Abstract
Skeletal muscle satellite cells are integral to muscle development and regeneration. Long non-coding RNAs (lncRNAs), a significant class of non-coding RNAs, participate in numerous biological processes and play critical roles in myogenic differentiation. However, research on the regulation of porcine satellite cell (PSC) proliferation and differentiation remains limited. To uncover key lncRNAs involved in PSC regulation, we performed strand-specific RNA-seq to comprehensively profile lncRNA and mRNA expression in PSCs at different stages of proliferation and differentiation (P24h, P48h, D18h, D28h). On average, 61.62 million clean reads were obtained across all samples. Through stringent filtering, we identified 1950 novel lncRNAs, 9367 annotated lncRNAs and 57252 mRNA transcripts for further analysis. This dataset provides valuable insight into the role of lncRNAs in PSC differentiation and contributes to a more detailed understanding of the molecular mechanisms underlying porcine muscle development.
Similar content being viewed by others
Background & Summary
Skeletal muscle is a highly heterogeneous tissue, constituting approximately 40% of body weight, and is integral to the regulation of body movement, metabolism and homeostasis1,2,3. Porcine satellite cells (PSCs) are essential for skeletal muscle development and regeneration4. These quiescent cells reside between the basal lamina and plasma membrane of myofibers and, upon activation by injury or stress, proliferate and differentiate into myoblasts, thereby facilitating muscle repair and growth. Understanding the molecular mechanisms that govern PSC proliferation and differentiation is critical for the progression of animal agriculture, regenerative medicine, and research on muscular dystrophies.
Long non-coding RNAs (LncRNAs) have recently attracted considerable attention due to their complex roles in regulating various biological processes. LncRNAs are a class of RNA transcripts longer than 200 base pairs with less or no protein-coding potential, often exhibiting cell- or tissue-specific expression patterns. These molecules play crucial roles in various biological processes, including epigenetic regulation, cell differentiation, apoptosis, metabolism, signal transduction and immune response5,6,7. Emerging evidence indicates that certain lncRNAs are integral to skeletal muscle development. For instance, during early human myogenesis, lncFAM recruits HNRNPL to the MYBPC2 promoter, increasing its mRNA and protein level, thereby facilitating the differentiation of human myoblasts into myotubes8. In mice, the novel lncRNA lncMREF interacts with Smarca5, thereby promoting the binding of p300/CBP/H3K27ac to myogenic regulatory elements, which accelerates muscle regeneration9. Additionally, the mouse lncMGPF, homologous to pig lncRNA AK394747 and human lncRNA MT510647, facilitates muscle differentiation by asponging miR-135a-5p, resulting in increased MEF2C expression. In pigs, lncRNA H19 regulates porcine satellite cells by sponging miR-140-5p and binding to DBN110. Overall, lncRNAs are integral to skeletal muscle formation. Despite their prevalence and functional significance, the molecular mechanisms of lncRNAs in various species, including pigs, are not well understood.
In this study, we isolated PSCs in vitro and induced their differentiation, collecting cells at two time points during proliferation (P24h, P48h) and two time points during differentiation (D18h, D28h) (Fig. 1). We then performed cell line-specific RNA sequencing and bioinformatics analysis to characterize both lncRNA and mRNA expression profiles (Fig. 2). This comprehensive datasets is a valuable resource to exploring the molecular mechanisms underlying muscle development and regeneration, with implications for both animal agriculture and human health.
Proliferation and differentiation of PSCs. (a) The state of PSCs during proliferation and differentiation. P24h: PSCs proliferated for 24 hours, P48h: PSCs proliferated for 48 hours, D18h: PSCs differentiated for 18 hours, D28h: PSCs differentiated for 28 hours. (b) Statistics of the number and proportion of PAX7 + positive cells. (c) Expression of MYHC in PSCs at different stages of proliferation and differentiation. (d) Differentiation ability of PSCs detected by MyHC immunofluorescence.
Methods
Animals
Three seven-day-old male Large White piglets, all full-sib offspring, were used in this study. All animal care and experimental procedures were conducted in accordance with the National Research Council Guide for the Care and Use of Laboratory Animals and were approved by the Animal Care and Use Committee of Xinyang Normal University (XYEC-2019-009).
