Abstract
The CLAVATA3 (CLV3)/EMBRYO SURROUNDING REGION (ESR)–RELATED (CLE) gene family encodes a large number of polypeptide signaling molecules involved in the regulation of shoot apical meristem division and root and vascular bundle development in a variety of plants. CLE family genes encode important short peptide hormones; however, the functions of these signaling polypeptides in cotton remain largely unknown. In the current work, we studied the effects of the CLE family genes on growth and development in cotton. Based on the presence of a conserved CLE motif of 13 amino acids, 93 genes were characterized as GhCLE gene family members, and these were subcategorized into 7 groups. A preliminary analysis of the cotton CLE gene family indicated that the activity of its members tends to be conserved in terms of both the 13-residue conserved domain at the C-terminus and their subcellular localization pattern. Among the 14 tested genes, the ectopic overexpression of GhCLE5::GFP partially mimicked the phenotype of the clv3 mutant in Arabidopsis. GhCLE5 could affect the endogenous CLV3 in binding to the receptor complex, comprised of CLV1, CLV2, and CRN, in the yeast two-hybrid assay and split-luciferase assay. Silencing GhCLE5 in cotton caused a short seedling phenotype. Therefore, we concluded that the cotton GhCLE gene family is functionally conserved in apical shoot development regulation. These results indicate that CLE also plays roles in cotton development as a short peptide hormone.
Similar content being viewed by others
Introduction
Intercellular communication is essential for the development of tissues and organs. In plants, peptide-receptor signaling modules play an important role in mediating intercellular communication and interactions during development, as well as responding to environmental stimuli1. One of the most well explored gene families encoding small peptide ligands is the CLAVATA3 (CLV3)/EMBRYO SURROUNDING REGION (ESR)–RELATED (CLE) gene family1,2,3,4,5. CLE gene family peptides are involved in short- and long-distance signal transduction6,7.
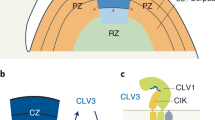
The WUSCHEL-CLAVATA (WUS-CLV) feedback loop of Arabidopsis thaliana maintains stem cell homeostasis in shoot apical meristems (SAMs)8,9,10. Plant SAMs are the sources of all the above-ground parts of the plant11. They achieve this by slowly dividing as stem cells and constantly transferring daughter cells to the surrounding marginal area, where they are incorporated into the primordia of the leaves or flowers12. In order to maintain the function of SAMs, a balance needs to be struck between the generation of new meristematic cells through division and the separation of cells from the meristem through differentiation5,9. The WUS encodes a homeodomain transcription factor that actively regulates the activity of meristems and is essential for maintaining stem cell populations during the later stages of embryo development13,14. wus mutants show SAM growth arrest, and then axillary buds resume growth14, showing undulations and phenotypes similar to those in conditions of mild-to-moderate overexpression of CLV315. CLV3 is a member of the CLE gene family, which contains a conserved C-terminal CLE motif of 12 to 13 amino acids16. The interaction between WUS and CLV3 produces a negative feedback regulation that balances stem cell maintenance and cell differentiation in shoot apical meristems5,15,17,18. CLV1 belongs to the leucine-rich repeat–receptor-like kinase (LRR-RLK) subfamily, which contains 21 LRR- and 223 RLK-domain members19,20. One of the differences between CLV1 and CLV2 is that there is no kinase domain in CLV219,21,22. CLV1 homodimers play a role in parallel to the CLV2-CORYNE heterodimer, in which CLV2, with its extracellular domain, interacts with CORYNE, which has an intracellular kinase domain to transduce the CLV3 signal23,24.
In addition to maintain the homeostats of SAM, other CLE gene family members also play important roles in multiple plant organ development control25. CLE14 is one of the main regulator of root apical meristem differentiation26. CLE9/10 and CLE25 plays roles both in xylem formation and stomatal development27,28. CLE19 is reported to regulate the cotyledon and endosperm development in Arabidopsis29. Two Lotus japonicus CLE peptides are reported to be involved with nodule nodule organ formation30. A homolog of AtCLV3 in Brassica napa controls the multilocular silique development31.
Cotton (Gossypium) is one of the most important cash crops and provides fiber for the textile industry. Cotton is also a model polyploid plant used to study whole-genome duplication events32. GhWUS is an important regulator of somatic embryogenesis and bud regeneration33. Previously, 55 CLE genes were extracted from the close D5 donor genome of allotetraploid cotton species, wild diploid cotton Gossypium raimondii2. The role of CLE genes in the upland cotton cultivar is still unknown, however. In the present study, we carried out a functional analysis of CLE genes in the upland cotton (Gossypium hirsutum) genome. By means of a genome-wide study, translational analysis, protein interaction assay, and virus-induced gene silencing (VIGS) technology, we identified members of the cotton CLE gene and screened out the cotton GhCLE5 with ectopic effects in the reassembly of the Atclv3 mutant.
Results
The CLE gene family in upland cotton
Based on the sequences of CLE genes in the Arabidopsis genome, homologous alignment of the psi-BLAST and G. hirsutum Texas Marker-1 (TM-1) genome sequences was performed. In total, 93 genes containing the CLE motif were obtained and named sequentially according to their distribution on the Upland cotton TM-1 chromosomes (Fig. 1A).
