Abstract
Iodine deficiency represents a public health problem worldwide. To increase the amount of iodine in the diet, biofortification strategies of plants have been tried. They rely on the exogenous administration of iodine to increase its absorption and accumulation. However, iodine is not stable in plants and can be volatilized as methyl iodide through the action of specific methyltransferases encoded by the HARMLESS TO OZONE LAYER (HOL) genes. The release of methyl iodide in the atmosphere represents a threat for the environment due to its ozone depletion potential. Rice paddies are among the strongest producers of methyl iodide. Thus, the agronomic approach of iodine biofortification is not appropriate for this crop, leading to further increases of iodine emissions. In this work, we used the genome editing CRISPR/Cas9 technology to knockout the rice HOL genes and investigate their function. OsHOL1 resulted a major player in methyl iodide production, since its knockout abolished the process. Moreover, its overexpression reinforced it. Conversely, knockout of OsHOL2 did not produce effects. Our experiments helped elucidating the function of the rice HOL genes, providing tools to develop new rice varieties with reduced iodine emissions and thus more suitable for biofortification programs without further impacting on the environment.
Similar content being viewed by others
Introduction
Rice (Oryza sativa L.) is among the oldest cultivated plants and one of the most important cereal, still representing a staple food crop for about 2.5 billion people worldwide1. Its cultivation covers the 1% of the global land area2, and, with maize and wheat, it contributes to bring half of the calories consumed by the human population3. As a result, ingestion of rice may represent an important pathway for nutrients into human diet and biotechnologies have been developed to enrich rice varieties of essential vitamins and microelements4.
Iodine represents one of the most important micronutrients for human health, being necessary in the thyroid gland to synthesize the hormones triiodothyronine and thyroxine5. Iodine deficiency is one of the most serious public health problems worldwide, affecting almost one-third of the human population6. Usually cultivated crops and vegetables are poor sources of iodine. This is mainly due to the low content of the element in the soil, especially in rural areas distributed in mountainous and other inland zones7, where plants cannot find adequate amounts of the element8. The range of iodine in grain crops, for example, is estimated between 2 and 30 µg/kg, which is too low to meet the daily demand, being the iodine requirement for an adult around 150–200 µg per day9.
Iodine deficiency affects both developing and well-developed countries but is particularly severe in areas where rice represents the main staple food, being this species generally poor of iodine. In inland areas and in flooded fields, where rice is often cultivated, the iodine present in the soil is generally leached away, thus reducing its content in plants10. They, indeed, absorb this element mainly through the roots, being less than 0.2% the amount taken up by the atmosphere11. Moreover, polished rice contains only 0.055% of the element absorbed by the plant, and more than 70% is found in the straw11. A reduced phloematic transport is partially responsible of the low iodine amount in the seeds, but, remarkably, the process of iodine volatilization, that releases methyl iodide (CH3I) in the gaseous form, may also strongly contribute to it8. As a matter of fact, due to these emissions, the iodine foliar concentration in rice decreases exponentially with a half-life of 14 days12. Consequently, the element stored in the plant, potentially prone to be translocated to the edible grains, is drastically reduced.
Methyl halides, including CH3I, are produced from natural and anthropogenic activities and represent an important source of halogens in the atmosphere. Their breakdown products are involved in catalytic heterogeneous chemical reactions, which play an important role in the tropospheric chemistry and in the stratospheric ozone loss13. CH3I has a short lifetime compared to other methyl halides, and therefore a lower ozone depletion potential. However, iodine produced from methyl iodide can strongly influence the oxidizing capacity of the troposphere and induce a radiative effect affecting new particle formation14. The global production of CH3I is around 260–610 kt/year, and approximately 5% of this, about 16–29 kt/year, comes from rice paddies worldwide15,16: rice cultivation has therefore a major impact on the chemistry of the atmosphere. CH3I is mainly emitted during the early stages of rice growth, before maximum tillering, and particularly when rice is grown in soils with elevated halide concentrations17. Temperature is also important: a 1˚C temperature rise can increase the CH3I emission from rice by 10%16.
