Abstract
Multi-echo-time (MTE) diffusion MRI (dMRI) offers several advantages over conventional single TE dMRI, including disentanglement of microstructural and compositional differences, reduction of bias in microstructural properties, and addition of sub-cellular T2 measures. However, MTE methods require additional data acquisition and complex model fitting. In this work, we share a comprehensive MTE dMRI dataset acquired from three healthy subjects in ten TE sessions (each with eight TEs: 62-132 ms and repeated measures at the shortest and longest TEs). The dataset includes two b-values (700 and 2000 s/mm2) with 30 gradient directions for each b-value and four b = 0 images, with diffusion times fixed (δ/Δ = 15.2/25.2 ms) across b-values and TEs. Preprocessing steps include denoising, corrections for B0 inhomogeneity, eddy current and motion correction, and aligning the DWIs and b-vectors to the first TE session. The dataset quality is validated by SNR and head motion assessments. The usage of the dataset is shown with microstructure metrics and orientation distribution functions across TE sessions, which may facilitate investigation in combined diffusion-relaxometry.
Similar content being viewed by others
Background & Summary
Diffusion magnetic resonance imaging (dMRI) is a noninvasive technique widely used to probe the microstructure of biological tissues1. Conventional dMRI acquisitions employ a single echo time (TE)2,3,4,5,6, which imposes fixed T2 weighting across all measurements and have been shown to cause variations of dMRI metrics when the TE changes. For instance, changes with TE have been reported in the apparent diffusion coefficient (ADC)7,8, the diffusion tensor imaging (DTI) metrics including the fractional anisotropy (FA), axial diffusivity (AD), and radial diffusivity (RD) in the white matter regions9,10, as well as the compartmental signal fractions from the neurite orientation dispersion and density imaging (NODDI). This TE-dependence of dMRI metrics limits the interpretability of conventional studies.
More recent multi-echo-time (MTE) dMRI studies address this limitation by collecting multi-dimensional data that enhances sensitivity to both microstructural and compositional differences within tissue compartments11,12,13,14,15,16. With such data, it is possible to model both diffusion and T2 effects in tissue compartments. This combined diffusion-T2 modeling not only reduces biases in microstructural parameters but also provide additional sub-cellular T2 measures, offering potential for more specific biomarkers.
Despite these valuable insights, methods that incorporate multiple TEs are not widely accessible and therefore remain underutilized. Several factors attribute to this limitation: (1) acquiring dMRI data at multiple TEs necessitates additional scan time, which can be impractical in clinical settings; (2) longer TEs often result in reduced signal-to-noise ratio (SNR), complicating data acquisition in brain regions with short T2 relaxation time; and (3) echo-planar imaging (EPI), the predominant sequence used for dMRI, is prone to distortions that intensify at longer TEs because of increased susceptibility to B0 field inhomogeneities.
To bridge this gap, we present a 3 T dMRI dataset collected from three subjects across ten sessions, incorporating a diverse range of TEs. This dataset includes eight different TE sessions, ranging from 62 ms to 132 ms, with two b-values (700 and 2000 s/mm2) across 30 gradient directions and diffusion time fixed at each b-value and TE session. The multi-shell acquisition makes the data applicable to more complex models17,18. Additionally, we repeated the measurements at TE = 62 and 132 ms, considering that the shortest and longest TE sessions have been shown to critically impact the fitting of MTE model11.
By openly sharing this dataset, we aim to facilitate research in combined diffusion-relaxometry modeling, reproducibility analysis, and optimization of minimal acquisition protocols. This dataset further enables exploration of tissue-specific variations in MTE-derived microstructural properties across the brain, advancing the development of robust biomarkers for clinical research.
Methods
Participants
Data were collected from three healthy volunteers: subject 1 (male, 24 years), subject 2 (female, 27 years), and subject 3 (male, 28 years). The participants were recruited through printed advertisements. The written informed consent for data collection and data sharing was obtained from all participants and approved by the local research Ethics Committee of Zhejiang University (ref. 2018376).