The GhCLE gene family in the upland cotton genome. (A) The chromosome distribution of GhCLE genes. A01-13 and D01-13 indicate the chromosome number of upland cotton. The GhCLE genes are labeled on the relative locations on the corresponding chromosome. (B) The sequence logos of CLE motif peptides for seven GhCLE gene family groups. (C) Heat maps representing the Group 1 and Group 2 GhCLE gene expression patterns in the cotton seed, cotyledon, root, leaf, torus, petal, stamen, pistil, calycle, and ovule tissues 0 day post anthesis (0 DPA) and in 10 DPA fibers.
Forty-two genes were in the AT subgroup of G. hirsutum and 42 were in the DT subgroup (Fig. 1A, Supplemental Table 1). The distribution map showed that chromosomes A02 and ChrD03 had no CLE genes. The CLE genes were evenly distributed on the other chromosomes. Most of the CLE genes were composed of a single exon (Supplemental Figure 1). Through collinear analysis, we identified a total of 35 pairs of homeologous genes in the A and D subgenomes (Supplemental Table 2). The average Ka/Ks value was 0.54, and the Ka/Ks values of three pairs of direct homologous genes exceeded 1 (Supplemental Table 2), indicating that the CLE gene family had a relatively fast evolutionary speed.
The CLE family members have a highly conserved amino acid sequence containing 12 to 13 amino acids, named the CLE motif. Previous studies in the model plant Arabidopsis have reported that CLE family members can be classified into four or five groups according to the diversity of their CLE motif sequences, which represent unique biologic functions34. We adapted the analysis of the CLE motifs and classified the cotton CLEs into seven categories, named Groups 1 to 7 (Fig. 1B, Supplemental Figures 2 and 3). The CLE motif of Group 1 showed the highest sequence similarity to that of the CLV3 motif. That of Group 6 showed the highest similarity to the B-type TDIF CLE35.
GFP fusion signal illustrating the subcellular localization of the selected GhCLE genes from seven groups using transient expression in tobacco leaves. (A) Schematic chart showing the construct structure of the 35S::GhCLE::GFP vector. (B) GhCLEs::GFP confocal micrographs at 488 nm excitation. Each examination included five biologic replications.
The phenotype of Arabidopsis thaliana with ectopic expression of GhCLE5. (A) The 6-week-old plant. (B) The stems of the GhCLE5 transgenic Arabidopsis plants from multiple T1 lines. (C) The photo images of 4-week-old plants. Bar = 2 cm. (D) The vertical view of florescence. Bar = 5 mm. (E) The bird’s-eye view of the florescence. Bar = 1 mm. F: The scanning electron microscope (SEM) images of the shoot apical meristem (SAM). Bar = 100 μm.
The seven GhCLE groups showed distinct expression patterns in the cotton root, cotyledon, leaf, stem, torus, petal, stamen, pistil, calyx, ovule, and fiber tissues. Groups 1, 2, 6, and 7 showed ubiquitous expression patterns in most of the cotton tissues (Fig. 1C, Supplemental Figure 4). Genes in Group 3 were predominantly expressed in the hypocotyl and cotyledons of seedlings and stems (Supplemental Figure 4), whereas genes in Group 5 were specifically expressed in flower tissues, including torus, petal, and stamen tissues (Supplemental Figure 4). Some members of Groups 2 and 5 had consistently low expression patterns in most of the cotton tissues; this could indicate that these are stress-responsive genes.
To validate the CLE genes characterized in allotetraploid upland cotton of G. hirsutum, we further performed a comparison between the CLEs from the D subgenome in allotetraploid (Gh) and diploid D genome (Gr) previously reported2 (Supplement Table 3). Of the 55 Gr-genome CLE genes reported by Goad et al., 42 orthologs are found among our Gh CLE genes. In addition, 10 CLE genes from Goad are not present in the D subgenome of Gh, and another 3 CLE genes from Gr only have orthologs in the A subgenome of Gh. Finally, 2 CLE genes from the D subgenome in our report have not been identified previously in Gr. These results validate the CLE genes we characterized in Gh and also imply the dynamics of CLE gene evolution after polyploidization in allotetraploid species.
Unprocessed GhCLE proteins were predominantly localized to the cell perimeter
To investigate the translational capacity of the cotton CLE genes, we performed subcellular localization tests. Two candidate genes from each cotton CLE group on average (Supplemental Table 4) with relatively high expression activity were selected for this study. GFP was fused to the 3′ end of the candidate genes with a 35S promoter (Fig. 2A). The GFP fusion constructs were transiently expressed in tobacco leaves, and fluorescence was observed. Six proteins, GhCLE12, GhCLE20, GhCLE23, GhCLE58, and GhCLE65, GhCLE77 were located in the cell membrane and nucleus (Fig. 2B). Eight proteins, GhCLE5, GhCLE32, GhCLE39, GhCLE59, GhCLE73, GhCLE79, GhCLE83, and GhCLE93, were located in the cell membrane (Fig. 2B). Previous studies have shown that the CLE proteins can be processed into short peptides that can be secreted across the cell membrane. The GhCLEs::GFP fusion signal represented the proteins after translation but before they were processed. In Fig. 2, cotton CLE proteins with a GFP signal can be seen to be predominantly distributed in the cell membrane regions in comparison with the 35S::GFP signal. Although the GFP fused construct might affect the CLE protein process to some extent, the CLE::GFP localization exhibited a unique pattern distinct from the GFP itself, which could be a ready-to-go position for the processed peptide to undergo transmembrane movement. These subcellular localization signals indicate that the cotton CLEs are highly likely to be secretory proteins similar to those found in Arabidopsis.