Methyl iodide emissions in plants involve enzymatic reactions catalyzed by halide methyltransferases (HMTs) or halide/thiol methyltransferases (HTMTs), which both have an S-adenosyl-L-methionine (SAM)-dependent methyl transferase activity and generate S-adenosyl-L-homocysteine as the by-product18,19,20. The identification of these enzymes in several higher plants21 indicates that their activity is widespread. However, the versatility shown for different substrates makes their physiological role still unclear. In an in vivo study performed in the model species Arabidopsis thaliana, Rhew et al.22 showed that methyl halides do not play a fundamental role in plant development. However, in soils with high salinity, like the salt marshes, the production of methyl halides can be considerably higher than in other ecosystems23,24. Thus, in halotolerant plants, methyl halides could be produced to maintain homeostatic levels of halide ions in the cytoplasm and to detoxify the plant from them, being their potential phytotoxicity widely demonstrated8,25. Moreover, in some groups of plants, such as the Brassicaceae, some HTMTs show a particular ability to methylate thiocyanate or bisulfide ions, produced from the hydrolysis of glucosinolates20. In such species, HTMTs could be involved in the detoxification of sulfur compounds20,22, and, in this case, halomethanes may simply be by-products of metabolism26.
Plant HMTs and HTMTs are encoded by small families of genes. The HOL1 (HARMLESS TO OZONE LAYER1) gene, identified in Arabidopsis and thus named for the loss of function of the mutant enzyme, is the main responsible for methyl halide emissions in this species22. Other two homologous genes were identified in Arabidopsis, but their proteins seemed to play minor roles in vivo27. Homologous of these enzymes are widespread in the plant kingdom, including dicots, monocots, and unicellular algae27. In Oryza sativa, two genes were identified, OsHOL1 and OsHOL2, which show, respectively, a 51% and a 57% similarity with Arabidopsis HOL128. In in vitro studies, recombinant OsHOL proteins showed SAM-dependent methyltransferase activities towards halide and thiocyanate ions, with the highest effects exerted with iodide, suggesting a major involvement in the metabolism of iodine28.
Here, transgenic rice plants for the genes encoding the enzymes involved in iodine volatilization were produced to analyze the impact of each gene knockout, obtained by CRISPR/Cas9 technology, as well as of gene overexpression, on methyl iodide emissions. OsHOL1 and the relative protein were identified as the main players involved in the process. To deepen our knowledge on the biochemical mechanisms underlining the methyl halides production, the subcellular localization of OsHOL1 and its ability to take part to multiprotein complexes were studied. We developed an effective mechanism to reduce methyl iodide emissions from rice plants and propose a possible strategy to enrich the iodine content of rice crops without further impacting on the environment.
Results
In rice plants, the halide methyl transferase genes are mainly expressed in leaves
The two genes of rice identified as homologous of AtHOL122, OsHOL1 and OsHOL228, belong to different chromosomes. OsHOL1 (Os03g62670) is located on chromosome 3 and organized into seven exons and six introns, which can be alternatively spliced in three predicted different isoforms (Fig. S1A). The longest full-length coding sequence (cds) (Os03g62670.1) corresponds to an open reading frame of 741 bp, coding for a 246 amino acid-long polypeptide. OsHOL2 (Os06g06040) is located on chromosome 6 and organized into eight exons and seven introns, producing seven putatively different isoforms, with the longest full-length cds (Os06g06040.1) of 753 bp, encoding a protein of 250 amino acids (Fig. S1B).
The proteins OsHOL1 and OsHOL2 predicted from the two longest transcript isoforms are identified as “S-adenosylmethionine-dependent methyltransferases (SAM- or AdoMet-MTase), class I”, on the basis of their main conserved structural domains (Fig. S2A, B). They well aligned with other plant HMTs and HTMTs previously identified (Fig. S2C), even if showing some extra amino acid short sequences. OsHOL2 showed an 18 amino acid pre-sequence in the N-terminus, whereas OsHOL1 presented additional short stretches in the conserved central SAM-MTase domain, within the SAM binding site (Fig. S2C).
