Abstract
Quantitative metabolomic analysis was performed for eleven tissues of freshwater fish pike-perch (Sander lucioperca), including gill, heart, liver, kidney, spleen, muscle, brain, milt, lens, aqueous (AH) and vitreous (VH) humors with the use of NMR spectroscopy. The absolute values of concentrations were determined for more than 65 most abundant metabolites in every tissue. It was found that from the metabolomic viewpoint, kidney and gill are the most similar tissues, while the metabolomic compositions of ocular tissues—lens, AH, and VH significantly differ from that of other tissues. The combinations of intracellular osmolytes and antioxidants are specific for every tissue. In particular, the concentration of antioxidant ovothiol A in the lens is much higher than in any other tissue, while the brain enjoys the elevated level of ascorbate. The most abundant osmolyte in the fish spleen, muscle, and heart is taurine, and in the brain, gill, and lens—myo-inositol. Other important osmolytes specific for particular tissues are N-acetyl-histidine, N-acetyl-aspartate, betaine, threonine-phosphoethanolamine, and serine-phosphoethanolamine. The quantitative data obtained in the present work can be used as the baseline metabolite concentrations in the fish tissues to evaluate the influence of seasonal, ecological and other factors on the fish metabolism.
Similar content being viewed by others
Introduction
Metabolomics is the youngest branch of “-omics” sciences emerged at the beginning of twenty-first century. The complete set of small-molecular-weight compounds in tissue, i.e. its metabolome, includes hundreds of metabolites and reflects the actual metabolic processes occurring in the tissue. Pathological processes cause significant changes in the tissue metabolomic composition; therefore, metabolomic analysis is considered as one of the most promising approaches for the early diagnosis and monitoring of a wide spectrum of diseases1,2,3,4,5. For that reason, the metabolomes of many human tissues are studied in details6,7,8,9,10,11,12, including both quantitative metabolomic profiling and changes induced by various diseases. However, animal tissues are studied to a lesser extent, and the metabolomic composition is known for very limited number of animal tissues13,14,15.
Different tissues consist of different types of cells. Although many metabolic cycles are shared between cell types, the contributions of these cycles into the total metabolic activity are different. Metabolites play an important role in maintaining the cell homeostasis and cellular protection, the protection against oxidative and osmotic stresses in particular. Different cell types may contain different combinations of antioxidants and osmolytes. Thus, one can expect considerable difference between metabolomic compositions of tissues, and it would be of fundamental importance to determine the levels of major metabolites in different tissues of the same species. That will bring a better understanding of mechanisms of cellular protection and of roles of specific metabolic cycles in different tissues. In our recent work15, we compared the metabolomic compositions of lenses and gills of freshwater fish—pike-perch (Sander lucioperca) and Siberian roach (Rutilus rutilus lacustris). It was found that the sets of antioxidants and osmolytes in lenses and gills are different, and they undergo significant seasonal variations. We have also reported the discovery of very powerful natural antioxidant—ovothiol A (OSH) in the fish lens12. The origin of OSH in the lens remains uncertain, and it is interesting to monitor this antioxidant throughout the fish tissues.
This work is devoted to the quantitative metabolomic analysis of a large set of tissues belonging to the same species, S. lucioperca, including gill, heart, liver, kidney, spleen, muscle, brain, milt, lens, aqueous (AH) and vitreous (VH) humors. The major goal of the study is to establish the baseline metabolite concentrations in these tissues. The comparison of metabolomic compositions of different tissues will also help to determine the major osmolytes and antioxidants, and to evaluate the roles of metabolic processes specific for every particular tissue.
Results
Figure 1 shows an example of the NMR spectrum of protein-free lipid-free extract from the fish brain. Similar spectra were obtained for other fish tissues: gill, heart, liver, kidney, spleen, muscle, milt, lens, aqueous and vitreous humors; raw data are available at the MetaboLights repository, study identifier MTBLS1763 (https://www.ebi.ac.uk/metabolights/MTBLS1763). The spectra of S. lucioperca lens and gill have recently been published15 by our group. For the majority of metabolites, the identification was performed according to their NMR spectra available in literature2 and in our in-house library9,10,11,15,16 without additional confirmation. In some cases, the signal assignment was unobvious; in these cases, the identification was confirmed by spiking the extract with commercial standard compounds.