Ectopic overexpression of GhCLE5 mimicked the Atclv3 phenotype
The 14 selected GhCLE candidate genes were transformed into the wild type of A. thaliana Col-0 for further functional study. The ectopic expression of GhCLE5 caused the highest variation in phenotype in the Arabidopsis transgenic lines. Among the 34 T1 positive lines of 35S::GhCLE5 transgenic Arabidopsis (Supplemental Figures 5 and 6), 12 showed phenotypes in the shoot that clearly differed from the wild type (Fig. 3, Supplemental Table 5). The phenotypic traits included dwarf plants, clv3-like florescence, fasciated stems, and club-shaped siliques (Fig. 3B to F, Supplemental Table 5). A representative line, GhCLE5-23, is shown in Fig. 3A,C, with curved rosette leaves and bushy stems in the bolting plants. The siliques were shorter than the wild type (Col-0) but not as short as clv3 (Fig. 4A–C). GhCLE5-23 exhibited florescence similar to that of clv3 (Fig. 3D,E). The flower primordium and floral bud number were obviously more in Atclv3 and GhCLE5 transgenic lines than that of Col-0 (Fig. 3F). Other lines, such as GhCLE5-10, GhCLE5-20, GhCLE5-24, and GhCLE5-27, showed fasciated stems (Fig. 3B, Supplemental Table 5). GhCLE5-19 showed an extreme phenotype of rosette leaf regeneration on the stem (Fig. 3B) with infertile flowers. 35S::GhCLE5 did not have any consistent impact on the root growth of the transgenic lines (Supplemental Figures 7 and 8).
The interaction between AtCLV1, AtCLV3, and GhCLE5. (A) The amino acid sequence alignment of GhCLE5, AtCLE12, AtCLE13, and AtCLV3. The conserved CLV3 motif is indicated. The black box is the putative signal peptide (http://smart.embl-heidelberg.de/). (B) The schematic chart showing the protein structure of AtCLV1 and the design for the yeast two-hybrid assay. (C) and (D) Photo images for the yeast two-hybrid assay that tested the interaction between AtCLV3, GhCLE5, and AtCLV1.
Validation of GhCLE5 binding activity with the CLV1, CLV2, CRN, and CLV2/CRN heterodimers. (A) The split-luciferase assay to detect the GhCLE5 binding activity with AtCLV1, AtCLV2, and AtCRN, respectively. The SGT1a-Nluc and RAR1-Cluc are used as positive controls for the binding protein. (B) The split-luciferase assay to detect the GhCLE5 binding activity in parallel with the AtCLV2 and AtCRN together. Each examination included three biologic replications.
The phenotype of cotton seedlings following VIGS to suppress the expression of GhCLE5. (A) VIGS-treated cotton seedlings. The population sizes of the pTRV2:GhCLE5 and pTRV2:00 treatments were both 31. The pTRV2:CLA was the positive control group for VIGS showing bleached leaves. (B) Histogram indicating the differences in plant height between the pTRV2:GhCLE5 and pTRV2:00 groups derived from the VIGS-treated plants shown in panel A (Student’s t test). Error bar = Std. (C) Histogram showing the relative expression of GhCLE5 in the pTRV2:GhCLE5 and pTRV2:00 groups derived from the VIGS-treated leaf tissues shown in panel A (Student’s t test). Error bar = Std. (D) Histogram showing the GhCLE5 expression pattern in the cotyledon (Cot), hypocotyl (Hyp), root, stem, leaf, ovule, and fiber tissues of upland cotton using RNA-seq RPKM values. DPA, days post anthesis. (E) The imagines show the GUS staining of the tobacco leaves transient expressed with the GUS reporter driven by GhCLE promoter (pGhCLE5:GUS). The negative control is GV3101 bacterial injection buffer with and without the pBI121 vector containing a GUS driven by 35S promoter (p35S:GUS). Three biological replicates were applied for each test group. Scale bar = 1 cm.
The phenotypes observed in the 35S::GhCLE5 transgenic lines partially resembled the Atclv3 phenotype in terms of the morphology of the florescence and fasciated stems. AtCLE12/13 was reported to lead to a dwarf plant with short silique, which mimics the Atclv3 phenotype at overexpression34,36. The ectopic expression of AtCLE1-7 showed almost identical phenotype with Atclv337. The amino acid alignment shows that the CLE motif of GhCLE5 is similar to AtCLE12, AtCLE13, and AtCLV3 (Fig. 5A). Therefore, the ectopic expression of GhCLE5 interrupted the AtCLV3 function in a way that may be similar to the ectopic expression of AtCLE12. We speculated that GhCLE5 could behave as a signal either by competing with the AtCLV3 peptide or blocking the downstream signaling transduction.
GhCLE5 interacted with the CLV3 receptor complex
GhCLE5 contained a predicted signal peptide on the N-terminus (http://smart.embl-heidelberg.de/) (Fig. 5A). The active translation of GhCLE5 was further confirmed by GFP signal examination (Supplemental Figure 6B). GhCLE5::GFP signals were observed in root, leaf, and stem tissues (Supplemental Figure 6B). As previously reported, the CLE proteins must be processed into short peptides containing the CLE motif before they carry out their function. Therefore, the GhCLE5::GFP proteins observed were unprocessed proteins with no function. However, the GhCLE5::GFP signal indicated that GhCLE5 is highly likely to be a transmembrane protein that is ready to be digested and secreted across the cell membrane. If GhCLE5 was processed into a peptide to occupy the position of the CLV3 motif as a hormonal molecule, the GhCLE5::GFP protein should have undergone protein processing and the GFP degradation would render it undetectable. Furthermore, we did not observe any GFP signal in the SAM tissue of any of the phenotypic transgenic GhCLE5::GFP plants. Therefore, we speculate that GhCLE5 is processed into a short peptide that could compete with the CLV3 motif in SAM development regulation.