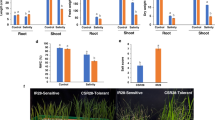
To assess the expression levels of the two genes in rice under our experimental conditions, a quantitative real-time polymerase chain reaction (RT-qPCR) was performed analyzing leaves, stems, and roots sampled from young plants grown in soil. Transcripts of both genes were identified in all the organs analyzed, although at very different levels (Fig. 1A). OsHOL1 appeared to be expressed more than OsHOL2 and the difference was particularly high in leaves, where OsHOL1 resulted mostly expressed. In roots and stems, both genes showed lower transcription rates.
Expression of OsHOL genes in rice plants and CRISPR/Cas9 mutagenesis of the same genes. (A) RT-qPCR analysis of OsHOL1 and OsHOL2 in rice plant. Transcript levels were measured in leaves, stems and roots collected from 2-week-old plants. OsHOL expression levels were normalized to the reference gene OsGAPDH. Data points represent means ± s.d. of three biological replicates. T-test analysis was performed between OsHOL1 and OsHOL2 within each organ. Differences are indicated with asterisks. **** mean P ≤ 0.0001. (B) Schematic diagrams showing the exon–intron structures corresponding to the longest coding sequences of OsHOL1 (Os03g62670.1) and OsHOL2 (Os06g06040.1). Blue rectangles represent exons, connecting lines represent introns, yellow triangles indicate the position of the target sequence recognized by the gRNA produced for that gene, whose sequence is reported, with the protospacer adjacent motif (PAM) colored in red. (C) Schematic diagram of the T-DNA contained in the expression vector used in the study, the Ph-Ubi-CAS9-7-sgRNA vector. LB, left border; Hyg, hygromycin resistance gene; OsCas9, Cas9 gene sequence optimized for Oryza sativa; ZmpUbi, Zea mays Ubiquitin1 promoter; OsU3, Oryza sativa U3 promoter; sgRNA, single-guide RNA sequence; RB, right border. Sequence of OsHOL1 (D) and OsHOL2 (E) around the gRNA target sequence (highlighted) in wild type Nipponbare plants and in CRISPR/Cas9 mutated lines generated in the study. The type of mutation is reported at the end of each edited sequence. In each sequence the PAM is colored in red.
Mutagenesis of the halide methyl transferase genes by CRISPR/Cas9 technology
To mutagenize OsHOL1 and OsHOL2 through the CRISPR/Cas9 technology, candidate target sequences for specific gRNAs were identified and, according to the position along the genes and trying to minimize the likelihood of off-target effects, a gRNA targeting the third exon of OsHOL1 and a gRNA targeting the second exon of OsHOL2 were finally selected (Fig. 1B). Being the two genes characterized by multiple splicing isoforms (Fig. S1), the targeted exons were chosen among those present in all the predicted alternative transcripts. Each of the two gRNAs was assembled, cloned in a pOs-sgRNA entry plasmid, and then recombined with the Ph-Ubi-Cas9-7-sgRNA binary destination vector29.
Scutellum-derived rice calli were transformed with Agrobacterium cultures carrying the binary vectors with the CRISPR/Cas9-gRNA cassettes (Fig. 1C). Starting from a hundred rice seeds for each transformation, 54 and 24 in vitro regenerated plants were produced, respectively, for the mutagenesis of OsHOL1 and OsHOL2. The presence of the T-DNA was attested in these plants by the PCR amplification of the hygromycin resistance gene contained in the genetic cassette (Fig. 1C, Fig. S3A). For each transgenic line, a 567 or 624 bp genomic fragment surrounding the gRNA target sequence in OsHOL1 or OsHOL2, respectively, was then amplified (Fig. S3B) and sequenced. This allowed to distinguish between wild type and mutated copies of the genes, and homozygous, heterozygous and biallelic mutations, as well as chimeric mutations, were identified (Table S1).
