Abstract
Understanding the population dynamics of fish is vital to maintaining healthy ocean ecosystems and the human communities that depend on them. However, most fish population models lack crucial information about early life stages. Even when fish embryos and larvae (collectively, ichthyoplankton) are sampled, similar morphological features make species-level visual identification difficult, especially for embryos. So, although the Northeast US continental shelf is well-studied, existing ichthyoplankton datasets generally lack species-level information on embryos. Unlike visual methods, DNA barcoding can readily discriminate between morphologically indistinguishable specimens. Thus, our dataset of 2,294 DNA barcoded fish embryos and larvae, collected near New York (USA), contributes valuable insight into local ichthyoplankton assemblages. This dataset contains 50 unique species. Most have previously been documented in our region, but one was novel: we identified an embryo of Bathyanthias mexicanus, a species not known to inhabit or spawn in the sampled area. More broadly, this dataset can be used to address questions about fish habitat across life stages and to help ensure that local fisheries are sustainably managed.
Similar content being viewed by others
Background & Summary
Fish are the most abundant and diverse vertebrate taxa on Earth, giving rise to modern tetrapods. Studying fish diversity and distribution is critical to understanding biodiversity in aquatic ecosystems. Fish also play crucial roles in marine food webs. Changes in fish populations can alter ecosystems by disrupting predators, including marine birds and mammals, and impact nutrient cycling1.
Moreover, fish are vital to human life, possessing social, cultural, recreational, and economic significance1,2. They are a global protein source, particularly in developing countries, and contribute largely to global and local economies3. In its 2022 report to Congress, the National Oceanic and Atmospheric Administration (NOAA) stated that recreational and commercial marine fisheries contributed $321 billion to the American economy and supported 2.3 million jobs. According to a 2018 survey developed by the Suffolk County Department of Economic Development and Planning, marine employment on Long Island, New York (USA), generated approximately 34,000 jobs with an estimated $1 billion in wages. Additionally, the NOAA Fisheries database indicates that in 2022, the Montauk and Shinnecock ports brought in a combined 12.5 million pounds of fish while generating $24 million in revenue.
Understanding fish population dynamics is essential to maintaining these important ecological, social, and economic benefits. Considerable resources are devoted to monitoring fish, including regular surveys (e.g., refs. 4,5,6,7) and tagging and tracking studies (e.g., refs. 8,9,10,11). These efforts primarily gather data on adult fish. Such data are used to develop population dynamics models, which in turn inform stock assessments and management guidelines. However, fish population dynamics models often lack data on their early life stages – especially the distributions and abundances of fish embryos. This knowledge gap contributes to uncertainty in stock assessments12, hindering effective marine fish species management.
Fish have complex life histories. Many species begin their life cycle as plankton, residing within 200 meters of the ocean surface and exhibiting limited swimming capabilities13. Small size and lack of mobility make planktonic fish embryos and larvae – collectively referred to as ichthyoplankton – vulnerable to various elements. Survival through these fragile early stages can determine overall population dynamics13. As fish mature into juveniles, some shift from pelagic waters to the benthos14. Adults may migrate long distances to spawn or recruit, a strategy to compensate for the differing habitat requirements during the various life history stages of a species14. Therefore, monitoring both ichthyoplankton and adult fish is important to gain a complete understanding of fish population dynamics.
However, studying the early life history stages of fish species, especially the embryonic stages, has historically been challenging due to the high degree of morphological similarities between species. Our inability to visually identify fish embryos at the species level leads to extensive knowledge gaps regarding their spatial and temporal abundances and distributions, which further contributes to uncertainty in overall estimates of population dynamics.
DNA barcoding is an effective solution to this challenge as it allows for the easy and reliable species-level classification of ichthyoplankton. Using the polymerase chain reaction (PCR), a specific genetic region is amplified, most commonly the mitochondrial cytochrome c oxidase subunit 1 (CO1) gene. The CO1 sequence is compared to reference databases, such as the Barcode of Life Database System (BOLD). As of October 2024, over 19.3 million samples have been identified through this database, including 524,359 bony fish (class Actinopterygii).
