Abstract
The liver is capable of regeneration, regaining mass through compensatory hypertrophy even after resecting 70% of its mass. However, the underlying regenerative mechanism remains poorly understood. Thus, in this study, microarray data were procured over time, from 0 to 14 days post-70% partial hepatectomy (PH) in mice. This dataset was correlated with the body weight and liver weight measured at the time of liver harvest, allowing us to determine the progress of compensatory hypertrophy in each sample. Furthermore, the impact of individual differences in regenerative capacity was analyzed by preparing three biological replicates at each time point. The microarray data were confirmed to be of high quality based on the RNA integrity number and QC stats of the dataset. The reliability of the dataset was further supported by the results of hierarchical clustering and the expression variation of cell cycle–related genes. This microarray data will contribute to the elucidation of the regenerative mechanism in the 70% PH model.
Similar content being viewed by others
Background & Summary
The liver has a unique regenerative capacity. After the surgical removal of 70% of the liver mass, the remaining tissue enlarges as hepatocytes proliferate to compensate for the lost tissue and function1,2. Hepatocyte proliferation is critical for liver regeneration because hepatocytes, which account for 80% of the liver mass, regain their weight by hypertrophy and proliferation after 70% partial hepatectomy (PH)3,4. In previous studies, intravenous glucose supplementation5, complementation of altered metabolic capacity6, and suppression of lipid accumulation7 inhibit PH-induced hepatocyte proliferation as a control mechanism for hepatocyte proliferation. Thus, these changes have been considered as necessary triggers for hepatocyte proliferation. However, these studies have investigated the specific phenomena and genes that contribute to the proliferation of hepatocytes during liver regeneration after 70% PH. There have been few comprehensive studies linking each mechanism; thus, the regulatory mechanism of hepatocyte proliferation remains poorly understood.
In recent years, large-scale gene expression analysis has become important in biology and tissue regeneration research. In particular, microarray analysis has long been used to examine gene expression with known nucleotide sequences. In addition, the platform match would allow direct comparative analysis with considerable microarray data accumulated in previous studies. In the 70% PH model, microarray analysis was used to compare normal livers with livers in the proliferative phase to extract genes that control hepatocyte proliferation8, as well as studies that examined gene expression over time in the same model9. However, the microarray data used in previous studies on the mechanism underlying liver regeneration lacked biological replicates, making it impossible to eliminate the effect of individual differences in regenerative capacity. In addition, these data were only associated with time information and were not linked to important liver weights in the 70% PH model.
In this study, a longitudinal microarray dataset of the liver was obtained during liver regeneration after 70% PH in mice. High-purity and high-quality RNA was collected from the liver after obtaining weight and liver weight information from three mice at each time point after 70% PH. High-quality microarray datasets were generated from the collected RNA, and the gene expression profiles were evaluated. The results indicated that the expression level of cell cycle–related genes significantly increased with compensatory hypertrophy of the liver. No genes with significantly altered expression were extracted after 14 days of resection, indicating that the data accurately reflected the state of liver regeneration.
Collectively, the microarray dataset containing sufficient biological replicates obtained in this study will be utilized to elucidate the mechanism underlying liver regeneration after 70% PH based on the body–liver ratio linked to each dataset to accurately determine the degree of regeneration.
Methods
70% PH
All animal experiments were performed in accordance with the Nihon University Animal Care and Use Committee and Nihon University Rules concerning Animal Care and Use. C57BL/6 N mice were purchased from the Charles River Laboratories. 70% PH was performed as previously described10. In brief, the mice (10 weeks old) were anesthetized with isoflurane (Wako Pure Chemical). After restraining the mouse, a 3-cm incision was created in the midline of the abdominal skin and muscle to expose the xiphoid process. The median and left lateral lobes were tied around the top of the lobes using a 4-0 silk surgical thread. Then, the lobes were removed surgically, thereby removing about 70% of the total liver weight. The abdomen of the mice was sutured closed. Afterward, the mice were weighed and sacrificed at 0 day before 70% PH and at 1, 2, 3, 4, 7, and 14 days after PH. The livers were harvested from the mice, and liver weights were measured. Three biological replicates were prepared for each sample category using different mice.