For silencing of OsHOL1, 34 independent transgenic lines, with 18 lines bearing homozygous or biallelic mutations, 15 chimeras, and one showing a heterozygous mutation, were produced, whereas, for silencing of OsHOL2 20 independent mutated lines were obtained, of which 12 with homozygous or biallelic mutations, seven chimeras, and one with a heterozygous mutation (Table S1). Most of the mutations identified were single or very short insertions or deletions (indels) (1, 2 or 4 nucleotides) (Table S1, Fig. 1D,E), producing frameshift mutations. Some indels were larger (20 or 29 nucleotides) and could have led as well to frameshift mutations (Table S1). Others (e.g., 6 or 24 nt deletions), being in-frame, could have produced loss of a few amino acids in the relative polypeptide chains. The rest of the T0 regenerated transformed plants showed wild type alleles in OsHOL1 and OsHOL2.
To exclude possible off-target effects, the gene OsHOL2 was amplified and sequenced in some independent OsHOL1 mutants. Indeed, considering the 8–10 bp seed region of the gRNA selected for OsHOL1 mutagenesis, the only predictable off-target could have been in OsHOL2, which contains a sequence homologous to the gRNA target (Fig. S4A). However, as shown in Fig. S4B, no OsHOL1 mutant line showed unintended mutations in the region of OsHOL2 surrounding and downstream of the possible gRNA off-target. As far as the possible off-targets for the gRNA selected for OsHOL2 mutagenesis, taking into the account the high specificity of its seed region we could exclude off-target effects in other rice genes, OsHOL1 included.
The growth and the morphology of the regenerated T0 plants with or without mutations was similar. Apart from some common developmental responses to acclimatize from the stressful in vitro conditions, they did not show any peculiar vegetative phenotype and many of the ex vitro plantlets grew in soil till flowering. However, the number of spikelets resulted low and many developing seeds aborted, likely as a consequence of stress during in vitro culture.
Knockouts in OsHOL1 affected methyl iodide emissions from leaves
Single CRISPR/Cas9-T0 lines carrying different homozygous frameshift mutations in OsHOL1 or OsHOL2 were selected and the CH3I emissions from their leaves were measured by GC–MS. T0 regenerated plants with wild type alleles of the two genes were used as controls. All the lines mutagenized in OsHOL1 showed very reduced emissions compared to control, whereas those mutagenized in OsHOL2 presented variable phenotypes, some of them exhibiting emissions similar to control and others lower than control (Fig. 2A).
Methyl iodide emissions in CRISPR/Cas9 mutated lines. CH3I emissions were measured from T0 (A) and T1 (B) plants. Air samples from leaves were analyzed 24 h after incubation with 0.5 mM KI. Data points are the mean ± s.d. of four biological replicates for each line and two technical replicates per each biological sample. Data were subjected to one-way analysis of variance (ANOVA). Values followed by the same letter are not significantly different (Tukey’s HSD post-hoc test, P ≤ 0.05).
Plants belonging to T1 progenies produced by T0 lines carrying homozygous + 1 insertion mutations in OsHOL1 or OsHOL2 (lines hol1-L9 and hol2-L3, respectively) were grown and analyzed to check the presence of the mutations in the genes of interest. In both the T1 progenies, homozygous + 1 insertion mutations in OsHOL1 or OsHOL2 were confirmed in all the plants, whereas the hol1-L9.3 and hol2-L3.3 lines segregated for the hygromycin resistance gene, indicating the loss of the T-DNA genetic cassette (Fig. S5). Two T1 plants obtained from seeds collected from the heterozygous T0 hol1-L57 line (+ 1 bp/0 OsHOL1 genotype) were also analyzed and found to contain homozygous + 1 insertion mutations in OsHOL1 as well.
A characterization of CH3I emissions was carried out in all these T1 plants. Leaf samples from hol1-L9.1 ÷ L9.6 and hol1-L57.1-L57.2 plants, mutagenized in OsHOL1, showed absence or very reduced levels of CH3I emissions, while plants carrying mutations in OsHOL2 (hol2-L3.1 ÷ L3.3) showed a phenotype not different from control (Fig. 2B).