To determine how the ectopic overexpression of GhCLE5 in Arabidopsis led to a phenotype partially resembling the Atclv3 phenotype, we further examined the binding capacity of GhCLE5 in each of the receptor complexes of CLV1, CLV2, and CRN/CLV2. AtCLV1 contains an ectodomain (ECD) on the N-terminus and a transmembrane domain on the C-terminus (Fig. 5B). A yeast two-hybrid assay was first conducted to detect the binding efficiency of GhCLE5 with the whole length of AtCLV3 receptors, AtCLV1, AtCLV2, and AtCRN. The assay did not show any direct interaction between GhCLE5 and AtCLV1, AtCLV2, or AtCRN (Fig. 5C). There was also no interaction observed between AtCLV3 and any of these proteins. This may be because the yeast system failed when used on proteins with a transmembrane structure. When we used the ECD of AtCLV1 on the pGBK, the yeast two-hybrid result showed clear binding of GhCLE5 and the ECD domain of AtCLV1(Fig. 5D). To confirm the binding capability between GhCLE5 and the AtCLV3 receptors, split-luciferase assays were performed. These assays no interactions between GhCLE5-Nluc and either Cluc-AtCLV2 or Cluc-AtCRN were detectable (Fig. 6A). However, there showed an interaction between GhCLE5-Nluc and Cluc-AtCLV1 (Fig. 6B). CLV3/CLV2/CRN can form a complex that regulates the SAM status in Arabidopsis. We hypothesize that GhCLE5 could replace CLV3 to form a complex with CLV2 and CRN.
GhCLE5 functional validation in cotton seedling height regulation
To investigate the function of GhCLE5 in cotton, we employed VIGS technology in upland cotton accession TM-1. The GhCLE5-silenced plants showed relatively slow development in terms of seedling height (Fig. 7A–C). The expression of GhCLE5 RNA was relatively high in the hypocotyl, root, and stem tissues of cotton seedlings (Fig. 7D). The promoter activity of GhCLE5 was validated by the transient expression assay using tobacco leaf assay (Fig. 7E). The expression pattern also supported a functional role for GhCLE5 in seedling height control. Plant height is under the regulation of multiple factors, including, but not limited to, the development of vascular tissue and the SAM status. Therefore, the phenotypes of ectopic GhCLE5 expression in Arabidopsis support the proposed endogenous function of GhCLE5 in cotton seedlings.
Discussion
Based on the presented data, we propose that GhCLE5 could occupy the CLV3 motif binding sites with CLV1, CLV2, and CRN in the Arabidopsis ectopic overexpression line (Fig. 8). Due to the sequence similarity of CLE motifs, CLV3 receptors can be engaged with ectopic GhCLE5. In this way, the signal mediated by the CLV3/CLV1 and CLV3/CLV2/CRN complexes could not be transmitted. The Arabidopsis ectopic GhCLE5 transgenic lines might have unstable protein-processing efficiency, because the ectopic expression of GhCLE5 led to a variety of developmental phenotypes, each of which partially mimicked the Atclv3 phenotype. This also indicates that the cotton GhCLE gene family could be a potential functional regulator for plant development.
CLE genes can derive short peptides that function as hormone-signaling molecules to direct the development of plant shoots and roots. The CLE gene sequences vary greatly from species to species. The conserved CLE motif is as short as 12 to 13 amino acids, which makes it difficult to identify CLE gene family members in alternative genomes. Cotton is a fiber plant and also a model plant for polyploid genome study. We identified 93 CLE genes in the upland cotton genome, with more than 40 members from each subgenome. This number is comparable with the CLE gene members identified in G. raimondii genome (55)2. The gene number is not significantly larger than that in Arabidopsis (45), although the cotton genome is about 10 times larger than that of Arabidopsis, due to multiple whole-genome duplication events38. The CLE gene family was not enlarged together with the whole-genome duplication events, which indicates its importance and the strength of the selection pressure on it.
Although the CLE gene family was not enlarged in cotton, the CLE motif was profoundly diversified. According to the CLE motif sequence, new groups of CLE patterns can be distinguished. However, no identical CLE motif of CLV3 was detected in the cotton genome. In addition, the homologous WUS gene in cotton is known to play a similar role in embryotic callus induction33,39,40. Given the conserved circuit of SAM maintenance, it seems that cotton has derived a unique CLE motif to fulfill this function. Here we demonstrated that GhCLE5 can interact with the CLV3 receptors in tobacco leaves. However, the main function of GhCLE5 in cotton is largely unknown. VIGS treatment did not have a dramatic impact on cotton SAMs; however, the efficiency of VIGS was not high. We cannot rule out the possibility that any other GhCLE member could also be involved at this stage, especially those from Group I. The endogenous receptors of GhCLE5 in cotton are still unknown.