Image acquisitions
All MRI scans were conducted using a Siemens 3 T Prisma scanner (Siemens, Erlangen, Germany) equipped with a 64-channel head-neck coil. The dMRI data were acquired with a repetition time (TR) of 5800 ms across ten sessions, each lasting approximately seven minutes. Imaging included eight different TEs: 62, 72, 82, 92, 102, 112, 122, and 132 ms. Two repetitions were collected at the shortest and longest TEs (62R2 and 132R2). Two b-values were used in the acquisition: b = 700 s/mm2 and b = 2000 s/mm2, applied in 30 gradient directions with uniform angular coverage, and the same directions were used for both b-values. A total of four b = 0 images were acquired: two in the anterior-to-posterior (A-P) direction and two in the posterior-to-anterior (P-A) direction. Diffusion times were fixed with gradient width and separation set at δ/Δ = 15.2/25.2 ms for all b-values and TEs. Other imaging parameters included: Field of View (FOV): 210 × 210 mm2; Number of slices: 48; Resolution: 2.5 × 2.5 × 2.5 mm³; Slice acceleration factor: 2; Phase acceleration factor: 2; Bandwidth: 2380 Hz/pixel.
Image preprocessing
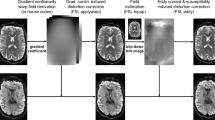
The data were organized according to the Brain Imaging Data Structure (BIDS) format. The preprocessing pipeline for diffusion-weighted images (DWIs) was performed separately for each TE session (Fig. 1). This included: (1) denoising using MRtrix3 with dwidenoise function (Marchenko-Pastur Principal Component Analysis denoising)19,20; (2) correcting for B0-inhomogeneity using TOPUP function21; (3) correcting for eddy current-induced distortion using EDDY22 functions in FSL. The effect of additional preprocessing steps, specifically Gibbs ringing, B1 field inhomogeneity and Rician bias correction, was assessed. However, considering the marginal improvement that these steps had, only the results with the first three steps are reported to keep preprocessing minimal.
Preprocessing pipeline for multiple TEs data. The left panel (Raw data) shows unprocessed b = 0 images for anterior-posterior (AP) and posterior-anterior (PA) phase encoding directions, with DWIs at b = 700 s/mm2 and b = 2000 s/mm2 across 30 directions. The right panel (Preprocessed data) displays the same data after applying preprocessing steps, including denoising, B0 inhomogeneity correction, eddy and motion correction, and aligning the DWIs and b-vectors to the first TE session.
To align the DWIs across multiple TE sessions, linear registration was performed using FLIRT in FSL to register mean b = 0 images from later TEs to the first TE. The affine matrix obtained from FLIRT was then applied to the eddy-corrected DWIs. Finally, the eddy corrected b-vectors of the later TEs were rotated again using the obtained affine matrix to align with the first TE.
Data Records
The dataset is available at Science Data Bank23 (https://doi.org/10.57760/sciencedb.o00133.00038). All personal identifiers were not included and the facial features were also not included as the FOV only covered the brain. The neuroimaging dataset is available in two formats: raw and preprocessed MRI data, both of which are available for all release subjects. For each subject, unprocessed and preprocessed images for dMRI are provided within each TE session (Fig. 2).
Data organization structure for dMRI datasets. Subjects (sub-01, sub-02, sub-03) are acquired with multiple echo times (TEs), including TE62, TE72, TE82, TE92, TE102, TE112, TE122, and TE132. Two repetitions are acquired at the shortest and longest TEs (TE62R2, and TE132R2). The data is stored in different folders: raw (containing A-P and P-A b = 0 images and DWIs with b-values and vectors); and dwi (unmasked average b = 0 image and DWIs with b-values and vectors).
Technical Validation
Visual inspection
The mean DWIs from each b-value and TE session from all subjects (Fig. 3) were shown with no significant ghosting or reconstruction artifacts by visual inspection. The increase in TE resulted in reduced signal intensities for every b-value, indicating the reduction of the signal to noise ratio at longer TEs.
Multi-echo diffusion-weighted brain MRI images from all subjects (A–C). In each panel, each row represents the mean diffusion-weighted images at b = 0 s/mm2, b = 700 s/mm2, and b = 2000s/mm2, and each column represents different TE sessions (TE62, TE62R2, TE72, TE82, TE92, TE102, TE112, TE122, TE132, and TE132R2).