Differently from T0 plants, all the CRISPR/Cas9 mutants belonging to the T1 generations could complete their life cycle: they flowered and developed spikelets and seeds. Due to the reduced number of T1 plants, a comparison with control Nipponbare plants in terms of grain yields or resistance to biotic or abiotic stresses could not be performed. However, the T1 mutant plants, either mutated in OsHOL1 or OsHOL2, grew similarly to control and continued to show no visible peculiar phenotypes (Fig. S6).
Overexpression of OsHOL1 in rice plants and consequent methyl iodide emissions
A full-length cds of 741 nucleotides, corresponding to the predicted isoform Os03g62670.1 of OsHOL1 (Fig. S7), was amplified from rice leaves. Several independent transgenic lines overexpressing OsHOL1 were produced after transformation of scutellum-derived rice calli with a genetic cassette expressing the cloned cds under the constitutive maize Ubiquitin1 promoter. As in the case of the CRISPR/Cas9 mutagenized lines, all the regenerated plants showed no peculiar phenotype, excluding a reduced number of spikelets and seeds. This prompted us to ascribe the general reduced fertility shown by the transgenics to the in vitro regeneration process, rather to a genotype-specific effect.
The level of overexpression of OsHOL1 was checked by RT-qPCR in a subset of transgenics. Most of the lines showed an expression of the gene higher than control (Fig. 3A), and, when measuring their CH3I leaf-emissions, they also resulted, in many cases, higher than in wild type plants (Fig. 3B). Interestingly, whereas from roots sampled from wild type plants no emissions of CH3I were found, CH3I emissions were measured from roots isolated from OsHOL1-overexpressing lines, even if lower than those observed in leaves (Fig. 3C). This result was not unexpected being the ectopic expression of OsHOL1 in the transgenic lines driven by a ubiquitous promoter.
RT-qPCR and methyl iodide emission analyses in OsHOL1 overexpressing lines. (A) Transcript levels of OsHOL1 were measured in 2-week-old plants and expressed as relative values in comparison to the expression of control wild type plants. Values represent the means ± s.d. from three biological replicates. Data were subjected to one-way ANOVA. Values followed by the same letter are not significantly different (Tukey's post-hoc test, P ≤ 0.05). CH3I emissions measured from leaves (B) and roots (C) collected from OsHOL1 overexpressing plants. Air samples from leaves or roots were analyzed 24 h after incubation with 0.5 mM KI. Data points are the mean ± s.d. of four biological replicates for each line and two technical replicates per each biological sample. Data were subjected to one-way ANOVA. Values followed by the same letter are not significantly different (Tukey’s HSD post-hoc test, P ≤ 0.05).
OsHOL1 is a dimeric protein with a complex subcellular localization
The GFP-OsHOL1 fusion protein, obtained from the expression of the cloned Os03g62670.1 cds, was expressed in Arabidopsis protoplasts. A subcellular localization in both cytosol and nuclei was observed, and the superimposition in the nuclei of the GFP fluorescence with the red fluorescence of an NLS-Cherry-RFP construct was evident (Fig. 4A).
Cell localization and BiFC assay of OsHOL1 proteins. (A) Subcellular localization of OsHOL1, transiently expressed in Arabidopsis protoplasts as fusion proteins with GFP. GFP, NLS-Cherry RFP and merged GFP + NLS-Cherry RFP fluorescence and bright field images of protoplasts are shown. (B) Protein–protein BiFC interaction analysis of OsHOL1. OsHOL1-YFPC + OsHOL1-YFPN fusion proteins, and YFPN + YFPC split proteins were co-expressed in Arabidopsis protoplasts, as indicated in the picture. BiFC was detected from reconstitution of the YFP signal. Individual and merged images of YFP and NLS-Cherry RFP as well as bright field images of protoplasts are shown. Scale bars = 10 μm.
Being known the multimeric nature of some MTs, able to produce homodimer or homotetramer complexes30, protein–protein interaction studies were carried out. A Bimolecular Fluorescence Complementation assay clearly showed the formation of dimers of OsHOL1, as attested by the reconstitution of the fluorescence of the YFP protein thanks to the reassociation of the two split YFP fragments fused to two different OsHOL1 proteins (Fig. 4B). Notably, the YFP fluorescence exhibited the same subcellular localization of the GFP-OsHOL1 fusion protein (Fig. 4A), further substantiating the double subcellular localization of OsHOL1.