GhCLE Might Undergo Protein Processing. Protein processing is critical for CLE proteins. A typical CLE protein is located on the membrane with a signal peptide on the extracellular surface. The CLE motif can be processed into a peptide that serves as a signaling molecule. The cellular localization assay demonstrated that cotton CLEs are predominantly located on the cell membrane. According to the predicted protein structure, GhCLE5 contains a signal peptide on the N-terminal. The C-terminal with the CLE motif is also predicted to be an outside arm. In 35S::GhCLE5::GFP transgenic Arabidopsis, GFP signals represented unprocessed or incompletely processed proteins. The GhCLE5-23 and GhCLE5-19 lines, which exhibited a strong phenotype, had weak GFP signals and strong RNA transcription activity. Statistical analysis was not conducted due to the very limited biologic replication numbers. However, observation indicated that the CLE5 processing efficiency could be high in those lines. We can take this as an indication that cotton employs the conserved short peptide–processing mechanism of CLE proteins.
Materials and methods
Retrieval of the CLE gene in upland cotton
The reference genome and annotation data of the upland cotton G. hirsutum were retrieved from the Cotton Research Institute, Nanjing Agricultural University (http://mascotton.njau.edu.cn/index.htm)41. The amino acid sequence of the Arabidopsis CLE gene was retrieved from the TAIR database (http://www.arabidopsis.org/). To identify CLE gene candidates in cotton, the Arabidopsis CLE genes were used as a query source for searching against the cotton genome using BLASTP41. Only genes with translated sequences with a CLE motif, signaling peptides in the N-terminal, and a molecular weight of less than 15 KD were considered as cotton CLE genes42.
Evolutionary analysis and structure prediction
The screened CLE protein sequences were aligned using ClustalX (http://www.clustal.org/) with default parameters. In constructing the phylogenetic tree, ModelGenerator (http://mcinerneylab.com/software/modelgenerator/) software43 was used to calculate the best alternative model. PhyML (http://www.atgc-montpellier.fr/phyml/) was used to construct the phylogenetic tree with the parameters Bootstrap100 and model JTT. A Gene Structure Display Server (GSDS, http://gsds.gao-lab.org/) was used to analyze the intron–exon structure of genes.
Chromosome mapping
According to the genomic annotation file, the CLE genes were tagged on the chromosome using the R package chromPlot (http://www.bioconductor.org/packages/release/bioc/html/chromPlot.html)44. Orthologous CLE genes and duplicated CLE genes were identified using MCScanX (http://chibba.pgml.uga.edu/mcscan2/) software with default parameters45.
CLE gene classification
The hormonal fragment of CLE proteins is a conserved 13-amino-acid sequence, namely, the CLE motif. CLAN software (https://omictools.com/clans-tool) was used to cluster the CLE genes based on the similarity of their translated CLE motifs. Next, CLE genes were clustered into seven groups, and the R package ggseqlogo46 was used for visualization of the motif of each group.
CLE gene expression analysis
RNA-seq data from cotton tissues (root, cotyledon, leaf, stem, torus, petal, stamen, pistil, calyx, ovule, and fiber) were downloaded from our previously released publication47,48 and data (PRJNA248163, PRJNA490626). Raw data were trimmed using Trimmomatic (http://www.usadellab.org/cms/?page=trimmomatic) software49. Data on the expression of genes were normalized into FPKM using TopHat2 and Cufflinks50. Next, the expression levels of CLE genes were visualized using the R language package Pheatmap (https://cran.r-project.org/web/packages/pheatmap/).
Vector construction and plant transformation
The total RNA was isolated from the Upland cotton (G. hirsutum) leaf samples using the Rapid RNA Extraction Kit (Zhong Ding Biology, RK2002). Full-length cDNA was synthesized using HiScript II Q RT SuperMix (Vazyme, R222-01) for qPCR. To generate GhCLE overexpression plants using A. thaliana Col-0 as a background, the full-length coding sequences of GhCLEs were first amplified using the primers listed in Supplemental Table 6, and then cloned into the cloning vector pBINPLUS.GFP4 via Sal I and BamH I restriction enzyme sites. Arabidopsis transformation was performed using a floral dipping protocol with the Agrobacterium tumefaciens GV3101 strain.
RT-PCR
Primer Premier (Primer Premier 5 1.0, https://macdownload.informer.com/primer-premier/) software was used to design RT-PCR primers (Supplemental Table 6) based on the sequences of the GhCLE5 genes. The A. thaliana reference gene was UBQ5, and the G. hirsutum reference gene was histone 3. Nanjing Prime Biotech Co., Ltd., performed the primer synthesis. Using the SYBR Green I dye method, the 20 μL reaction system in the PCR tube was mixed with 0.5 μL of each of the left and right primers, 1.5 μL of the cDNA template, and 7.5 μL of the ddH2O. RT-PCR was carried out using a Roche LightCycler 480 real-time PCR instrument. Expression level analysis was performed in triplicate using the minimum number of sample threshold cycles (Ct value) and 2−ΔΔCt methods.
Subcellular localization
Subcellular localization was performed by cloning the full length of the GhCLE coding sequences (Supplemental Table 6) into the pBINPLUS.GFP4 vector. The genes were fused in-frame with green fluorescent protein (GFP) for expression under the control of the CaMV35S promoter. The fused 35S::GhCLEs::GFP construct was transformed into tobacco (Nicotiana benthamiana) by infiltration with the A. tumefaciens GV3101 strain, and GFP signals were observed using a laser confocal microscope (Zeiss LSM780) with 488-nm excitation.