Cell isolation and culture
Porcine satellite cells were isolated from male Large white piglets euthanized via carotid artery incision. Hind leg muscles were rapidly collected and washed with PBS supplement with 1% antibiotic-antimycotic (AA; Gibco, 15240-096). After removing connective and adipose tissue, the muscle samples were minced into small pieces and digested with 300 U/mL type II collagenase (Gibco, 17101-015) at 37 °C in a water bash sharker for 2.5 hours. The digestion was terminated using high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Gibco, 10569-010) containing 10% fetal bovine serum (FBS; Gibco, 10099-141).
The cell suspension was sequentially filtered through 100, 70, and 40 µm cell strainer, and the pellets were collected and resuspended in PBS (Gibco, SH30256.01), RPMI-1640 (Gibco, A10491-01), or complete culture medium after centrifugations. Finally, cells were resuspended and cultured in complete culture medium containing 20% FBS, 0.5% Chicken Embryo Extract (CEE; Gemini, 100–163 P), 1% GlutaMax (Gibco, 35050-061), 1% Non-Essential Amino Acids (NEAA; Gibco, 11140-050), 1% AA, 2.5 µg/L basic fibroblast growth factor (bFGF; Invitroogen, 13256-029) and RPMI-1640 at 37 °C in 5% CO2.
Fibroblasts were removed from the mixed cell population due to differential adhesion properties by incubating cells on uncoated plates for 2.5 hours. Purified satellite cells were then transferred to Matrigel-coated plates (BD Biosciences, 356234) for proliferation. Once porcine satellite cells researched 80–90% confluence, theywere transferred into differentiation medium (DMEM supplemented with 5% horse serum (HS; Gibco, 26050-070) and 1% AA) to induce myotubes formation at 37 °C with 5% CO211.
Total RNA isolation and RT-qPCR
Total RNA was extracted from satellite cells using TRIzol (Invitrogen, 15596-026) following the manufacturer’s protocol. Briefly, satellite cells were lysed with 1 mL of TRIzol, and supernatants were collected after centrifugation at 4 °C, 13000 rpm for 15 minutes. Then chloroform was used to separate the supernatants from proteins and DNA, and total RNA was precipitated with isopropyl alcohol. Finally, RNA pellets were washed with ethanol and dissolved in RNase-free water. RNA purity and concentration were assessed using a NanoDrop 2000 spectrophotometer (Thermo Scientific, USA), with 260/280 absorbance ratios ranging from 1.90 to 2.00. RNA integrity and contamination were verified via 1% agarose gel electrophoresis.
Complementary DNA (cDNA) was synthesized using the PrimeScript RT Reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa, RR047A). Read-time quantitative polymerase chain reaction amplification (RT-qPCR) was performed with TB Green® Premix Ex Taq™ II (TaKaRa, RR820A) on a Bio-Rad CFX-96 Real-Time PCR detection system (Bio-Rad, USA). Relative gene expression level were calculated using the 2−ΔΔCT method, with 18S ribosomal RNA (18S rRNA) as an internal control for normalization.
Western blot
Total protein was extracted from porcine satellite cells using RIPA buffer (Beyotime, P0013B) and a protease inhibitor mix (Beyotime, P1008) after washing the cells three times with PBS. Protein concentration was determined using the BCA Protein Assay Kit (Beyotime, P0012), following standard protocols. A total of 20 μg of protein was loaded and separated on a 12% SDS polyacrylamide gel, followed by transfer to a polyvinylidene fluoride (PVDF) membrane (Millipore, IPVH00010). The membrane was blocked with 5% skimmed milk at room temperature for 2 hours, then incubated overnight at 4 °C with antibodies (1:1000) against MYHC (Santa, sc-32732) and β-tubulin (Beyotime, AF2835). After washing with Tris Buffered Saline with Tween (TBST), goat anti-mouse IgG-HRP (Abmart, M21001) or goat anti-rabbit IgG-HRP (Abmart, M21002) secondary antibodies were applied. Protein detection was performed using an ultrasensitive ECL chemiluminescence kit (Beyotime, P0018M) and visualized using the ChemiDoc™ MP Imaging System (Bio-Rad, California, USA).
Strand-specific RNA-seq library preparation & sequencing
We prepared a strand-specific RNA-seq library for each sample. First, ribosomal RNA (rRNA) was removed from 2 μg total RNA using Ribo-ZeroTM Gold Kits (Epicentre, USA). Sequencing libraries were then prepared using the NEBNext® UltraTM Directional RNA Library Prep Kit for Illumina (NEB, Ispawich, USA) according to the manufacturer’s recommendations with different index label. Finally, PCR products were purified using the AMPure XP system, and library quality was assessed with the Agilent Bioanalyzer 2100 system. Libraries were sequenced on an Illumina NovaSeq 6000 platform to generate 150 bp paired-end reads.