Temporal signal to noise ratio
The temporal signal-to-noise ratio (tSNR) was calculated for each voxel as the ratio of the mean to the standard deviation derived from the sigma noise map obtained in the denoising preprocessing step. White matter (WM) and gray matter (GM) masks were generated using FAST24 function in FSL based on the average b = 0 image for each subject. The average tSNR within both masks was computed for each subject by averaging tSNR values across all voxels in the masks. The increase in TE led to a substantial reduction in average tSNR (Fig. 4), which was consistent with greater T2-related signal decay at longer echo times.
Head motion
Head motion was estimated using the EDDY tool during DWI preprocessing. Translation and rotation parameters relative to the first DWI volume were extracted from the *.eddy_parameters file (Fig. 5A and B). Additionally, two summary measures of volumetric head motion were calculated as the root mean square (RMS) displacement: one representing displacement relative to the first volume and the other representing displacement between adjacent volumes, as listed in the *.eddy_restricted_movement_rms file (Fig. 5C). The mean and standard deviation of these head motion measurements across all subjects were reported in Table 1 (Translation), Table 2 (Rotation), and Table 3 (RMS).
Head motion measurement estimated from the rigid-body relative to the first DWI volume of a representative subject, quantified in the translation (A) along the X, Y, and Z-axis, as well as rotation (B) (X-axis: pitch, Y-axis: roll, Z-axis: yaw), and (C) the RMS displacement to the first and previous volumes.
Impact of TE on Diffusion Model Parameters
To demonstrate the effects of TE on diffusion model parameters, we fitted the data from a representative subject to the DTI, NODDI, and the multi-shell multi-tissue constrained spherical deconvolution model (MSMT-CSD)18.
This assessment demonstrates the usage of the data for microstructural and fiber orientation mapping.
DTI
The DTI model25, a widely utilized model in clinical studies that requires data from only a single b-shell, was separately fitted to the dataset (b = 700 s/mm2) for each TE session using DTIFIT in FSL. Whole-brain analysis (Fig. 6A) demonstrated that progressive increases in TE were associated with the decrease in FA and AD, and increase in MD and RD.
To further illustrate TE-dependent effects, regions of interest (ROIs) analysis was performed on WM structures extracted from the Johns Hopkins University (JHU) WM atlas26, including the body (BCC), genu (GCC), splenium (SCC) of the corpus callosum, and the superior longitudinal fasciculus (SLF). ROI analysis (Fig. 6B) revealed that when increasing TE, FA decreased in the BCC, whereas other regions exhibited minimal percentage change. Similarly, the decrease of AD also displayed in the BCC and SCC with increasing TE but remained relatively stable in other regions. However, RD demonstrated a decline in the GCC and SCC with the increase in TE. Lastly, the mean diffusivity (MD) reduced in the SCC.
NODDI
The NODDI model, a more complex representation of tissue microstructure that requires data from both low and high b-values with multiple gradient directions, which provides three metrics such as neurite density index (NDI), orientation dispersion index (ODI), and free water (FW), was fitted to the dataset using the NODDI toolbox executed in MATLAB 2018a (MathWorks, Natick, MA, USA) with default settings for each TE session separately. Our results (Fig. 6C and D) indicated that an increase in TE led to an increase in NDI and FW, while ODI remained relatively stable in the whole brain and across all ROIs.
Maps of dMRI metrics for every echo time session for DTI model (A), and NODDI model (B), and percentage change of dMRI metrics across brain regions at different TEs compared with the first TE (TE = 62 ms) of a representative subject. Line plots illustrate TE-dependent effects on: (C) DTI metrics, FA, MD AD, and RD; (D) NODDI metrics, NDI, ODI, and FW. Negative values denote reductions, while positive values indicate increases relative to baseline (TE = 62 ms).
Comparison of effect size
The effect size analysis (Fig. 7) revealed substantial advantages for NODDI model, with NDI, ODI, and FW metrics demonstrating effect sizes 2-3 times larger than traditional DTI measures across multiple brain regions. These findings support the adoption of advanced dMRI models as complementary tools to standard DTI analysis, and may provide enhancement for detecting microstructural differences in both research and clinical applications.