RNA collection, isolation, and quality check
The collected livers were homogenized in TRIzol solution and mixed with an equal volume (300 μL) of the supernatant treated with chloroform. RNA was extracted using RNeasy, and genomic DNA was removed. A NanoDrop ND-1000 spectrophotometer was used to quantify the isolated RNA (NanoDrop Technologies, Wilmington, DE). The intactness of RNA was determined by using the Agilent RNA 6000 Nano Kit on the Agilent 2100 Bioanalyzer system and evaluated by using RNA integrity number (RIN) software algorithm.
Microarray hybridization and data acquisition
Fragmented and labeled cRNA samples were prepared from 250 ng of total RNA using the GeneChip® 3′ IVT PLUS Reagent Kit. cRNA was hybridized to the Mouse Genome 430 2.0 Array at 45 °C for 16 h (60 rpm). After washing, the array was scanned using a GeneChip® Scanner 3000 7 G.
QC stats
Affymetrix raw CEL files were preprocessed using the robust multichip average algorithm11, which includes background correction, quantile normalization, median polish summarization, and log base 2 transformations. The quality of the array data was evaluated by using the Bioconductor R packages “affy” and “simpleaffy,” and QC stats was performed.
Hierarchical clustering analysis
In evaluating the similarity of the expression profiles among the samples, hierarchical clustering analysis12 was performed using R (version 4.0.2) with the expression matrix data as input. In measuring the similarity between each sample, Manhattan was used, and the ward D2 method was used to measure clustering.
Extraction of differentially expressed genes
Differentially Expressed Genes (DEGs) were extracted by focusing on genes with a twofold or more increase or decrease and a P-value of less than 0.05. Student’s t-test was used to test for significant differences. Volcano plots were visualized using TIBCO Spotfire Desktop version 7.9.0 (TIBCO Software).
Statistical analysis
The body–liver ratio data were presented as mean ± SEM. Comparisons between several groups were analyzed using Tukey’s test, and a P-value of 0.01 was considered statistically significant.
Data Records
The dataset of liver microarray data from the 70% PH model mouse was deposited in the Gene Expression Omnibus (GEO) database with the accession number GSE27861813 and is available for public access.
The data can be accessed through https://identifiers.org/geo/GSE278618.
The GEO accession numbers for individual samples are listed in Table 1.
Technical Validation
Liver regeneration after PH
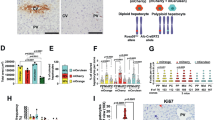
The body weight and liver weight of the mice were measured to evaluate whether the liver regains weight after 70% PH because of compensatory hypertrophy of the remaining liver (Table 1). The ratio of the liver weight to the body weight (body–liver ratio) significantly increased from 2 to 4 days after resection, indicating that the liver had recovered to a level with no significant difference compared with that before resection by 4 days (Fig. 1).
Assessment of RNA integrity
High-quality RNA for microarray analysis must be obtained to ensure the accuracy of gene expression studies. The purity of the total RNA of all samples was evaluated by measuring A260/280 using a NanoDrop ND-1000 spectrophotometer.
The A260/280 values of all samples ranged from 1.90 to 2.10, indicating high purity (Table 2). The quality of the total RNA of all samples was measured on an Agilent 2100 Bioanalyzer system using the Agilent RNA 6000 Nano Kit and evaluated using the RIN software algorithm. The images (Fig. 2a) and electropherograms (Fig. 2b) showed peak signals of 18 S and 28 S ribosomal bands in each sample, indicating that the RNA from all samples was of high quality and suitable for microarray analysis.
Evaluation of microarray data
The quality of the microarray data obtained from all the extracted RNA samples is important for microarray analysis. The QC stats evaluation showed no significant differences in mean background, scale factor, % present value, and 3′–5′ ratio among the microarray data (Fig. 3).
Quality check of the microarray data. Affymetrix quality control parameters for all microarrays: The numbers in the first column indicate present call (41.17%–46.86%), and the numbers below indicate background (30.62–38.82). The horizontal line indicates the scale factor, with all samples inside the 3 × dotted line. Circles and triangles indicate the 3′ 5′ ratio of GAPDH to actin.