Transcript assembly and novel lncRNA transcripts prediction
The workflow for transcript assembly and novel lncRNA transcripts prediction is shown in Fig. 2. Sequence quality was evaluated using FastQC (version 0.11.8)12. Low-quality reads, adapter sequences, and reads containing poly-N were removed from raw sequencing reads using Trimmomatic (version 0.36) to obtain clean data13.
To ensure the accuracy of lncRNA identification, potential ribosomal RNA (rRNA) contamination was systematically addressed. Porcine rRNA reference sequences were retrieved from Silva (release 138.2), Ensembl (Sscrofa11.1) and NCBI RefSeq (assembly GCF_000003025.6). Cleaned data were aligned to these composite rRNA references using Bowtie2 (version 2.5.4) with stringent parameters (--norc).
The clean reads were then mapped to the Sus Scrofa reference genome (Sscrofa11.1) using HISAT2 (version 2.1.0) with default parameters14,15. Subsequently, transcript de novo assembly was performed using StringTie (version 2.0)15,16 with default parameters, guided by the Sus Scrofa reference annotation (Sscrofa11.1). The merge function of StringTie is used to merge all sample assembled transcripts with reference annotation into a single annotation file. This merged annotation file was used to reassemble transcripts and obtain the FPKM values of all genes.
To ensure the uniformity of sequencing, we analyzed 5′-3′ read coverage uniformity across all transcripts using Deeptools (version 3.5.5)17. Transcript coordinates were divided into 100 bins, and coverage was normalized by RPKM.
Novel lncRNA transcripts were identified using the following criteria: (1) transcripts unannotated in the genome; (2) FPKM > 0.5 in at least one sample; (3) transcripts comprising multiple exons; (4) transcript exceeding 200 bp in length; (5) transcripts that neither overlap with protein-coding genes exons nor are within 2 kb of protein-coding genes; and (6) trascripts with coding potential predicted by CPC and CNCI, where a CPC score and CNCI score below 0 indicate non-coding transcript18,19. Using these stringent criteria, we screened a total of 1950 novel lncRNA transcripts.
Then we performed a Blastn alignment (e-value < 1e-5) of our novel lncRNAs against the ALDB database20. None of the transcripts showed significant homology (identity >90%), confirming their novelty.
Differential expression of lncRNAs and mRNAs
First, read counts for all coding genes and lncRNAs, including annotated lncRNAs and novel lncRNAs, were obtained from SAM files using featureCounts (v2.0.0)21. Reads count were then used to perform differential expression analyses between different PSC stages using the R package DESeq 2 (v1.36.0)22. Significantly differentially expressed mRNAs and lncRNAs were identified based on |Log2 (fold change) | > 1 and adjusted P-value < 0.05. Volcano plots of differentially expressed mRNAs and lncRNAs were generated using the R package ggplot2 (v3.4.2)23, and heatmaps of their expression were produced using the R package pheatmap (v1.0.12).
Technical Validation
Isolation and identification of porcine satellite cells
PSCs were isolated from 7-day-old piglets using an enzymatic digestion method. The morphological characteristics of PSCs during proliferation and differentiation were evaluated at 24 and 48 hours of proliferation (P24, P48) and 18 and 28 hours of differentiation (D18, D28), respectively. As shown in Fig. 1a, PSCs exhibited a fusiform shape during proliferation and became progressively elongated following differentiation induction. Muscle tubes formed through cell fusion by D18, with both the number and thickness of myotubes increasing by D28. PAX7, a marker of quiescent and proliferative skeletal muscle satellite cells, was used to assess purity. Purity monitoring revealed that more than 95% of isolated cells were PAX7 positive (Fig. 1b), indicating high purity of the adherent PSCs, thereby making them suitable for subsequent studies. The differentiation potential of the PSCs was evaluated using Myosin Heavy Chain (MYHC) as a maker, with expression levels measured at various time points via Western Blot. The results demonstrated that MYHC was expressed at D18 and D28, but not during the proliferative phase of the PSCs (Fig. 1c). Immunofluorescence analysis at D28 further confirmed the myogenic differentiation potential (Fig. 1d). These findings are consistent with previous studies24, confirming that the isolated PSCs are appropriate for further experimental applications.