However, while DNA barcoding of ichthyoplankton can fill essential knowledge gaps in our understanding of fish early life history, the technique has not been widely applied to samples collected on the Northeast US continental shelf. In this region, ichthyoplankton have been studied for decades15,16 – including via NOAA’s plankton surveys, conducted regularly since the 1970s. These studies typically rely on visual methods and as a result do not provide species-level information on embryos, which are virtually impossible to distinguish morphologically. Lewis et al. (2016) used DNA barcoding to identify 1,495 (93.26%) embryos collected from 2002–201217, but this pilot project has not been continued since.
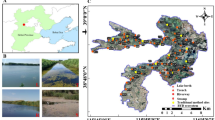
Thus, we present our DNA barcoding dataset to contribute key missing information on ichthyoplankton distribution and abundance. We sampled across the New York Bight subregion of the Northeast US continental shelf, from the south shore of Long Island to the continental shelf break (Fig. 1), generating a large, comprehensive dataset of species-level identification of early life stages of fish. Concurrent with biological sampling, we collected environmental data using a conductivity, temperature, and depth (CTD) unit. Our dataset contains results from quarterly sampling from 2021–2023. We isolated 2,294 individual ichthyoplankton, including 1,344 embryos. We have nearly doubled the number of available species-level identifications of fish embryos in our commercially important region.
Map of the New York Bight region surveyed between spring 2021 and fall 2023. The pie charts reflect our sampling effort: the diameter of the circles represents the frequency at which a location was sampled, and the colored slices show the seasonal breakdown. The westernmost transect was sampled in the initial survey design as the starting point of survey cruises. A slight change in the survey design moved the stations eastward.
This dataset provides valuable insights that can address several critical questions related to species presence, as well as the spatial and temporal dynamics of fish spawning. It can also be used to detect shifting species ranges. Additionally, relationships between ichthyoplankton abundance and abiotic factors, including temperature and stratification, and potential impacts of climate change can be explored with our data. These results can also inform the development of conservation methods necessary for local fish populations to sustain and preserve ecosystem stability. Our dataset is especially significant given that information on fish embryos present in the New York Bight is generally lacking, despite the importance of fishing to local economic and recreational activities.
Methods
Sampling protocols
From 2021 to 2023, we sampled fixed stations in the New York Bight subregion of the Northeast US continental shelf, which spans from the mouth of the Hudson River to Montauk Point and extends to the continental shelf break (Fig. 1). Cruises were conducted seasonally, although adverse weather, mechanical difficulties, and COVID-related disruptions sometimes shortened sampling efforts. In 2023, the station design was altered slightly, and winter sampling was discontinued. Ichthyoplankton samples were collected with a vertical plankton tow of a 60 cm diameter, 333 μm mesh net at a 25 m maximum depth. Collections were permitted by the New York State Department of Environmental Conservation (scientific permit #1145). After collection, samples were stored in 95% ethanol. At the same station, we collected water column profiles of temperature, salinity, fluorometry, dissolved oxygen (DO), photosynthetic active radiation (PAR), and pH using a Seabird 911 + CTD with a rosette carrying eight 10 L Niskin bottles. This data was processed using Seabird’s data processing software.
Ichthyoplankton analyses
Samples were stored at 4 °C prior to analyses and then diluted to a 2:1 ethanol:biovolume ratio. Samples were sorted in their entirety under a Nikon SMZ1270 stereo zoom microscope. Ichthyoplankton individuals were isolated and imaged using a DFK 33UX174 camera from the Imaging Source and IC Imaging Control software. We used ImageJ to measure the diameter of embryos and the body length of larvae. We specifically report the notochord length, measuring from the end of the tail, excluding the finfold, to the tip of the nose. For bent larva, we measured adjacent segments following the spine to estimate the length. We recorded no available measurement when the specimen was severed or folded onto itself.
After imaging, the isolated individuals were placed in a 96-well PCR plate containing 30 μL of 95% ethanol per well. One well was omitted as a negative control. The plate was shipped to the Canadian Centre for DNA Barcoding at the University of Guelph (Canada), where standard protocols were followed, as detailed below.