A Split-Luciferase Complementation assay also showed a strong interaction between the OsHOL1 partners of two Renilla reniformis luciferase split proteins, which were able to reconstitute a functional luciferase enzyme, again indicating the formation of homodimers of OsHOL1 (Fig. 5A, B). The specificity of the interaction between the two OsHOL1 proteins was confirmed by the lack of interaction of the same polypeptide with another protein, the Arabidopsis ERF-VII transcription factor RAP2.12, not functionally related with iodine metabolism but known to localize in cytosol and nuclei as well31 (Fig. 5B).
Split-Luciferase Complementation assay of OsHOL1 proteins. (A) Scheme of the Split-Luciferase Complementation assay designed to study the possible interactions between OsHOL1 and AtRAP2.12 proteins with the expected outcomes. (B) Quantitative result of the Split-Luciferase Complementation assay performed in Arabidopsis protoplasts through the expression of the fusion proteins NRLuc-OsHOL1, CRLuc-OsHOL1, NRLuc-AtRAP2.12, and CRLuc-AtRAP2.12. Different combinations of fusion proteins were analyzed, as reported in the picture. Data were expressed as luminescence units (RLuc) normalized to the protein content. One-way ANOVA followed by Tukey’s HSD post-hoc test (n = 4 biological replicates ± s.d.; P ≤ 0.05) was performed.
Discussion
Iodine is traditionally not considered an essential element for higher plants, even if very recent studies have indicated its involvement in plant metabolism as a micronutrient32. Although several experiments showed how high concentrations of this element can be toxic for plants8,33, at low concentrations iodine can act as a growth enhancer, positively influencing growth and nutritional parameters33,34,35,36. It was also hypothesized that, by inducing an antioxidant response, low levels of iodine may protect plants from both biotic and abiotic stress conditions, such as salinity or heavy metals8,37. The impact of iodine on vegetables strongly depends on the species, the iodine form, the application method, and the properties of the substrate where plants are grown36,37. It is, however, undoubted that plants are able to take up iodine from the soil through the root system and from the air through the leaves, and that the quantity taken up is usually dependent on the amount of iodine available in the environment8,37.
Plants, however, do not only absorb iodine but also emit considerable amounts of it in the form of methyl iodide, thus reducing its content likely to avoid possible phytotoxic effects. However, in this way, they impact on the integrity of the stratosphere, being CH3I an ozone-depleting gas13. The CH3I volatilization process is mediated in plants by the HOL genes. There is only scattered evidence on possible tissue-specific expression patterns of these genes. The TMT1 HOL gene of Brassica oleracea, for example, showed high levels of expression in roots, stems and leaves38, and in Arabidopsis AtHOL1 seemed to be ubiquitously expressed in the plant, including seedlings27. In Raphanus sativus, on the other hand, the activity of the HMT codified by RsHOL was found high in leaves but absent in roots21. Thus, it is still not clear whether plants can produce and volatilize methyl halides from all their tissues. In the present study, the expression of the two HOL genes of rice was analyzed in plants grown in soil without any exogenous iodine administration. Young plants were used, since previous data indicated that rice mostly volatilizes CH3I during the early stages of growth13. The RT-qPCR analysis we performed showed that the transcription levels of OsHOL1 and OsHOL2 were quite different, and that OsHOL1 was the most expressed of the two genes, with a particularly high transcription rate in leaves. Our results agreed with the data reported in previous studies28, and also with those present in the Rice Expression Profile Database39, which show a sustained OsHOL1 expression in leaves and a low transcription of OsHOL2, without preferential locations in the plant (Fig. S8). It seems therefore that shoots represent the main source of CH3I volatilization from rice plants.