Representative 1H NMR spectra of protein-free lipid-free extract from S. lucioperca brain with the metabolite assignment: Asc Ascorbate, GABA gamma-Amino-butyrate, Glc Glucose, Gl-PhCholine Glycerophosphocholine, i-But Isobutyrate, Lac Lactate, myo-In myo-Inositol, NAA N-acetyl-Aspartate, NAH N-acetyl-Histidine, OSH Ovothiol A, Pant Pantothenate, PhCholine Phosphocholine, scyllo-In scyllo-Inositol, Ser-PETA Serine-phosphoethanolamine, Tau Taurine, Thr-PETA Threonine-phosphoethanolamine, Unk Unknown. For amino acids and nucleotides standard 3-letter symbols are used.
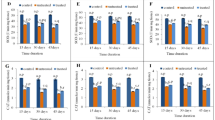
The metabolite quantification was performed by the NMR signal integration relatively to the internal standard DSS followed by the calculation of metabolite concentrations in tissue (in nmoles per gram of the tissue wet weight). Typically, 65–70 compounds were identified in every sample; however, the NMR signals from some compounds were either too weak or strongly overlapped by other signals, which made the quantification of these compounds unreliable. For that reason, the final set of metabolites studied in this work was restricted to 66 compounds. For every tissue, the measurements were performed for 4–5 samples obtained from different species, the results were averaged. The obtained data are collected in Table 1 (mean ± std). Figure 2 demonstrates the graphical representation of the abundances of several major metabolites in the fish tissues.
Figure 3 shows the PCA plots for the 1st versus 2nd principal components built for all eleven tissues studied in the present work. It demonstrates that the lens metabolomic composition most drastically differs from metabolomes of other tissues: the 1st principal component is almost completely determined by the metabolomic difference between the lens and other tissues (provided mostly by amino acids, N-acetyl-histidine (NAH), N-acetyl-aspartate (NAA), nicotinamide adenine dinucleotide (NAD), adenosine triphosphate (ATP), and OSH; Fig. 3B). All other tissues in Fig. 3A vary mostly along the 2nd principal component with AH and VH being at the very bottom of the plot and liver—at the top. The data for muscle, heart, kidney, spleen, brain, gill, and milt are concentrated in the same region. Apparently, that does not mean that the metabolomic compositions of these tissues are almost identical; that rather indicates that the difference between the lens and these tissues is less pronounced than the difference between the lens and AH, VH, and liver.
Scores (A, left) and loadings (B, right) plots of principal component analysis (PCA) of metabolomic profiles of S. lucioperca tissues. The data are auto scaled. Colored ovals indicate 95% confidence regions. Variance explained by the first (PC1) and second (PC2) principal components are indicated on the axis of scores plot.
To compare the metabolomic profiles of muscle, heart, kidney, spleen, brain, gill, and milt, the PCA was performed for these tissues only (Fig. 4). The scores plot (Fig. 4A) shows that among these tissues, muscle and brain have the most distinct metabolomic compositions, while the profiles of gill, kidney, milt, and spleen are relatively similar. The loadings plot (Fig. 4B) demonstrates that the difference between brain and other tissues is mostly provided by the high abundance of NAH, NAA, glutamine, gamma-amino-butyrate (GABA), and serine-phosphoethanolamine (Ser-PETA), while specific for muscle metabolites are creatine, histidine, glycine, and betaine.
Scores (A, left) and loadings (B, right) plots of principal component analysis (PCA) of metabolomic profiles of selected S. lucioperca tissues (AH, VH, liver, and lens are excluded). The data are auto scaled. Colored ovals indicate 95% confidence regions. Variance explained by the first (PC1) and second (PC2) principal components are indicated on the axis of scores plot.
The analysis of similarities of metabolomic compositions of different tissues can be performed with the use of hierarchical clustering (Fig. 5). Hierarchical clustering analysis (HCA) as opposed to PCA was performed for non-scaled data with the use of Euclidian distance and Ward’s linkage as the clustering parameters. HCA shows that all samples are grouped first of all by tissues indicating low dispersion of concentrations in each tissue and noticeable difference between tissues (with the exception of AH and VH). AH and VH have very similar metabolomic compositions: as it will be discussed below, the metabolomes of both fluids originate from the blood plasma metabolome. The metabolomic composition of milt is closest to that of AH and VH. In Fig. 5, one can observe the following groups of tissues with relatively similar metabolomes: kidney and gills; heart and spleen. That is in a good agreement with the PCA results (Figs. 3, 4). According to HCA, liver and muscle are characterized by the most unique and distinct metabolomic compositions.
Discussion
In the present work, we performed quantitative metabolomic analysis for eleven biological tissues of S. lucioperca. The advantage of the quantitative approach over commonly used semi-quantitative measurements is that the obtained data on the metabolite concentrations expressed in nmoles per gram of a tissue can be directly used by any researcher as a reference to the baseline level of metabolites in that tissue. Quantitative data also allow for the comparing the tissues with very dissimilar metabolomic compositions.