PCR Protocols
The samples were broken down with standard lysis buffer containing proteinase K (Thermo Fisher Scientific Inc., Waltham, MA), and DNA extraction was performed using an automated protocol. PCR primers cocktails C_FishF1t1 (made up of VF2_t1: TGTAAAACGACGGCCAGTCAACCAACCACAAAGACATTGGCAC and FishF2_t1: TGTAAAACGACGGCCAGTCGACTAATCATAAAGATATCGGCAC18 ) and C_FishR1t1 (FishR2_t1: CAGGAAACAGCTATGACACTTCAGGGTGACCGAAGAATCAGAA and FR1d_t1: CAGGAAACAGCTATGACACCTCAGGGTGTCCGAARAAYCARAA18), and a PCR mix of 6.25 μL of 10% trehalose, 1.25 μL 10 × PCR buffer, 0.625 μL (2.5 mM) MgCl2, 0.625 μL (10 mM) deoxyribonucleotide triphosphates (dNTPs), 0.625 μL Platinum Taq polymerase (Thermo Fisher Scientific Inc.), 3 μL H20 and 1 μL of DNA template amplified the ~658 base pair CO1 gene. This target region was chosen so that our results align with the previous ichthyoplankton barcoding study conducted in our region17. This mixture was heated in cycles of 1 min at 94 °C; 5 cycles of 30 s at 94 °C; 40 s at 55 °C; 1 min at 72 °C; 35 cycles of 30 s at 94 °C; 40 s at 55 °C; 1 min at 72 °C; 10 min at 72 °C. The mixture underwent gel electrophoresis through the E-Gel system (Thermo Fisher Scientific Inc.). The amplified DNA fragments were sequenced through BigDye Terminator version 3.1 and reviewed on an Applied Biosystems 3730xl DNA Sequencer (Thermo Fisher Scientific Inc.). Trace files were put through Codon Code Aligner software (CodonCode Corp., Centerville, MA) to produce continuous sequences of DNA. Species identifications were obtained through comparison with BOLD’s database of published sequences. Resulting sequences were shared on Genbank in addition to BOLD.
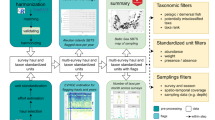
In the rare instances when DNA barcoding or sequencing failed, we employed one of two alternative methods for identification. For one plate with a particularly low success rate, we reran PCRs using an alternative forward primer (AquaF2, ATCACRACCATCATYAAYATRAARCC18) and processed the fragments and sequences as above. Additionally, in some cases, sequence information was low quality but salvageable; in these instances, we used the DNA Subway toolkit19 to clean the sequence and identify the individual via a BLAST search. When PCR and/or sequencing failed entirely, and no sequence information was available, we attempted visual identification of larvae. To do so, we consulted both our own images of larvae that we had successfully identified through BOLD and ref. 15, which details the unique features of Western North Atlantic species throughout their larval development. Figure 2 describes our full workflow, from collection to identification. We also note that the elimination of winter sampling should not impact the utility of this dataset, as only a small minority of the resident fish species spawn in winter. When we sampled in the winter of 2022, for example, we only isolated one individual from seven stations.
Flow chart representing the steps from collection to species-level identification of ichthyoplankton. (A) is a Northern Searobin (Prionotus carolinus) embryo collected during October 2023. (B) is a Gulfstream Flounder (Citharichthys arctifrons) embryo collected during October 2023. These two embryos cannot be distinguished visually but are readily identifiable genetically.
For a small number of closely related species, we made assumptions based on known species ranges. For example, some embryos’ sequences matched Atlantic Menhaden (Brevoortia tyrannus) and Gulf Menhaden (Brevoortia patronus). Our study area overlaps with the range of Atlantic but not that of Gulf Menhaden, and therefore, we assumed the embryos belonged to the Atlantic species (note also that recent genetic evidence does not categorically support the historic species distinction, instead suggesting they might be separate populations of a single species20). Similarly, we inferred that embryos were Atlantic, not Gulf, Butterfish (Peprilus triacanthus, not Peprilus burti) and Northern, not Blackwing, Searobin (Prionotus carolinus, not Prionotus rubio). Finally, we deduced that sequences that matched both Bullet and Frigate Tuna (Auxis rochei and Auxis thazard) matched with sequences previously misidentified as the latter species. We, therefore, counted them all as Bullet Tuna. We did not use visual information to distinguish between these closely related species.
In total, we isolated 2,294 individual ichthyoplankton – 1,344 embryos and 950 larvae – from 7 offshore cruises. Of these, 2,175 (95%) were identified at the species level using one of the techniques described above, finding 50 unique species in our region and sampling period (Table 1).