Quality control of RNA integrity
The quality of total RNA was assessed using NanoDrop2000 and agarose gel electrophoresis. All samples exhibited high RNA integrity, with concentrations ranging from 200 to 500 ng/µL, OD260/280 values between 1.90 and 2.00, and OD260/230 values between 1.80 to 2.00. These metrics confirmed that the RNA samples were of sufficient quality for further sequencing.
RNA-Seq data quality
FastQC was used to evaluate the quality of raw sequencing data. As illustrated in Fig. 3, the reads exhibited consistently high-quality scores (Fig. 3a), with GC content approximating 50% (median: 46%), which aligns closely with the theoretical GC content of mammalian coding regions (45~50%) as reported in genome-wide studies of Sus Scrofa25,26 (Fig. 3b). Approximately half of the reads were uniquely mapped (Fig. 3c). Additional FastQC metrics confirmed that the data were suitable for downstream analysis.
Quality monitoring results of RNA-seq. (a) Representative quality score distribution for all 150 bp bases. (b) Representative distribution of GC content of per sequence. (c) Representative distribution of unique reads and duplicate reads. (d) The distribution density of transcript length among new assembly lncRNAs, annotated lncRNAs and protein_coding RNAs. (e) Cumulative distribution of FPKM value among new assembly lncRNAs, annotated lncRNAs and protein_coding RNAs. (f) The distribution density of coding probability among new assembly lncRNAs, annotated lncRNAs and protein_coding RNAs.
And the alignment rates to rRNA sequences were consistently below 0.01% across all samples (Table S2) which demonstrates the effectiveness of our library preparation protocol in minimizing ribosomal RNA carryover, thereby ensuring that subsequenc analyses focus specifically on non-coding RNA species without interference from abundant rRNA fragments.
Furthermore, sequencing reads were uniformly distributed across both chromosomes and genome strands (Fig. 4a). Subsequently, mapping to the reference genome S. scrofa 11.1 yielded a mapping rate exceeding 97% across all samples (Table 1). Gene expression levels were also analyzed, revealing that the overall expression profiles of all transcripts were consistent across all samples (Fig. 4b). Moreover, principal component analysis (PCA) and hierarchical clustering revealed distinct expression patterns between different time points, indicating variability between groups and high reproducibility within each group (Fig. 4c,d).
To ensure the uniformity of sequencing, we analyzed 5′-3′ read coverage uniformity across all transcripts. Coverage plots for all transcripts were generated using Deeptools plotProfile, which demonstrate consistent read distribution patterns across the transcript bodies, with no significant positional bias (Fig. S2).
Identification of novel lncRNAs
A total of 1,950 novel lncRNAs were identified through a rigorous screening process (Fig. 2). We further analyzed and compared the length distribution, expression levels, and coding potential of novel lncRNAs, annotated lncRNAs, and mRNAs. Similar to annotated lncRNAs, novel lncRNAs exhibited shorter transcript lengths, lower coding potential, and reduced expression levels compared to mRNAs (Fig. 3d–f). These findings confirm that the identified novel lncRNAs are reliable and can be used for differential expression analysis.
Differential expression of lncRNA and mRNA
We analyzed the expression profiles of lncRNAs and mRNAs in porcine satellite cells (PSCs) across various stages of proliferation and differentiation. Gene expression analysis of PSCs at distinct temporal points revealed significant variations. Compared to P24 stage, 565 lncRNAs and 1,173 mRNAs were upregulated in P48 PSCs, while 116 lncRNAs and 820 mRNAs were downregulated (Fig. 5a,e). In contrast, when comparing the D18 stage to the earlier time point, 407 lncRNAs and 973 mRNAs were upregulated, while 315 lncRNAs and 1,203 mRNAs were downregulated (Fig. 5b,f).
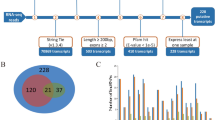
Differential expression analysis of lncRNAs and mRNAs. (a–c) Volcano plot showing differential expression results of lncRNAs in different stages of PSCs proliferation and differentiation. (d) Venn diagram showing that 16 lncRNAs were differentially expressed during the proliferation and differentiation of PSCs. (e–g) Volcano plot showing differential expression results of mRNAs in different stages of PSCs proliferation and differentiation. (h) Venn diagram showing that 136 mRNAs were differentially expressed during the proliferation and differentiation of PSCs.
However, when analyzing gene expression differences between the two stages of differentiated PSCs (D18 and D28), fewer differentially expressed lncRNAs and mRNAs were observed. Specifically, compared to D18, only 40 lncRNAs and 171 mRNAs were upregulated in D28, while 23 lncRNAs and 170 mRNAs were downregulated (Fig. 5c,g). Across all time points, 16 lncRNAs and 136 mRNAs exhibited differential expression (Fig. 5d,h).