The metabolomic analysis performed in the present work demonstrates that although the majority of metabolites are common for all tissues, their concentrations in tissues may vary at a large scale. Moreover, there are some tissue-specific compounds with very high abundance in only 1–2 types of tissue. The examples of such metabolites are glycine, histidine, creatine, and betaine in muscle, ovothiol A in lens, NAA in lens and brain, glucose in liver. Apparently, these compounds are important for biological functions specific for these particular tissues.
Two groups of metabolites, osmolytes and antioxidants, play the key role in the cell protection against osmotic and oxidative stresses. In this work, the following compounds were conventionally assigned to osmolytes: taurine, myo-inositol, NAH, NAA, betaine, threonine-phosphoethanolamine (Thr-PETA), and Ser-PETA. Obviously, this assignment is rather arbitrary: some of these compounds, besides osmotic protection, perform other cellular functions, including cell signaling, providing substrate for biosynthesis, and so on17,18,19. At the same time, the tissues under study contain metabolites with concentrations of the same level or even higher than the concentrations of compounds assigned to osmolytes: lactate, glucose, acetate, creatine. These metabolites are mostly related to the reactions of cellular energy generation, and their concentrations should strongly depend on the fish activity. For that reason, in this work we did not include them into the list of osmolytes. Figure 6 shows the concentrations of osmolytes in different fish tissues (excluding acellular tissues AH and VH), and demonstrates that the composition of osmolytes in tissues strongly depends on the cell type.
We found in the fish tissues the following compounds with antioxidative properties: glutathione (GSH), ascorbate, OSH, and NADH. The detection of minor amounts of one more well-known thiol antioxidant, ergothioneine, in the gills of another freshwater fish, R. rutilus lacustris, has recently been reported15; however, in the present work ergothioneine was not found neither by NMR nor by LC–MS in any of the studied tissues of S. lucioperca. NADH was found only in NMR spectra of liver, muscle, heart, and lens, and its concentration in these tissues does not exceed 15 nmol/g. GSH, OSH, and ascorbate are present in much higher concentrations in the majority of the fish tissues, so these three compounds play the main role in the cellular defense against the oxidative stress. The presence and the relative abundance of GSH and OSH in the fish tissues were confirmed by LC–MS data.
The metabolomic features of particular fish tissues are discussed below.
AH and VH
AH and VH are acellular fluids with minimal metabolic activity. AH is produced in the ciliary epithelium through both the active secretion and the passive diffusion/ultrafiltration of blood plasma20,21,22,23. Consequently, the metabolomic composition of AH is similar to that of plasma10. VH is also connected with blood via the hematoophthalmic barrier24 and with AH, and one can see (Table 1, Fig. 2) that the metabolomic compositions of AH and VH are close to each other. Thus, it is safe to assume that the levels of metabolites in AH and VH reflect their levels in blood plasma, which circulates through the majority of fish tissues. Significant deviations of metabolite levels in tissue as compared to plasma should be attributed to the intracellular metabolic activity specific for this particular tissue.
Lens
The eye lens is one of the most anatomically isolated tissues. The lens mostly consists of metabolically inert fiber cells without nuclei and organelles with the exception of metabolically active epithelial monolayer. The data present in Table 1 indicate that the lens contains very high levels of proteinogenic amino acids: for some amino acids (for example, branched-chain amino acids, glutamine, aspartate) their levels in the lens are more than ten-fold higher than that in AH. Moreover, the concentrations of the majority of amino acids in the lens are higher than in any other fish tissue. The elevated levels of amino acids in the lens has been noticed many years ago25, and it was attributed to the active amino acid transport from AH to lens26,27,28,29. These amino acids are presumably needed to synthesize high protein content (up to 40% of the total lens weight), which is in turn needed to provide high refraction coefficient.
The fish lens contains a unique set of osmolytes and antioxidants. The lens osmolytes are myo-inositol, NAH, NAA, Thr-PETA, and Ser-PETA. We have previously shown15 that the concentrations of osmolytes in the fish lens undergo significant seasonal variations. At the late winter time, when the fish was caught for this study, the most abundant lens osmolyte is myo-inositol. High concentrations of this compound are also found in other fish tissues, including brain, gill, and spleen. Thr-PETA and Ser-PETA are also among the most abundant metabolites in the majority of the fish tissues. In opposite, NAH and NAA are present in high concentrations only in the fish lens and brain. At the same time, the concentration of taurine, which is the most abundant osmolyte in all other fish tissues, in the lens is rather low.