Scanning electron microscopy (SEM)
The shoot apical tissue of 5-week old Arabidopsis were harvested for the SEM sample preparation. The sample was first fixed with 2.5% glutaraldehyde in phosphate buffer (0.1 M, pH7.0) for more than 4 h; washed three times in the phosphate buffer (0.1 M, pH7.0) for 15 min at each step; then post fixed with 1% OsO4 in phosphate buffer for 1–2 h and washed three times in the phosphate buffer (0.1 M, pH7.0) for 15 min at each step. The sample was first dehydrated by a graded series of ethanol (30%, 50%, 70%, 80%, 90% and 95%) for about 15 min at each step, then dehydrated two times by alcohol for 20 min at each step or stored in alcohol. The sample was dehydrated in Hitachi Model HCP-2 critical point dryer. The dehydrated sample was coated with gold–palladium in Hitachi Model E-1010 ion sputter for 4–5 min and observed in Hitachi Model SU-8010 SEM.
The application of synthetic peptides to Arabidopsis
The CLE motif of GhCLE5 was selected for chemical synthesis (GL Biochem Ltd., Shanghai, China) (RLVPTGPNPLHH, purity: 94.1%, 10 mg/tube). The synthesized peptides were fully dissolved with ddH2O. After complete dissolution, this stock buffer was filtered and sterilized with a 0.22 μm sterilizer. The 1/2MS medium was sterilized by autoclaving for 15 min, cooled, and then different amounts of peptide stock buffer were added to configure plates with the concentration gradient assigned in the corresponding tests. The five treatments were 0 nmol/L, 10 nmol/L, 102 nmol/L, 103 nmol/L, and 104 nmol/L. Arabidopsis seeds were cultivated under a 16/8 h light and dark cycle. Root lengths were examined and photographed at two weeks after germination.
Yeast two-hybrid assay
The full-length coding sequences of AtCLV2, AtCLV1, AtCRN and AtCLV1-ECD were each fused to the Gal4 DNA binding domain of pGBKT7. The full-length coding sequences of AtCLV3 and GhCLE5 were each fused to the Gal4 activation domain in pGADT7. The constructed bait and prey composition was co-transformed into the yeast strain gold yeast. Two days after growth on SD-Leu/-Trp plates, the interaction between the bait and the prey was observed on the SD-Leu/-Trp/-His selective medium. Yeast strains containing pGBKT7_AtCLV3 and pGBKT7_GhCLE5 and a negative pGADT7 vector were used as negative controls. The constructed bait and prey composition was co-transformed into the yeast strain gold yeast following the kit manufacturer’s instructions (Frozen-EZ Yeast Transformation II Kit, Zymo Research). All primer information can be found in Supplemental Table 6.
Luciferase complementation for protein–protein interactions
We adapted a split-luciferase assay method51 to determine the protein–protein interactions. The full-length coding sequences of AtCLV2, AtCLV1, and AtCRN were each fused to pCAMBIA1300-cLUC. The full-length coding sequences of AtCLV3 and GhCLE5 were each fused to pCAMBIA1300-nLUC. The fused constructs were transformed into tobacco (N. benthamiana) by infiltration with the A. tumefaciens GV3101 strain. The Agrobacterium was shaken overnight on a shaker at 28 °C until the bacterial solution turned orange–yellow. Following overnight culture, the solution was centrifuged at 4000 rpm for 10 min, and the cells were collected. The inoculation dye solution was resuspended to an OD600 of 1. The prepared bacterial solution was allowed to stand in the incubator at 28 °C for 2 h in the dark. The N-LUC and C-LUC solutions were mixed at a ratio of 1:1 and then injected into the back of the tobacco leaves. The tobacco plants were covered with a black plastic bag to prevent direct light from reaching them and were placed in a light incubator at 23 °C for 48 h. After the dark treatment, the black plastic bag was removed, and the tobacco plants were placed in a light incubator at 28 °C. After 16 h of light exposure, the LUC activity was measured. One mL of luciferin was added to the leaves, and the materials were kept in the dark for 8 min to quench the fluorescence. A low-light-cooled CCD imaging apparatus (Tanon 5200) was used to capture the LUC image. Primer information is shown in Supplemental Table 6.
Virus-induced gene silencing
VIGS primers were designed and amplified by PCR based on the full-length coding sequence of the GhCLE5 gene. The PCR product was connected to a pTRV2:00 empty carrier. pTRV2:GhCLE5 was introduced into A. tumefaciens GV3101 using a freeze–thaw method. The positive strain and agrobacteria containing the plasmids pTRV1, pTRV2:00, and pTRV2:CLA52 were expanded and cultured. After suspension for 3 h, the pTRV1 was mixed with pTRV2:00, pTRV2:GhCLE5, and pTRV2:CLA at a volume ratio of 1:1. Cotton seedlings with cotyledons that had just flattened were injected with a fungus solution at the back of two thick cotyledons. All cotton seedlings were placed in a light incubator and cultured at 21 to 25 °C for 15 days. The albino phenotype of the silent pTRV2:CLA plants was observed 7 to 8 days after injection. The plants lost their green color from the first true leaf, and there was no difference between the injected no-load and noninjected negative controls. The height from the cotyledon to the growing point of the cotton was measured, and Student’s t tests were used to determine significant differences between experimental and control plants.