Noteworthy findings
Of the 50 species identified, 32 (64%) were also found in an earlier study that used DNA barcoding to identify fish eggs collected across the Northeast US continental shelf – an area including but not limited to our study region17. Of the remaining 18 species, 8 were found exclusively at a shelf break station, adjacent to the Slope Sea, that we sampled in September 2022. An additional species (Bothus robinsi) was also found at the shelf break in September 2022 and at one other station we sampled. Of the nine species uniquely found in September 2022, eight were previously found in a 1988 study of larval fish in the New York Bight and described as belonging to a ‘slope assemblage’16. Of note, the September 2022 cruise occurred between Hurricanes Fiona (Category 4) and Ian (Category 5) – suggesting that the transport of these uncommon species may have been storm-related.
There were nine taxa not found in refs. 16,17, nor on the shelf break during September 2022. Of these, 4 are regularly identified in routine larval sampling by NOAA: Ophidion sp., Opisthonema oglinum, Vinciguerria attenuata, and Rhomboplites aurorubens21. Of the remaining 5, larvae of 4 species – Antigonia capros, Astroscopus guttatus, Rachycentron canadum and Stenotomus chrysops – are extremely rare in routine offshore ichthyoplankton sampling, with fewer than 10 records in decades of bimonthly effort (Dr. David Richardson, personal communication). Scup (Stenotomus chrysops) is of particular interest as this is a commercially important species whose early life history is poorly understood. We found eight scup embryos in our study, which would not have been identified via traditional visual methods. Cobia (Rachycentron canadum) supports a valuable recreational fishery, and it has been hypothesized that this species is undergoing a climate change-induced northward range shift22,23 – a hypothesis our results support.
Bathyanthias mexicanus is the one ichthyoplankton species we found that, to our knowledge, had not been previously identified in our region. This species was first described from samples collected in the Gulf of Mexico in the 1950s24. It is described as a deep water Serranid of the subfamily Epinephelinae, ranging from the Guianas and Venezuela to the Florida east coast, but is most common in the Gulf of Mexico25.
None of the species included in our results are considered endangered, threatened, or protected.
Data Records
All data records are available on Figshare26. This includes records of each species identification (scientific and common name), cruise ID, collection date, time, longitude, and latitude, and links to images27. We included information on whether species identification was determined via BOLD, BLAST, or visually, as well as temperature and salinity measurements from CTD casts. Along with the CTD measurements, we report the longitude, latitude, date, and time of each CTD cast. Additionally, we have shared folders of specimen images organized by the 96-well PCR plate number submitted to CCDB and the corresponding BOLD process IDs28.
Technical Validation
Species identifications were obtained through BOLD Systems with a strict identification threshold of 99% homology. Additionally, we estimated our sorting error rate, i.e., the probability that we failed to isolate a fish embryo or larva from the other zooplankton in our sorting process, at 5.6%. We derived this estimate by selecting a random 10% of our sampled volume for re-sorting. Two people conducted re-sorting simultaneously to increase the likelihood of finding any individuals missed during the initial round. For most re-sorted samples, we found zero individuals; in the minority, we isolated one or two. However, we did notice one anomalous sample in which a large number of individuals had been missed in the first round. We re-sorted 14% of this sample volume and found 12 individuals, while 17 had been found previously. We thus determined that there was a more systematic issue with this particular sample – likely related to poor ship-board storage – and decided to re-sort it in its entirety. This is ongoing work, and the databases will be updated to reflect new results once they are available.
Any individuals isolated during the quality control re-sorting were barcoded, and their results are included in our current dataset. No new species were identified in this process.
Usage Notes
In addition to Figshare, data are available on BOLD Systems as a public dataset29. Although this study is ongoing, data reported in BOLD, Genbank, and Figshare reflect the work completed at the time of publication of this manuscript. Added data will be associated with a new version of the respective datasets.
Code availability
No custom code was used in this study.
References
Nissar, S. et al. A review of the ecosystem services provided by the marine forage fish. Hydrobiologia 850, 2871–2902, https://doi.org/10.1007/s10750-022-05033-1 (2023).
