectively. The authors would like to sincerely acknowledge Ms. Laura Caddy and Mr. Justin Ngo from the University of British Columbia, and Dr. Mats Havström and Dr. Stina Weststrand from Gothenburg Botanical Garden, in assisting the preparation and logistics of research materials that enhanced our study on Hyacinthus orientalis L. The authors also want to thank Miss Stacey Shun-Kei Tsang in assisting post-sequencing analyses and revising this manuscript. The acknowledgements are in no particular order.
Author information
Authors and Affiliations
Contributions
D.T.W.L. acquired funding supports, supervised the project, conceived and design the study. J.H.L.H. and P.C.S. supervised the project and contributed to the study design. H.Y.W. collected the samples, conducted RNA extraction and contributed to the study design and data validation. C.W.Y.S. and J.H.L.H. provided guidance and technical supports in post-sequencing analysis. K.H.W. conceived and designed the study, grew and collected the samples, conducted extraction of RNA and floral pigments, prepared herbarium specimens, documented photos and RHS Colour Chart measurements, curated and analysed the data, and drafted the manuscript. All authors revised and contributed to the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wong, KH., Wu, HY., Shum, C.WY. et al. Transcriptome profiling of Hyacinthus orientalis L. cultivars in floral pigmentation. Sci Data 12, 689 (2025). https://doi.org/10.1038/s41597-025-04977-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-04977-y
This article is cited by
-
Whole genome duplication drives transcriptome reprogramming in response to drought in alfalfa
Plant Cell Reports (2025)