The major antioxidant of the fish lens is OSH12. It has been shown that the level of OSH in S. lucioperca lens vary from 3 µmol/g at autumn to 1.5 µmol/g at winter15, which is in a good agreement with our present data (Table 1). The concentration of the second most abundant lens antioxidant, GSH, is 3–4 times lower than that of OSH. Taking into account the properties of OSH30,31,32,33, it has been proposed12,34 that OSH is a primary protector against the oxidative stress, while the main function of GSH in the lens is the maintenance of OSH in the reduced state. It should be noticed that although OSH was also found in other fish tissues (Table 1, Fig. 2), its concentration in these tissues is significantly lower than in the lens. Therefore, in respect to fish, OSH can truly be called “lenticular antioxidant”.
High concentrations of amino acids, osmolytes, antioxidants and some other compounds in the lens indicate that these metabolites are either synthesized in metabolically active epithelial cells, or pumped into the lens from AH against the concentration gradient with the use of specific transporters also located in the epithelial layer. The fiber cells of the lens are metabolically passive, and fresh metabolites can appear in these cells only due to the diffusion from the epithelial layer toward the lens center. Therefore, one can expect that the concentrations of the most important metabolites decrease from the lens cortex toward the lens nucleus. To check this assumption, we measured the metabolomic profiles for cortex and nucleus separately. The measurements were performed for three lenses; then the ratios of the metabolite concentrations in the cortex to that in the nucleus were calculated and averaged. The results of the calculations are shown in Fig. 7 (only for metabolites with the highest and the lowest cortex/nucleus ratios) and Supplementary Table S2 (for all metabolites). Indeed, the levels of the majority of metabolites in the lens nucleus are significantly lower than in the cortex. For five metabolites, namely ATP, NAA, inosinate, ADP, and GSH the difference exceeds the factor of thirty; that means that these compounds are almost completely depleted during their diffusion toward the lens nucleus.
Brain
The brain tissue similarly to the lens is isolated from the vascular system by means of the hematoencephalic barrier. However, in opposite to the lens, brain is very metabolically active tissue, as in particular indicated by the high level of lactate (14 µmol/g). Similar lactate concentrations were found only in muscle and heart (Table 1). Besides lactate, the most abundant metabolites of the fish brain are osmolytes myo-inositol, Ser-PETA, taurine, Thr-PETA, NAH, and NAA; the concentrations of these compounds in brain are in the range from 2.5 to 13 µmol/g (Table 1). The brain tissue also contains high levels of glutamate and creatine, which are used by brain cells for the cellular energy generation. The level of antioxidant ascorbate in brain (400 nmol/g) is significantly higher than in other fish tissues, which indicates the importance of ascorbate for the brain correct operation. Besides ascorbate, the brain tissue also contains OSH and GSH, but at significantly lower concentrations (100–200 nmol/g).
Blood-rich organs: liver, spleen, milt, muscle, heart, gill, kidney
Figure 4 demonstrates that from the metabolomic viewpoint, gill, kidney, milt, and spleen are the most similar tissues. However, the quantitative analysis indicates significant differences. In particular, spleen does not contain measurable by NMR amounts of antioxidants OSH and GSH. The levels of osmolytes are also different: the concentration of taurine in spleen is threefold higher than in gill and milt, while the level of myo-inositol in milt is much lower than in spleen and gill (Fig. 6). Significant differences are also found for some amino acids (alanine, creatine), organic acids (lactate, GABA), and nucleosides (ATP, ADP, AMP, inosine).
One of the important liver functions is the maintaining the glucose level in blood regulated by producing glucose from stored glycogen. Correspondingly, the level of glucose in liver is extremely high (40 µmol/g), which is higher than in any other tissue by at least an order of magnitude. Liver also contains elevated (as compared to other tissues) concentrations of threonine, glutamate, succinate, fumarate, AMP, and nicotinamide.
The biological functions of muscle and heart are relatively similar; however, the metabolomic compositions of these tissues differ significantly. The main osmolyte in muscle cells is taurine (30 µmol/g), while in heart the osmotic protection is shared between taurine (15 µmol/g) and Ser-PETA (12 µmol/g). Muscle contains very high levels of glycine and histidine. Glycine is known to protect muscles from wasting under various wasting conditions35,36, while histidine and histidine-related compounds were reported to play the role of intracellular proton buffering constituents in vertebrate muscle37. Very likely that both glycine and histidine also participate in the osmotic protection of the muscle cells. The level of creatine—the energy source—in muscle (23 µmol/g) is five-fold higher than in heart.