Promoter activity assay using tobacco leaves
The GhCLE5 promoter was cloned using G. hirsutum acc. TM-1 genomic DNA and ligated into the pBI121 vector (primer sequences shown in Supplemental Table 6). Using Agrobacterium tumefaciens GV3101 as the mediating bacteria, the plasmid was transformed into Agrobacterium by heat transformation. The resulting Agrobacterium GV3101 single clone was cultivated in liquid LB medium containing kanamycin (50 µg/mL) and rifampin (50 µg/mL) at 28 °C overnight with shaking at 200 rpm. The bacteria were then collected by centrifugation at 4000 rpm/min for 10 min, and resuspended in injection buffer (10 mM MgCl2, 10 mM MES, pH 5.7, 150 µM acetosyringone) to a concentration of OD600 = 0.8–1. The bacterial injection buffer was incubated in the dark at 28 °C for 3 h and then injected into six-week-old tobacco leaves. After a 72 h cultivation in the dark, the tobacco leaves were harvested for GUS staining. Staining was carried out at 37 °C for 2–3 h, and then the leaves washed with fixative (70% ethanol: glacial acetic acid (V/V) = 9:1). The experiment was repeated three times, with three biological replicates each time.
References
Nardmann, J., Chandler, J. W. & Werr, W. Stem cell fate versus differentiation: the missing link. Trends Plant Sci. 21, 725–727. https://doi.org/10.1016/j.tplants.2016.07.002 (2016).
Goad, D. M., Zhu, C. & Kellogg, E. A. Comprehensive identification and clustering of CLV3/ESR-related (CLE) genes in plants finds groups with potentially shared function. New Phytol. 216, 605–616. https://doi.org/10.1111/nph.14348 (2017).
Hastwell, A. H., Gresshoff, P. M. & Ferguson, B. J. Genome-wide annotation and characterization of CLAVATA/ESR (CLE) peptide hormones of soybean (Glycine max) and common bean (Phaseolus vulgaris), and their orthologues of Arabidopsis thaliana. J. Exp. Bot. 66, 5271–5287. https://doi.org/10.1093/jxb/erv351 (2015).
Clark, S. E., Running, M. P. & Meyerowitz, E. M. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121, 2057–2067 (1995).
Fletcher, J. C., Brand, U., Running, M. P., Simon, R. & Meyerowitz, E. M. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science (New York, N.Y.) 283, 1911–1914. https://doi.org/10.1126/science.283.5409.1911 (1999).
Kucukoglu, M. & Nilsson, O. CLE peptide signaling in plants—the power of moving around. Physiol. Plant. 155, 74–87. https://doi.org/10.1111/ppl.12358 (2015).
Takahashi, F. et al. A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 556, 235–238. https://doi.org/10.1038/s41586-018-0009-2 (2018).
Aichinger, E., Kornet, N., Friedrich, T. & Laux, T. Plant stem cell niches. Annu. Rev. Plant Biol. 63, 615–636 (2012).
Somssich, M., Je, B. I., Simon, R. & Jackson, D. CLAVATA-WUSCHEL signaling in the shoot meristem. Development 143, 3238–3248 (2016).
Su, Y. H. et al. Integration of pluripotency pathways regulates stem cell maintenance in the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. U. S. A. 117, 22561–22571. https://doi.org/10.1073/pnas.2015248117 (2020).
Han, H., Liu, X. & Zhou, Y. Transcriptional circuits in control of shoot stem cell homeostasis. Curr. Opin. Plant Biol. 53, 50–56. https://doi.org/10.1016/j.pbi.2019.10.004 (2020).
Whitewoods, C. D. et al. CLAVATA was a genetic novelty for the morphological innovation of 3D growth in land plants. Curr. Biol. 28, 2365-2376.e5. https://doi.org/10.1016/j.cub.2018.05.068 (2018).
Leibfried, A. et al. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438, 1172–1175. https://doi.org/10.1038/nature04270 (2005).
Laux, T., Mayer, K. F., Berger, J. & Jurgens, G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96 (1996).
Brand, U., Fletcher, J. C., Hobe, M., Meyerowitz, E. M. & Simon, R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. https://doi.org/10.1126/science.289.5479.617 (2000).
Schoof, H. et al. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644 (2000).
Brand, U., Fletcher, J. C., Hobe, M., Meyerowitz, E. M. & Simon, R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science (New York, N.Y.) 289, 617–619. https://doi.org/10.1126/science.289.5479.617 (2000).
Fletcher, J. C., Brand, U., Running, M. P., Simon, R. & Meyerowitz, E. M. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914 (1999).
Jeong, S., Trotochaud, A. E. & Clark, S. E. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11, 1925–1934. https://doi.org/10.1105/tpc.11.10.1925 (1999).
Clark, S. E., Running, M. P. & Meyerowitz, E. M. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119, 397–418 (1993).
Trotochaud, A. E., Hao, T., Wu, G., Yang, Z. & Clark, S. E. The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a rho-related protein. Plant Cell 11, 393–405 (1999).
Kayes, J. M. & Clark, S. E. CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125, 3843–3851 (1998).
Muller, R., Bleckmann, A. & Simon, R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20, 934–946 (2008).
Hu, C. et al. A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis. Nat. Plants 4, 205–211. https://doi.org/10.1038/s41477-018-0123-z (2018).
Fletcher, J. C. Recent advances in Arabidopsis CLE peptide signaling. Trends Plant Sci. 25, 1005–1016. https://doi.org/10.1016/j.tplants.2020.04.014 (2020).
Meng, L. & Feldman, L. J. CLE14/CLE20 peptides may interact with CLAVATA2/CORYNE receptor-like kinases to irreversibly inhibit cell division in the root meristem of Arabidopsis. Planta 232, 1061–1074. https://doi.org/10.1007/s00425-010-1236-4 (2010).