MSMT-CSD
The fiber orientation distributions (FODs) of a representative subject from dMRI data at different TE sessions was reconstructed using multi-shell multi-tissue (MSMT-CSD) with dhollander algorithm in MRtrix3 (Fig. 8). FODs resolved crossing fibers, with variations observed across TE sessions (TE62–TE132R2), underscoring influence of TE on microstructural sensitivity.
Fiber orientation distributions (FODs) from a representative subject, reconstructed using multi-shell multi-tissue constrained spherical deconvolution (MSMT-CSD) with the dhollander algorithm in MRtrix3. The left panel highlights a region of interest (red box) on a T2-weighted slice overlaid with the FODs, and the panels on the right display the zoom-in of the FODs within the red box at the crossing fiber region across multiple echo time (TE) sessions.
Usage Notes
The current dataset, which includes healthy controls, complementing prior patient-focused studies. Unlike the recently published dataset27—acquired with 4–16 gradient directions per b-value (b = 400, 800, 1600, 3200 s/mm2)—the current dataset employs 30 gradient directions per b-value, enabling robust validation of healthy control populations. This enhanced parameterization also facilitates detailed exploration of the interplay between diffusion and T2 relaxation effects, particularly for advanced modeling in tissue microstructure characterization and compartment analysis. However, the current dataset is limited by small sample size (N = 3) as the acquisition time is very long. With this dataset as a testbed for evaluating accelerated protocols, future studies can acquire larger dataset with an accelerated protocol. The current dataset doesn’t include higher b-shells considering that including them would have made the minimal TE much longer on the clinical system; a shorter minimal TE could potentially improve compartmental T2 estimation11,16. In addition, this dataset may benefit free-water elimination methods for DTI and DKI fitting.
Code availability
The MRtrix3 (version 3.0.4) software for denoising and reconstructing the FODs is publicly available (https://www.mrtrix.org/download/). The FMRIB Software Library (version 6.0.5.1) used in the diffusion data pre-processing (TOPUP and EDDY), and fitting the DTI model is publicly available (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FslInstallation). The NODDI Matlab toolbox is publicly available (http://mig.cs.ucl.ac.uk/index.php?n=Tutorial.NODDImatlab).
References
Le Bihan, D. Looking into the functional architecture of the brain with diffusion MRI. Nat Rev Neurosci 4, 469–480 (2003).
Yao, J. et al. Both noise‐floor and tissue compartment difference in diffusivity contribute to FA dependence on b ‐value in diffusion MRI. Human Brain Mapping 44, 1371–1388 (2023).
Crombé, A., Nicolas, R., Richard, N., Tourdias, T. & Hiba, B. High B-value diffusion tensor imaging for early detection of hippocampal microstructural alteration in a mouse model of multiple sclerosis. Sci Rep 12, 12008 (2022).
Hutchinson, E. B. et al. Analysis of the effects of noise, DWI sampling, and value of assumed parameters in diffusion MRI models. Magnetic Resonance in Med 78, 1767–1780 (2017).
Afzali, M. et al. The sensitivity of diffusion MRI to microstructural properties and experimental factors. Journal of Neuroscience Methods 347, 108951 (2021).
Wongkornchaovalit, P., Feng, M., He, H. & Zhong, J. Diffusion MRI With High to Ultrahigh b-Values: How It Will Benefit the Discovery of Brain Microstructure and Pathological Changes. Investig Magn Reson Imaging 26, 200 (2022).
Buckley, D. L. et al. The effect of ouabain on water diffusion in the rat hippocampal slice measured by high resolution NMR imaging. Magn Reson Med 41, 137–142 (1999).
Vestergaard-Poulsen, P., Hansen, B., Ostergaard, L. & Jakobsen, R. Microstructural changes in ischemic cortical gray matter predicted by a model of diffusion-weighted MRI. J Magn Reson Imaging 26, 529–540 (2007).
Qin, W. et al. Effects of echo time on diffusion quantification of brain white matter at 1.5 T and 3.0 T. Magn Reson Med 61, 755–760 (2009).
Lin, M. et al. Effect of myelin water exchange on DTI-derived parameters in diffusion MRI: Elucidation of TE dependence. Magn Reson Med 79, 1650–1660 (2018).