Materials and methods
Chemicals
Chloroform and methanol HPLC grade were purchased from Panreac (Spain). D2O 99.9% was purchased from Armar Chemicals (Switzerland). All other chemicals were purchased from Sigma-Aldrich (USA). H2O was deionized using an Ultra Clear UV plus TM water system (SG water, Germany) to the quality of 18.2 MOhm.
Fish sample collection
The study was conducted in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and the European Union Directive 2010/63/EU on the protection of animals used for scientific purposes, and with the ethical approval from International Tomography Center SB RAS. No special permission from the national or local authorities is required. S. lucioperca (males, body weight 800–1500 g) were caught in the ice-covered Ob reservoir with the use of a winter fishing rod at the beginning of April 2019 (n = 5). The fish were killed with a concussive blow to the head immediately after the catching, the tissues were cut from the fish, frozen and kept at − 70 °C until analyzed.
Fish tissue preparation
The sample preparation was performed as described in Ref.15. Each fish tissue was weighted prior to homogenization. The typical sample weight was: for lens—230 mg; for heart—240 mg; for spleen—230 mg; for liver—200 mg; for muscle—200 mg; for brain—140 mg; for kidney—130 mg; for milt—170 mg; for gill—210 mg. The typical sample volumes were 300 μl for AH and 250 μl for VH. Only one lens from each fish was used for the analysis. The fish gill was divided into arch and filaments, only gill filaments were used for the analysis. Muscle tissue was excised from the area under the anal fin. Heart tissues were not subjected to the blood washing procedure.
Each fish tissue (except AH) was placed in a glass vial and homogenized with a TissueRuptor II homogenizer (Qiagen, Netherlands) in 1600 µL of cold (− 20 °C) MeOH, and then 800 µL of water and 1600 µL of cold chloroform were added15. The mixture was shaken well in a shaker for 20 min and left at − 20 °C for 30 min. Then the mixture was centrifuged at 16,100g, + 4 °C for 30 min, yielding two immiscible liquid layers separated by a protein layer. The upper aqueous layer (MeOH–H2O) was collected, divided into two parts for NMR (2/3) and LC–MS (1/3) analyses, and lyophilized. Since AH contains very low levels of proteins and lipids, it was extracted with MeOH/H2O solvent (1600/800 µL) without chloroform. The obtained protein-free and lipid-free extracts were used for NMR and LC–MS measurements.
To compare the metabolomic composition of the lens cortex and nucleus, the nucleus was separated from the cortex by coring the lens with a 3-mm home-made stainless steel borer followed by cutting off of approximately 1 mm from each end of the core9. The procedure was performed with lenses taken from storage at − 70 °C and warmed up to − 18 °C for easier cutting, all tools were cooled down to − 18 °C, and all manipulations were performed in a cold room at − 5 °C. The nucleus and the cortex (the combined doughnut-shaped outer remainder of boring and the ends of the core) were weighed and then extracted in the same way as other tissues.
NMR measurements
The extracts for NMR measurements were re-dissolved in 600 μL of D2O containing 6 × 10–6 M sodium 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) as an internal standard and 20 mM deuterated phosphate buffer to maintain pH 7.2. The 1H NMR measurements were carried out at the Center of Collective Use “Mass spectrometric investigations” SB RAS on a NMR spectrometer AVANCE III HD 700 MHz (Bruker BioSpin, Germany) equipped with a 16.44 T Ascend cryomagnet as described in Ref.15. The concentrations of metabolites in the samples were determined by the peak area integration respectively to the internal standard DSS.
LC–MS measurements
In this work, LC–MS data were used only for the confirmation of data obtained by NMR method. The extracts for LC–MS analysis were re-dissolved in 100 μL of water. The LC separation was performed on an UltiMate 3000RS chromatograph (Dionex, Germering, Germany) using a hydrophilic interaction liquid chromatography (HILIC) method on a TSKgel Amide-80 h (Tosoh Bioscience, Griesheim, Germany) column (4.6 × 250 mm, 5 μm) as described earlier16. The chromatograph was equipped with a flow cell diode array UV–Vis detector (DAD) with 190–800 nm spectral range. Solvent A consisted of 0.1% formic acid solution in H2O, solvent B consisted of 0.1% formic acid solution in acetonitrile. The gradient was (solvent B): 95% (0–5 min), 95–65% (5–32 min), 65–35% (32–40 min), 35% (40–48 min), 35–95% (48–50 min), 95% (50–60 min); the flow rate was 1 mL/min, the sample injection volume was 10 μL. After the DAD cell, a home-made flow splitter (1:10) directed the lesser flow to an ESI-q-TOF high-resolution hybrid mass spectrometer maXis 4G (Bruker Daltonics, Bremen, Germany). The mass spectra were recorded in a positive mode with 50–1000 m/z range.