To date, the HMTs of rice have not been fully characterized in vivo at the protein level. In our study, GFP-imaging studies indicated that GFP-tagged OsHOL1 proteins localized both in the cytosol and in the nuclei of plant cells. This represented an unexpected result and was not in accordance with previous hypothesis on a possible localization of these enzymes in the chloroplasts, where they may play a role in the detoxification of excess ions produced upon sulphate reduction20. More in accordance with our data, a study carried out in Arabidopsis thaliana and in Brassica oleracea detected HOL activity in the cytosol and not in the chloroplasts40. Based on our results, we can speculate that OsHOL1 proteins may mediate iodine detoxification processes in the cytoplasm as well as in the nuclei, where they may be necessary, for example, to prevent toxic actions of iodide on nucleic acids, or even their iodination41,42.
The HMTs of rice belong to the group of SAM-dependent MTs, enzymes implicated in a plethora of different essential cellular activities, including signal transduction, biosynthetic reactions, protein repair, chromatin regulation and gene silencing30. They are classified according to their structural folds in five classes. Most of the MTs currently known, including the two HMTs of rice, belong to the class I, the “Rossmann-fold” class43. Three-dimensional structures of some class I MTs revealed the presence of homodimeric or homotetrameric complexes, even if most of them should act as monomers30. We have shown that OsHOL1 was able to interact with itself in vitro and this suggests that it can form homodimers in vivo. As far as we know, this is the first evidence indicating a possible dimeric structure of an HMT in plants, since previous studies characterized as monomeric other HMTs, included AtHOL119,44,45. The formation of homodimers was highlighted in other classes of plants MTs, for example in some of the type-1 family of SAM-dependent O-methyltransferases46. These enzymes are characterized by an auxiliary N-terminal domain that is required for the homodimerization and their structure consists of two monomers self-associating to form a homodimer, in which the central core is represented by the two dimerization domains, with the dimer interface contributing to the substrate binding, while the SAM binding domains are located externally47. Further studies are necessary to confirm the tridimensional structure of OsHOL1 in vivo, its possible dimeric nature, and, eventually, the specific domains involved in the dimerization. However, based on what already known in other MTs, dimerization could be of functional importance for the catalytic activity of the enzyme and its substrate discrimination.
In this study, we confirmed how the CRISPR/Cas9 system was an effective tool to mutagenize rice genes, thus representing a powerful tool which can contribute to the genetic improvement of this crop. The two major limits of this genome editing technology are the possible unfavorable editing at other sites in the genome48 and the on-target efficiency49. However, also possible on-target unexpected effects, such as large deletions, inversions, and insertions, were recently described50. The results we obtained showed the efficiency of both the selected gRNAs and the CRISPR/Cas9 vectors used. Gene knockouts were successfully generated in both OsHOL1 and OsHOL2 from T0 plants, and, in many cases, they consisted in single or very short indels inducing homozygous frameshift mutations. We did not extensively analyze off-target effects in the T0 edited plants, except for the predictable one in OsHOL2 sequence in CRISPR/Cas9 OsHOL1 mutants. This was excluded in three independent lines. The relevant number of independent T0 mutants produced for each gene, all showing phenotypes very similar among them and to wild type controls, did not suggest the existence of additional mutations. However, in the lines which will be finally maintained as reference stable knockout mutants for the two genes of interest, it would be appropriate to perform an in-depth whole-genome sequencing to definitively exclude such effects.
In our study a significant percentage of T0 plants resulted homozygous or biallelic for the mutated genes. As expected for the Cas9 enzyme and as already observed in rice plants, most of these mutations consisted of short indels (1 ~ 6 bp) in the close proximity of the protospacer motif, confirming that this kind of mutations frequently occur at an early stage of T0 rice plant development51. Single heterozygous mutations were also identified. This genotype, with the compresence of a mutated and a wild type allele, can be observed when mutations affect only one copy of the target gene in the embryogenic cell. However, it is also possible that mutations occurred after the division of the transformed embryogenic cell: in this case, the plant regenerated may be chimeric51. In such occurrence, heterozygote genotypes can also derive from chimeric tissues where some cells are homozygous for the mutation and others maintain the wild type genome. Many chimeras were produced in this study and identified in the regenerated T0 plants, confirming previous reports for CRISPR/Cas9-edited rice plants51. Based on the length of the indels obtained, most of which producing frameshifts, we could conclude that many homozygous mutations generated in T0 plants led to a loss of function of the genes of interest.