Expression profiles of the microarray data
Hierarchical clustering was performed to validate the microarray data (Fig. 4a). Hierarchical clustering showed that the livers at 1 to 3 days postresection were classified into different clusters compared with the livers at 0 day. In addition, the three biological replicates in all samples except for 2d-2 and 7d-1 formed one cluster or small clusters in close proximity. This phenomenon is due to individual differences related to the rate of regeneration, as the samples were classified into clusters of consecutive samples. In evaluating whether the gene expression profile returned to normal after PH, the number of differentially expressed genes was extracted by comparing the gene expression profile of each time point with that of the normal liver (Fig. 4b). The gene expression profile returned to normal 14 days after resection, as the genes that were significantly (P < 0.05) altered by more than twofold increase or decrease compared with the normal liver were not extracted after 14 days of resection. The expression level of cell cycle–related genes, such as Mcm2, Mcm3, Mcm4, Mcm6, Mcm7, Cdk1, Ccnb1, Ccnb2, and Mki67, increased significantly from 1 to 4 days after resection, which is consistent with the time of liver hypertrophy, indicating that the microarray data in this study were reliable. This result indicates the reliability of the microarray data in this study.
Gene expression profiles of the microarray data. (a) Dendrogram of signals normalized by robust multichip average for all 21 samples created using hierarchical clustering. The numbers above the dendrogram indicate the approximately unbiased (au) P-values and bootstrap probability (bp). All samples in the dendrogram are colored the same for each day. (b) The volcano plot of genes between 0 day and each cluster. DEGs were extracted for genes with more than twofold change in expression values and P-values less than 0.05 compared with 0 day. Fold change is shown in the x-axis. The P-values are shown in the y-axis. Dots indicate each gene, and numbers indicate the DEG number (red, upregulation; blue, downregulation). Labeling is a gene that promotes the cell cycle.
Code availability
All the software programs used in this study (data processing and analysis) are listed with the version in the Methods section. In the absence of details on parameters, the programs were used with the default settings. No custom code was generated or used for the analysis of the data presented.
References
Michalopoulos, G. K. Liver regeneration. J. Cell Physiol. 213, 286–300 (2007).
Michalopoulos, G. K. & DeFrances, M. Liver regeneration. Adv. Biochem. Eng. Biotechnol. 93, 101–134 (2005).
Thorgeirsson, S. S. Hepatic stem cells in liver regeneration. FASEB J. 10, 1249–1256 (1996).
Su, A. I., Guidotti, L. G., Pezacki, J. P., Chisari, F. V. & Schultz, P. G. Gene expression during the priming phase of liver regeneration after partial hepatectomy in mice. Proc. Natl. Acad. Sci. USA 99, 11181–11186 (2002).
Kohjima, M. et al. Delayed liver regeneration after partial hepatectomy in adipose differentiation related protein-null mice. J. Hepatol. 59, 1246–1254 (2013).
Weymann, A. et al. p21 is required for dextrose-mediated inhibition of mouse liver regeneration. Hepatology 50, 207–215 (2009).
Miyaoka, Y. et al. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr. Biol. 22, 1166–1175 (2012).
Hu, J. et al. Endothelial cell-derived angiopoietin-2 controls liver regeneration as a spatiotemporal rheostat. Science 343, 416–419 (2014).
Otu, H. H. et al. Restoration of liver mass after injury requires proliferative and not embryonic transcriptional patterns. J Biol Chem 282, 11197–11204 (2007).
Mitchell, C. & Willenbring, H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat. Protoc. 3, 1167–1170 (2008).
Irizarry, R. A. et al. Exploration, normalization, and summaries of high-density oligonucleotide array probe level data. Biostatistics 4, 249–264 (2003).
Suzuki, R. & Shimodaira, H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542 (2006).
NCBI GEO https://identifiers.org/geo/GSE278618 (2024).
Acknowledgements
This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (grant no. 19K06444 to K. Kano and no. 24K09289 to Y. Oki).
Author information
Authors and Affiliations
Contributions
T.N. and Y.O. conceived the study. T.N. prepared the RNA samples from the liver for microarray and weight data of the liver and mice. T.N., R.H., and Y.O. performed bioinformatics data analysis and data registration. All authors curated and validated the data and metadata analyzed in this study. T.N. wrote the original draft of the manuscript. All authors reviewed and edited the draft of the manuscript. Y.O. and K.K. contributed to funding acquisition. All the authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Nishi, T., Oki, Y., Hagiwara, R. et al. Time-course liver microarray data from 0 to 14 days in a 70% partial hepatectomy mouse model. Sci Data 12, 773 (2025). https://doi.org/10.1038/s41597-025-05128-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-05128-z