Data analysis
To explore the data and to display the general metabolomic features in the data, the principal component analysis (PCA) has been performed on a MetaboAnalyst 4.0 web-platform (www.metaboanalyst.ca38). PCA scores and loadings plots were constructed with the auto data scaling (mean-centered and divided by standard deviation of each metabolite concentration) to normalize the contributions of all metabolites.
Conclusions
The results of the present work demonstrate that NMR spectroscopy is an extremely useful tool for quantitative metabolomics; it allows for the determining the actual concentrations of up to hundred metabolites in biological fluids and tissues. The quantitative data indicate the most abundant compounds in every particular tissue, and, correspondingly, point to the most important and active metabolic processes in the tissue. It is important to notice that the metabolomic composition of tissues of fish and other marine and freshwater animals might be affected by a number of factors, including water temperature39,40, oxygen level15,41, and water contamination42,43,44,45. Therefore, the obtained quantitative data on the metabolite concentrations in the fish tissues correspond to rather specific conditions: species—Sander lucioperca, season—late winter, water temperature 4–7 °C, low level of dissolved oxygen in the ice-covered lake, moderate water pollution with the city waste. These data can be used as the baseline levels of metabolites for the analysis of peculiarities of metabolic processes in different species, and for the evaluation of influence of seasonal, ecological and other factors on the fish metabolism.
Data availability
The data obtained in this study including NMR raw data, metabolite concentrations and experimental protocols have been deposited in MetaboLights repository, study identifier MTBLS1057 (https://www.ebi.ac.uk/metabolights/MTBLS1057).
References
Wishart, D. S. et al. HMDB: The human metabolome database. Nucleic Acids Res. 35, D521-526. https://doi.org/10.1093/nar/gkl923 (2007).
Wishart, D. S. et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 46, D608–D617. https://doi.org/10.1093/nar/gkx1089 (2018).
Spratlin, J. L., Serkova, N. J. & Eckhardt, S. G. Clinical applications of metabolomics in oncology: A review. Clin. Cancer Res. 15, 431–440. https://doi.org/10.1158/1078-0432.CCR-08-1059 (2009).
Mishur, R. J. & Rea, S. L. Applications of mass spectrometry to metabolomics and metabonomics: Detection of biomarkers of aging and of age-related diseases. Mass Spectrom. Rev. 31, 70–95. https://doi.org/10.1002/mas.20338 (2012).
Wishart, D. S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 15, 473–484. https://doi.org/10.1038/nrd.2016.32 (2016).
Psychogios, N. et al. The human serum metabolome. PLoS ONE 6, e16957. https://doi.org/10.1371/journal.pone.0016957 (2011).
Bouatra, S. et al. The human urine metabolome. PLoS ONE 8, e73076. https://doi.org/10.1371/journal.pone.0073076 (2013).
Gowda, G. A. & Raftery, D. Quantitating metabolites in protein precipitated serum using NMR spectroscopy. Anal. Chem. 86, 5433–5440 (2014).
Tsentalovich, Y. P. et al. Metabolomic composition of normal aged and cataractous human lenses. Exp. Eye Res. 134, 15–23. https://doi.org/10.1016/j.exer.2015.03.008 (2015).
Snytnikova, O. A. et al. Metabolomics of the human aqueous humor. Metabolomics 13, 5. https://doi.org/10.1007/s11306-016-1144-0 (2016).
Snytnikova, O. A. et al. Quantitative metabolomic analysis of the human cornea and aqueous humor. Metabolomics 13, 152. https://doi.org/10.1007/s11306-017-1281-0 (2017).
Yanshole, V. V., Yanshole, L. V., Zelentsova, E. A. & Tsentalovich, Y. P. Ovothiol A is the main antioxidant in fish lens. Metabolites 9, 95. https://doi.org/10.3390/metabo9050095 (2019).
Yanshole, V. V., Yanshole, L. V., Snytnikova, O. A. & Tsentalovich, Y. P. Quantitative metabolomic analysis of changes in the lens and aqueous humor under development of age-related nuclear cataract. Metabolomics 15, 29. https://doi.org/10.1007/s11306-019-1495-4 (2019).
Cappello, T. et al. Baseline levels of metabolites in different tissues of mussel Mytilus galloprovincialis (Bivalvia: Mytilidae). Comp. Biochem. Physiol. Part D 26, 32–39. https://doi.org/10.1016/j.cbd.2018.03.005 (2018).