To further evaluate the reliability of RNA sequencing data, two differentially expressed lncRNAs were randomly selected, and their relative expression levels were assessed during PSCs proliferation and differentiation using RT-qPCR. As shown in Fig. 6, the RT-qPCR results were consistent with RNA-seq, confirming the reliability of the RNA-seq findings.
Data Records
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive27 in National Genomics Data Center28, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA01970429) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa. Detailed sample information and RNA-seq read statistics are provided in Table 1. The expression data, read counts files, DEG lists and the BED file of novel lncRNAs have been deposited in the Figshare database30.
Code availability
For this study, open-access software was employed as described in the Methods section using default parameters. The complete process from raw sequencing data to novel lncRNA transcript prediction is available in the lncRNA_identification.bash script.
1. FastQC (version 0.11.8) was used to check the quality of raw sequencing data: https://www.bioinformatics.babraham.ac.uk/projects/fastqc.
2. Trimmomatic (version 0.36) was used to remove adaptor sequence and low quality reads: https://anaconda.org/bioconda/trimmomatic.
3. Bowtie2 (version 2.5.4) was used to map the cleaned sequencing data to reference rRNA from Silva, Ensemble and NCBI: https://sourceforge.net/projects/bowtie-bio/files/bowtie2/.
4. HISAT2 (version 2.1.0) was used to map the cleaned sequencing data to reference genome: http://daehwankimlab.github.io/hisat2.
5. Deeptools (version 3.5.5) was used to calculated the read distribution bias across transcripts: https://test-argparse-readoc.readthedocs.io/en/latest/.
6. StringTie (version 2.0) was used to assemble transcripts and merge transcripts with reference annotation: https://ccb.jhu.edu/software/stringtie.
7. CPC was used to calculate the coding potential of transcripts: http://cpc2.gao-lab.org.
8. CNCI was used to calculate the coding potential of transcripts: https://github.com/www-bioinfo-org/CNCI.
9. FeatureCounts (v2.0.0) was used to calculate the reads count of transcripts: https://subread.sourceforge.net/featureCounts.html.
10. DESeq 2 (version 1.10.1) was used to identify differentially expressed genes: https://bioconductor.org/pack-ages/release/bioc/html/DESeq2.html.
11. Pheatmap (Version 1.0.12) was used to plot the heatmap: https://cran.r-project.org/web/packages/pheatmap/.
12. Ggplot2 (version 3.3.4) was used to generate the volcano plot: https://cran.r-project.org/web/packages/ggplot2/index.html.
References
Ma, J. et al. miR-205 Regulates the Fusion of Porcine Myoblast by Targeting the Myomaker Gene. Cells 12, https://doi.org/10.3390/cells12081107 (2023).
Luo, H. et al. Functional Non-coding RNA During Embryonic Myogenesis and Postnatal Muscle Development and Disease. Front Cell Dev Biol 9, 628339, https://doi.org/10.3389/fcell.2021.628339 (2021).
Jin, J. J. et al. Long noncoding RNA SYISL regulates myogenesis by interacting with polycomb repressive complex 2. Proc Natl Acad Sci USA 115, E9802–E9811, https://doi.org/10.1073/pnas.1801471115 (2018).
Li, Z. et al. LncIRS1 controls muscle atrophy via sponging miR-15 family to activate IGF1-PI3K/AKT pathway. J Cachexia Sarcopenia Muscle 10, 391–410, https://doi.org/10.1002/jcsm.12374 (2019).
Chen, M. et al. A novel long non-coding RNA, lncKBTBD10, involved in bovine skeletal muscle myogenesis. In Vitro Cell Dev Biol Anim 55, 25–35, https://doi.org/10.1007/s11626-018-0306-y (2019).
Wang, Y. et al. The gut microbiota reprograms intestinal lipid metabolism through long noncoding RNA Snhg9. Science 381, 851–857, https://doi.org/10.1126/science.ade0522 (2023).
Li, C.-c et al. Identification and functional prediction of long intergenic noncoding RNAs in fetal porcine longissimus dorsi muscle. Journal of Integrative Agriculture 20, 201–211, https://doi.org/10.1016/s2095-3119(20)63261-0 (2021).
Chang, M. W. et al. Enhanced myogenesis through lncFAM-mediated recruitment of HNRNPL to the MYBPC2 promoter. Nucleic Acids Res 50, 13026–13044, https://doi.org/10.1093/nar/gkac1174 (2022).