Gephart, J. A. et al. Scenarios for Global Aquaculture and Its Role in Human Nutrition. Reviews in Fisheries Science & Aquaculture 29, 122–138, https://doi.org/10.1080/23308249.2020.1782342 (2021).
Febrian, W. D., Sani, I., Wahdiniawat, S. A., Apriani, A. & Maulina, E. Sustainable Development Building With The Analytical Approach of Blue Economic And Food Security. Journal of Economics, Management, Entrepreneurship, and Business (JEMEB) 3, 54–60, https://doi.org/10.1016/j.ecolind.2021.108344 (2023).
Nieman, C. L., Pendleton, R. M., Kenney, G. H. & Solomon, C. T. Evaluation and optimization of a long-term fish monitoring program in the Hudson River. Ecological Indicators 133, 108344, https://doi.org/10.1016/j.ecolind.2021.108344 (2021).
Rastoin-Laplane, E. et al. Fluctuations of Galapagos mid-water and benthic reef fish populations during the 2015–16 ENSO. Estuarine, Coastal and Shelf Science 294, 108523, https://doi.org/10.1016/j.ecss.2023.108523 (2023).
Zhao, J. et al. Thirty years of change: Assessing the dynamics of fish communities in Daya Bay, a semi-enclosed coastal ecosystem of the South China sea. Water Biology and Security 3, 100268, https://doi.org/10.1016/j.watbs.2024.100268 (2024).
Bacheler, N. M. et al. Spatiotemporal dynamics and habitat use of red snapper (Lutjanus campechanus) on the southeastern United States Atlantic continental shelf. Fisheries Research 281, 107200, https://doi.org/10.1016/j.fishres.2024.107200 (2025).
Jensen, A. J. et al. Modeling ocean distributions and abundances of natural- and hatchery-origin Chinook salmon stocks with integrated genetic and tagging data. PeerJ 11, e16487, https://doi.org/10.7717/peerj.16487 (2023).
de Pontual, H. et al. Seasonal migration, site fidelity, and population structure of European seabass (Dicentrarchus labrax). ICES Journal of Marine Science 80, 1606–1618, https://doi.org/10.1093/icesjms/fsad087 (2023).
Goddard, B. K. et al. Half a century of citizen science tag-recapture data reveals stock delineation and cross-jurisdictional connectivity of an iconic pelagic fish. Rev Fish Biol Fisheries 34, 1433–1449, https://doi.org/10.1007/s11160-024-09880-0 (2024).
Marcotegui, P. et al. An independent stock of Engraulis anchoita in north Patagonian waters? Parasite tags as evidence. Fisheries Research 279, 107148, https://doi.org/10.1016/j.fishres.2024.107148 (2024).
Cadrin, S. X. Defining spatial structure for fishery stock assessment. Fisheries Research 221, 105397, https://doi.org/10.1016/j.fishres.2019.105397 (2020).
Houde, E. D. Fish Larvae. in Encyclopedia of Ocean Sciences (Second Edition) (ed. Steele, J. H.) 381–391, https://doi.org/10.1016/B978-012374473-9.00026-6 (Academic Press, Oxford, 2001).
Llopiz, J. et al. Early Life History and Fisheries Oceanography: New Questions in a Changing World. Oceanography 27, 26–41, https://doi.org/10.5670/oceanog.2014.84 (2014).
Fahay, M. P. Early Stages of Fishes in the Western North Atlantic Ocean. (Northwest Atlantic Fisheries Organization, Dartmouth, Nova Scotia, Canada, 2007).
Cowen, R., Hare, J. & Fahay, M. Beyond Hydrography: Can Physical Processes Explain Larval Fish Assemblages within the Middle Atlantic Bight? Bulletin of Marine Science 53, 567–587 (1993).
Lewis, L. A., Richardson, D. E., Zakharov, E. V. & Hanner, R. Integrating DNA barcoding of fish eggs into ichthyoplankton monitoring programs. Fishery Bulletin 114, 153–165, https://doi.org/10.7755/FB.114.2.3 (2016).
Ivanova, N. V., Zemlak, T. S., Hanner, R. H. & Hebert, P. D. N. Universal primer cocktails for fish DNA barcoding. Molecular Ecology Notes 7, 544–548, https://doi.org/10.1111/j.1471-8286.2007.01748.x (2007).