Tsentalovich, Y. P. et al. Seasonal variations and interspecific differences in metabolomes of freshwater fish tissues: Quantitative metabolomic profiles of lenses and gills. Metabolites 9, 264. https://doi.org/10.3390/metabo9110264 (2019).
Yanshole, V. V. et al. Metabolomics of the rat lens: A combined LC-MS and NMR study. Exp. Eye Res. 125, 71–78. https://doi.org/10.1016/j.exer.2014.05.016 (2014).
Son, H.-Y., Kim, H. & Kwon, Y. H. Taurine prevents oxidative damage of high glucose-induced cataractogenesis in isolated rat lenses. J. Nut. Sci. Vitaminol. 53, 324–330. https://doi.org/10.3177/jnsv.53.324 (2007).
Larner, J. D-chiro-inositol—Its functional role in insulin action and its deficit in insulin resistance. Int. J. Exp. Diabetes Res. 3, 47–60. https://doi.org/10.1080/15604280212528 (2002).
Ripps, H. & Shen, W. Review: Taurine: A “very essential” amino acid. Mol. Vis. 18, 2673–2686 (2012).
Macknight, A. D. et al. Formation of the aqueous humor. Clin. Exp. Pharmacol. Physiol. 27, 100–106. https://doi.org/10.1046/j.1440-1681.2000.03208.x (2000).
To, C., Kong, C., Chan, C., Shahidullah, M. & Do, C. The mechanism of aqueous humour formation. Clin. Exp. Optom. 85, 335–349. https://doi.org/10.1111/j.1444-0938.2002.tb02384.x (2002).
Gabelt, B., Kiland, A., Tian, B., Kaufman, P. In Duane’s Ophthalmology (eds Tasman, W. & Jaeger, E.A.), Vol. 6 (Lippincott Williams & Wilkins Publishers, Philadelphia, 2006).
Goel, M., Picciani, R. G., Lee, R. K. & Bhattacharya, S. K. Aqueous humor dynamics: A review. Open Ophthalmol. J. 4, 52–59. https://doi.org/10.2174/1874364101004010052 (2010).
Peiffer, R.L. & Petersen-Jones, S.M., In Small Animal Ophthalmology: A Problem-Oriented Approach (eds Peiffer, R.L. & Petersen-Jones, S.M.), 4th edn (Saunders Ltd., Edinburgh, 2008) ISBN 978-0-7020-2861-8.
Van Heyningen, R. Fluorescent glucoside in the human lens. Nature 230, 393–394. https://doi.org/10.1038/230393a0 (1971).
Kinsey, V. E. Amino acid transport in the lens. Investig. Ophthalmol. Vis. Sci. 4, 691–699 (1965).
Kern, H. L. & Ho, C. K. Transport of L-glutamic acid and L-glutamine and their incorporation into lenticular glutathione. Exp. Eye Res. 17, 455–462. https://doi.org/10.1016/0014-4835(73)90226-1 (1973).
Reddy, V. N. Transport of organic molecules in the lens. Exp. Eye Res. 15, 731–750. https://doi.org/10.1016/0014-4835(73)90007-9 (1973).
Lim, J., Lorentzen, K. A., Kistler, J. & Donaldson, P. J. Molecular identification and characterisation of the glycine transporter (GLYT1) and the glutamine/glutamate transporter (ASCT2) in the rat lens. Exp. Eye Res. 83, 447–455. https://doi.org/10.1016/j.exer.2006.01.028 (2006).
Holler, T. P. & Hopkins, P. B. Ovothiols as biological antioxidants. The thiol groups of ovothiol and glutathione are chemically distinct. J. Am. Chem. Soc. 110, 4837–4838. https://doi.org/10.1021/ja00222a057 (1988).
Weaver, K. H. & Rabenstein, D. L. Thiol/disulfide exchange reactions of ovothiol A with glutathione. J. Org. Chem. 60, 1904–1907. https://doi.org/10.1021/jo00111a065 (1995).
Zoete, V., Bailly, F., Catteau, J.-P. & Bernier, J.-L. Design, synthesis and antioxidant properties of ovothiol-derived 4-mercaptoimidazoles. J. Chem. Soc. Perkin Trans. 1, 2983–2988. https://doi.org/10.1039/A703741D (1997).
Mirzahosseini, A. & Noszál, B. Species-specific thiol-disulfide equilibrium constants of ovothiol A and penicillamine with glutathione. RSC Adv. 6, 26757–26764. https://doi.org/10.1039/C6RA01778A (2016).