Qian, P. et al. Author Correction: The CLE9/10 secretory peptide regulates stomatal and vascular development through distinct receptors. Nat. Plants 5, 238. https://doi.org/10.1038/s41477-018-0347-y (2019).
Qian, P. et al. The CLE9/10 secretory peptide regulates stomatal and vascular development through distinct receptors. Nat. Plants 4, 1071–1081. https://doi.org/10.1038/s41477-018-0317-4 (2018).
Xu, T. T., Ren, S. C., Song, X. F. & Liu, C. M. CLE19 expressed in the embryo regulates both cotyledon establishment and endosperm development in Arabidopsis. J. Exp. Bot. 66, 5217–5227. https://doi.org/10.1093/jxb/erv293 (2015).
Soyano, T., Hirakawa, H., Sato, S., Hayashi, M. & Kawaguchi, M. Nodule Inception creates a long-distance negative feedback loop involved in homeostatic regulation of nodule organ production. Proc. Natl. Acad. Sci. U. S. A. 111, 14607–14612. https://doi.org/10.1073/pnas.1412716111 (2014).
Fan, C. et al. A novel single-nucleotide mutation in a CLAVATA3 gene homolog controls a multilocular silique trait in Brassica rapa L. Mol. Plant 7, 1788–1792. https://doi.org/10.1093/mp/ssu090 (2014).
Guan, X., Song, Q. & Chen, Z. J. Polyploidy and small RNA regulation of cotton fiber development. Trends Plant Sci. 19, 516–528. https://doi.org/10.1016/j.tplants.2014.04.007 (2014).
Xiao, Y. et al. Effects of GhWUS from upland cotton (Gossypium hirsutum L.) on somatic embryogenesis and shoot regeneration. Plant Sci. 270, 157–165. https://doi.org/10.1016/j.plantsci.2018.02.018 (2018).
Strabala, T. J. et al. Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol. 140, 1331–1344. https://doi.org/10.1104/pp.105.075515 (2006).
Kondo, Y. & Fukuda, H. The TDIF signaling network. Curr. Opin. Plant Biol. 28, 106–110. https://doi.org/10.1016/j.pbi.2015.10.002 (2015).
Jun, J. et al. Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol. 154, 1721–1736. https://doi.org/10.1104/pp.110.163683 (2010).
Ni, J. & Clark, S. E. Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol. 140, 726–733. https://doi.org/10.1104/pp.105.072678 (2006).
Paterson, A. H. et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. https://doi.org/10.1038/nature11798 (2012).
Zheng, W. et al. AtWuschel promotes formation of the embryogenic callus in Gossypium hirsutum. PLoS ONE 9, e87502. https://doi.org/10.1371/journal.pone.0087502 (2014).
Bouchabke-Coussa, O. et al. Wuschel overexpression promotes somatic embryogenesis and induces organogenesis in cotton (Gossypium hirsutum L.) tissues cultured in vitro. Plant cell Rep. 32, 675–686. https://doi.org/10.1007/s00299-013-1402-9 (2013).
Zhang, T. et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. https://doi.org/10.1038/nbt.3207 (2015).
Oelkers, K. et al. Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biol. 9, 17–17 (2008).
Keane, T. M., Creevey, C. J., Pentony, M. M., Naughton, T. J. & Mclnerney, J. O. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol. Biol. 6, 29 (2006).
Oróstica, K. Y. & Verdugo, R. A. chromPlot: visualization of genomic data in chromosomal context. Bioinformatics 25, btw137 (2015).
Yupeng, W. et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49–e49 (2012).
Wagih, O. Ggseqlogo: A versatile R package for drawing sequence logos. Bioinformatics 33, 3645–3647 (2017).
Tianzhen, Z. et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537 (2015).
Hu, Y. et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 51, 739–748. https://doi.org/10.1038/s41588-019-0371-5 (2019).
Bolger, A. M., Marc, L. & Bjoern, U. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Cole, T. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Huamin, C. et al. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146, 368–376 (2008).
Gao, X., Britt, R., Shan, L. & He, P. Agrobacterium-mediated virus-induced gene silencing assay in cotton. J. Vis. Exp. https://doi.org/10.3791/2938 (2011).
Acknowledgements
We also thank Professor Honggui La from Nanjing Agricultural University, who provided advice on the luciferase assay.
Funding
This work was financially supported in part by grants from the National Natural Science Foundation of China (NSFC, 31971985, 32000379), the Leading Innovative and Entrepreneur Team Introduction Program of Zhejiang (2019R01002), China Postdoctoral Science Foundation (2019M652094) and the Fundamental Research Funds for the Central Universities.
Author information
Authors and Affiliations
Contributions
X.G. conceptualized this project. X.G. and K.W. designed the experiments. K.W., K.L., M.G., and Y.H. conducted the experiments. K.L., T.Z., and X.T. performed the bioinformatic analysis. X.G., K.W., M.G., D.Y. and G.X. analyzed the data. X.G. and K.W. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wan, K., Lu, K., Gao, M. et al. Functional analysis of the cotton CLE polypeptide signaling gene family in plant growth and development. Sci Rep 11, 5060 (2021). https://doi.org/10.1038/s41598-021-84312-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-021-84312-8
This article is cited by
-
Peptide hormones in plants
Molecular Horticulture (2025)
-
Integrative multi-omics approaches for crop abiotic stress tolerance
Discover Plants (2025)
-
Shining in the dark: the big world of small peptides in plants
aBIOTECH (2023)
-
CLE peptides: critical regulators for stem cell maintenance in plants
Planta (2022)