Marizzi, C. et al. DNA barcoding Brooklyn (New York): A first assessment of biodiversity in Marine Park by citizen scientists. PLOS ONE 13, e0199015, https://doi.org/10.1371/journal.pone.0199015 (2018).
Pozzobon, A. P. B. et al. Phylogenetic relationships, genetic diversity and biogeography of menhadens, genus Brevoortia (Clupeiformes, Clupeidae). Mol Phylogenet Evol 160, 107108, https://doi.org/10.1016/j.ympev.2021.107108 (2021).
NOAA Fisheries Northeast Fisheries Science Center. Zooplankton and ichthyoplankton abundance and distribution in the North Atlantic collected by the Ecosystem Monitoring (EcoMon) Project from 1977-02-13 to 202111-15 (NCEI Accession 0187513). NOAA National Centers for Environmental Information. https://www.ncei.noaa.gov/archive/accession/0187513 (2019).
Crear, D. P. et al. Contemporary and future distributions of cobia, Rachycentron canadum. Diversity and Distributions 26, 1002–1015, https://doi.org/10.1111/ddi.13079 (2020).
Crear, D. P., Watkins, B. E., Friedrichs, M. A. M., St-Laurent, P. & Weng, K. C. Estimating Shifts in Phenology and Habitat Use of Cobia in Chesapeake Bay Under Climate Change. Front. Mar. Sci. 7, https://doi.org/10.3389/fmars.2020.579135 (2020).
Schultz, L. P. Three New Serranid Fishes, Genus Pikea, from the Western Atlantic. Proceedings of the United States National Museum 108, 321–329, https://doi.org/10.5479/si.00963801.108-3405.321 (1958).
Bullock, L. H. & Smith, G. B. Seabasses: (Pisces: Serranidae) Volume 8, Part 2 of Memoirs of the Hourglass cruises. Florida Marine Research Institute 8, (1991).
Weisberg, S. NYOS_barcoding_data Figshare https://figshare.com/projects/NYOS_barcoding_data/251636 (2025).
Weisberg, S. NYOS_Barcoding_CTD_Data.xlsx Figshare https://doi.org/10.6084/m9.figshare.27984443.v4 (2025).
Hadjiargyrou, S. NY Offshore Ichthyoplankton Images Figshare https://doi.org/10.6084/m9.figshare.29230895 (2025).
Weisberg, S. Barcode of Life Data Systems https://doi.org/10.5883/DS-NYOS (2024).
Acknowledgements
Funding for this work was provided primarily by the New York State Department of Environmental Conservation (MOU #900286-02). We are grateful to the many scientists and ship crews who have been involved in the long-term NYOS monitoring since 2017. We especially thank Tyler Menz for collecting and processing the CTD data and generously supporting our work. We also thank Kailey Charles, Jason Marte, Olivia Bonilla, and Natasha Gonzales for their contribution to data collection and processing. We also thank several partner organizations for their funding support via student stipends: BioBus, Inc., NY Sea Grant, NY State Department of Environmental Conservation, the American Fisheries Society Hutton Scholars program, East Side Community School (part of NYC Public Schools), and the National Science Foundation’s GEOPaths program. Finally, we appreciate the support from Stony Brook University’s Simons STEM Scholars and WISE programs, which facilitated student participation in this project. We kindly thank the anonymous reviewers for their feedback, which greatly improved the manuscript.
Author information
Authors and Affiliations
Contributions
J.A.N. and S.J.W. directed the project, and S.J.W. collected samples. S.L.H., M.R., H.K., S.H., A.A.V.T., B.J.B., M.K.K., R.K., and S.J.W., processed plankton samples and isolated individual ichthyoplankton. S.L.H., H.K., M.R., M.K.K., B.J.B., and S.J.W. processed sequence data. H.K. led visual identification efforts. S.L.H. and S.J.W. drafted the manuscript with input from all authors. M.R. and S.J.W. performed data cleaning and quality control. All authors contributed feedback and helped shape the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hadjiargyrou, S.L., Rubin, M., Karkout, H. et al. Novel species-level database of fish embryos and larvae in New York offshore waters, 2021–2023. Sci Data 12, 1638 (2025). https://doi.org/10.1038/s41597-025-05536-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-025-05536-1