Gong, T. et al. MTE-NODDI: Multi-TE NODDI for disentangling non-T2-weighted signal fractions from compartment-specific T2 relaxation times. NeuroImage 217, 116906 (2020).
Benjamini, D. & Basser, P. J. Use of marginal distributions constrained optimization (MADCO) for accelerated 2D MRI relaxometry and diffusometry. Journal of Magnetic Resonance 271, 40–45 (2016).
Kim, D., Doyle, E. K., Wisnowski, J. L., Kim, J. H. & Haldar, J. P. Diffusion‐relaxation correlation spectroscopic imaging: A multidimensional approach for probing microstructure. Magnetic Resonance in Med 78, 2236–2249 (2017).
Lampinen, B. et al. Searching for the neurite density with diffusion MRI: Challenges for biophysical modeling. Human Brain Mapping 40, 2529–2545 (2019).
Slator, P. J. et al. Combined diffusion‐relaxometry microstructure imaging: Current status and future prospects. Magnetic Resonance in Med 86, 2987–3011 (2021).
Veraart, J., Novikov, D. S. & Fieremans, E. TE dependent Diffusion Imaging (TEdDI) distinguishes between compartmental T2 relaxation times. NeuroImage 182, 360–369 (2018).
Zhang, H., Schneider, T., Wheeler-Kingshott, C. A. & Alexander, D. C. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage 61, 1000–1016 (2012).
Jeurissen, B., Tournier, J.-D., Dhollander, T., Connelly, A. & Sijbers, J. Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data. NeuroImage 103, 411–426 (2014).
Veraart, J., Fieremans, E. & Novikov, D. S. Diffusion MRI noise mapping using random matrix theory. Magnetic Resonance in Med 76, 1582–1593 (2016).
Cordero-Grande, L., Christiaens, D., Hutter, J., Price, A. N. & Hajnal, J. V. Complex diffusion-weighted image estimation via matrix recovery under general noise models. Neuroimage 200, 391–404 (2019).
Andersson, J. L. R., Skare, S. & Ashburner, J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. NeuroImage 20, 870–888 (2003).
Andersson, J. L. R. & Sotiropoulos, S. N. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage 125, 1063–1078 (2016).
Wongkornchaovalit, P., Shao, B., Li, L., Chen, Y., He, H., Zhong, J., & Gong, T. Multi-TE Diffusion MRI Dataset for Exploring Combined Diffusion-Relaxometry Methods in Microstructure Imaging. https://doi.org/10.57760/sciencedb.o00133.00038 (2025).
Zhang, Y., Brady, M. & Smith, S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging 20, 45–57 (2001).
Basser, P. J., Mattiello, J. & LeBihan, D. Estimation of the effective self-diffusion tensor from the NMR spin echo. J Magn Reson B 103, 247–254 (1994).
MRI Atlas of Human White Matter. (Elsevier, Amsterdam Oxford, 2005).
Wu, Y., Liu, X., Huang, Y., Zhou, T. & Zhang, F. An open relaxation-diffusion MRI dataset in neurosurgical studies. Sci Data 11, 177 (2024).
Acknowledgements
This work was supported by the “Leading Goose” R&D Program of Zhejiang (2023C03094), National Natural Science Foundation of China (82372036, 81871428), and Open Research Fund of the State Key Laboratory of Cognitive Neuroscience and Learning (CNLZD2001).
Author information
Authors and Affiliations
Contributions
P.W. contributed to data analysis, validation, visualization, writing the original draft and editing; B.S., L.L. and Y.C. contributed to editing; H.H. contributed to conceptualization, funding, resources, editing, and supervision; J.Z. contributed to conceptualization, editing, and supervision; TG acquired the MRI data when she was at Zhejiang University, later contributed to conceiving the study, advising on data analysis, and editing the draft. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wongkornchaovalit, P., Shao, B., Li, L. et al. Multi-TE Diffusion MRI Dataset for Exploring Combined Diffusion-Relaxometry Methods in Microstructure Imaging. Sci Data 12, 1191 (2025). https://doi.org/10.1038/s41597-025-05544-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-05544-1