Lv, W. et al. Long noncoding RNA lncMREF promotes myogenic differentiation and muscle regeneration by interacting with the Smarca5/p300 complex. Nucleic Acids Res 50, 10733–10755, https://doi.org/10.1093/nar/gkac854 (2022).
Li, J. et al. Long Non-coding RNA H19 Regulates Porcine Satellite Cell Differentiation Through miR-140-5p/SOX4 and DBN1. Front Cell Dev Biol 8, 518724, https://doi.org/10.3389/fcell.2020.518724 (2020).
Cheng, X. et al. MEG3 Promotes Differentiation of Porcine Satellite Cells by Sponging miR-423-5p to Relieve Inhibiting Effect on SRF. Cells 9, https://doi.org/10.3390/cells9020449 (2020).
Brown, J., Pirrung, M. & McCue, L. A. FQC Dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 33, 3137–3139, https://doi.org/10.1093/bioinformatics/btx373 (2017).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120, https://doi.org/10.1093/bioinformatics/btu170 (2014).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37, 907–915, https://doi.org/10.1038/s41587-019-0201-4 (2019).
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11, 1650–1667, https://doi.org/10.1038/nprot.2016.095 (2016).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33, 290–295, https://doi.org/10.1038/nbt.3122 (2015).
Ramirez, F., Dundar, F., Diehl, S., Gruning, B. A. & Manke, T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42, W187–191, https://doi.org/10.1093/nar/gku365 (2014).
Kang, Y. J. et al. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res 45, W12–W16, https://doi.org/10.1093/nar/gkx428 (2017).
Sun, L. et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res 41, e166, https://doi.org/10.1093/nar/gkt646 (2013).
Li, A. et al. ALDB: a domestic-animal long noncoding RNA database. PLoS One 10, e0124003, https://doi.org/10.1371/journal.pone.0124003 (2015).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930, https://doi.org/10.1093/bioinformatics/btt656 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq 2. Genome Biol 15, 550, https://doi.org/10.1186/s13059-014-0550-8 (2014).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York (2016).
Moss, F. P. & Leblond, C. P. Nature of dividing nuclei in skeletal muscle of growing rats. J Cell Biol 44, 459–462, https://doi.org/10.1083/jcb.44.2.459 (1970).
Groenen, M. A. et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491, 393–398, https://doi.org/10.1038/nature11622 (2012).
Wang, Z., Feng, X., Tang, Z. & Li, S. C. Genome-Wide Investigation and Functional Analysis of Sus scrofa RNA Editing Sites across Eleven Tissues. Genes (Basel) 10, https://doi.org/10.3390/genes10070520 (2019).
Chen, T. et al. The Genome Sequence Archive Family: Toward Explosive Data Growth and Diverse Data Types. Genomics Proteomics Bioinformatics 19, 578–583, https://doi.org/10.1016/j.gpb.2021.08.001 (2021).
Members, C.-N. & Partners. Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucleic Acids Res 50, D27–D38, https://doi.org/10.1093/nar/gkab951 (2022).
Members, N. G. D. C. P. Genome Sequence Archive https://bigd.big.ac.cn/gsa/browse/CRA019704 (2025).
Bo, P. Genome Annotation. figshare https://doi.org/10.6084/m9.figshare.28481012.v1 (2024).
Acknowledgements
We thank our lab members for their discussion and comments. This work was funded by the National Natural Science Foundation of China (32202654, 31972537), the Department of Science and Technology in Henan Province of China (222102110013), Henan Provincial Natural Science Foundation of China (242300421337, 222300420272, 242300420509, 212300410230), Key scientific Research Projects for Higher Education Institutions in Henan Province (24A230015), Henan Association for Science and Technology (HNKJZK-2025-13B) and the Nanhu Scholars Program for Young Scholars of XYNU (Henan, China).
Author information
Authors and Affiliations
Contributions
C.C.L. designed the experiments and analyzed the data, X.F.C. and T.Y.L. conducted experiments, X.J.X, Q.X.L, F.C. and H.X.X. participated in experiments and data analysis, Y.J.X., and P.P.Z. conceived the project and designed the experiments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, C., Cheng, X., Li, T. et al. Long non-coding RNA and mRNA expression profiling of porcine satellite cells using strand-specific RNA-seq. Sci Data 12, 700 (2025). https://doi.org/10.1038/s41597-025-05041-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-05041-5