Castellano, I. et al. Anti-inflammatory activity of marine ovothiol A in an in vitro model of endothelial dysfunction induced by hyperglycemia. Oxid. Med. Cell Longev. 2018, 2087373. https://doi.org/10.1155/2018/2087373 (2018).
Koopman, R., Caldow, M. K., Ham, D. J. & Lynch, G. S. Glycine metabolism in skeletal muscle: Implications for metabolic homeostasis. Curr. Opin. Clin. Nutr. Metab. Care https://doi.org/10.1097/MCO.0000000000000383 (2017).
Caldow, M. K. et al. Glycine protects muscle cells from wasting in vitro via mTORC1 signaling. Front. Nutr. 6, 172. https://doi.org/10.3389/fnut.2019.00172 (2019).
Abe, H. Role of histidine-related compounds as intracellular proton buffering constituents in vertebrate muscle. Biochemistry 65, 757–765 (2000).
Chong, J. et al. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46, W486–W494 (2018).
Remø, S. C., Hevrøy, E. M., Breck, O., Olsvik, P. A. & Waagbø, R. Lens metabolomic profiling as a tool to understand cataractogenesis in Atlantic salmon and rainbow trout reared at optimum and high temperature. PLoS ONE 12, e0175491. https://doi.org/10.1371/journal.pone.0175491 (2017).
Sambraus, F. et al. Water temperature and dietary histidine affect cataract formation in Atlantic salmon (Salmo salar L.) diploid and triploid yearling smolt. J. Fish Dis. 40, 1195–1212. https://doi.org/10.1111/jfd.12594 (2017).
Allen, P. J. et al. Using 1-D 1H and 2-D 1H J-resolved NMR metabolomics to understand the effects of anemia in channel catfish (Ictalurus punctatus). Metabolomics 11, 1131–1143. https://doi.org/10.1007/s11306-014-0767-2 (2015).
Viant, M. R., Pincetich, C. A. & Tjeerdema, R. S. Metabolic effects of dinoseb, diazinon and esfenvalerate in eyed eggs and alevins of Chinook salmon (Oncorhynchus tshawytscha) determined by 1H NMR metabolomics. Aquat. Toxicol. 77, 359–371. https://doi.org/10.1016/j.aquatox.2006.01.009 (2006).
Cappello, T. et al. Advances in understanding the mechanisms of mercury toxicity in wild golden grey mullet (Liza aurata) by 1H NMR-based metabolomics. Environ. Pollut. 219, 139–148. https://doi.org/10.1016/j.envpol.2016.10.033 (2016).
Cappello, T., Maisano, M., Mauceri, A. & Fasulo, S. 1H NMR-based metabolomics investigation on the effects of petrochemical contamination in posterior adductor muscles of caged mussel Mytilus galloprovincialis. Ecotoxicol. Environ. Saf. 142, 417–422. https://doi.org/10.1016/j.ecoenv.2017.04.040 (2017).
Li, M.-H. et al. Metabolic profiling of goldfish (Carassius auratis) after long-term glyphosate-based herbicide exposure. Aquat. Toxicol. 188, 159–169. https://doi.org/10.1016/j.aquatox.2017.05.004 (2017).
Acknowledgements
Authors thank Ministry of Science and Higher Education of the RF for the access to NMR and MS equipment.
Funding
This research was funded by Russian Foundation for Basic Research, Grant numbers 18-29-13023 in sample preparation and 19-04-00092 in NMR measurements. The analysis of the antioxidant content in fish tissues was supported by the President of RF (project MК-2272.2020.3).
Author information
Authors and Affiliations
Contributions
Conceptualization, Y.P.T.; methodology, Y.P.T. and E.A.Z.; validation, E.A.Z., L.V.Y. and V.V.Y.; formal analysis, I.M.O., E.A.Z. and Y.P.T.; investigation, E.A.Z., L.V.Y., V.V.Y. and I.M.O.; data curation, Y.P.T.; writing—original draft, Y.P.T.; Writing—review & editing, V.V.Y.; supervision, Y.P.T.; project administration, Y.P.T.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tsentalovich, Y.P., Zelentsova, E.A., Yanshole, L.V. et al. Most abundant metabolites in tissues of freshwater fish pike-perch (Sander lucioperca). Sci Rep 10, 17128 (2020). https://doi.org/10.1038/s41598-020-73895-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-020-73895-3
This article is cited by
-
Metabolomics-based strategies for fish meat quality, authentication and food safety: a review
Food, Nutrition and Health (2026)
-
Is substrate choice an overlooked variable in ecotoxicology experiments?
Environmental Monitoring and Assessment (2023)