To determine if CRISPR/Cas9-mediated genome editing in OsHOL1 and OsHOL2 was heritable, we analyzed the T1 generations deriving from single T0 plants. First studies carried out in Arabidopsis thaliana reported that many mutations created by the CRISPR/Cas9 system occur in somatic cells and thus only a limited number of edited plants inherits the mutation in the subsequent generations52. In our study, inheritance of the homozygous mutations was observed in all T1 populations studied, suggesting that the CRISPR/Cas9 targeted modifications had occurred in single embryogenic cells. Moreover, all T1 plants deriving from T0 homozygous lines resulted homozygous for the same mutation identified in the progenitor plant, indicating that these mutations were stable and transmissible to the next generation, in accordance with the Mendelian laws of inheritance. Therefore, from these first analyses, it can be concluded that the mutations obtained in OsHOL1 and OsHOL2 T0 lines occurred in the germline cells and were inherited by T1 plants. Furthermore, in both groups of CRISPR/Cas9-edited plants, segregant T1 individuals, which lost the exogenous genetic cassette while maintaining the mutation in the gene of interest, were identified. Further analyses will be however necessary to ascertain the stability and heritability of these mutations in the following generations.
The most intriguing part of the study was the analysis of the effects of OsHOL1 silencing or overexpression on methyl halide production in planta. Methyl iodide emissions were reported in different plant species13,15,21 and previous studies focused on methyl iodide quantification in open fields13,15 or in vitro systems40. We used an experimental set-up that allowed quantification of emissions from single plants. Remarkably, the OsHOL1 homozygous knockout lines showed a significant reduction in CH3I emissions from their leaves, which, in some cases, were completely abolished. This phenotype was observed in all the different T0 lines analyzed and was also inherited from all the T1 plants bearing the same mutations. In the case of OsHOL2, the emission levels appeared variable among the different T0 homozygous mutant lines, but never eliminated, whereas in the T1 lines the CH3I production was identical to control. Since T0 plants, differently from their offspring, appeared somehow stressed as a consequence of the in vitro regeneration process, we assumed that comparison between control and mutant lines was more reliable when T1 plants were analyzed. Based on that, we concluded that the knockout of OsHOL2 did not have a significant impact on the volatilization of methyl iodide from rice leaves.
As a natural completion of the study, transgenic lines overexpressing OsHOL1 were produced and many of them showed higher CH3I emissions from leaves compared to control. Furthermore, in some lines methyl iodide emissions were also measured from roots, differently from wild type roots, where CH3I levels appeared negligible. These results confirmed the positive correlation between OsHOL1 gene expression and methyl halide production.
The higher expression levels and the lower emissions measured when mutagenized, as well as the higher emissions measured when overexpressed, make of OsHOL1 the main player in the iodine volatilization from rice. Our data did not support for the moment a possible active role of OsHOL2 in the methyl iodide production, at least in leaves. On the other hand, the absence of CH3I emission from wild type roots makes not plausible to hypothesize a possible activity of OsHOL2 in these organs as well. However, since the HMT activity of OsHOL2 was measured in vitro28, it is still possible that this enzyme carries out its action in other tissues of the plants or in specific developmental stages.
All the results here obtained were consistent with previous analyses performed in Arabidopsis, in which Athol1 mutants showed dramatically lower methyl halide emissions compared to wild type plants, in spite of the existence of other two HOL genes, which thus appeared to play only marginal roles in vivo22,27,53. Furthermore, exogenous iodine administrations resulted able to increase AtHOL1 transcription53. If this is also the case of rice plants, a possible increase of OsHOL1 transcription, for example as a consequence of higher iodine availability in the environment, would lead to a further increase in CH3I emissions from leaves, similarly to what observed in the OsHOL1 overexpressing lines. As a result, iodine-biofortification programs based on the use of exogenous iodine may lead to a further increase in methyl iodide production from rice crops, and therefore to an even more severe threat to the atmosphere.
The strong depressing effect of OsHOL1 knockout on CH3I emissions represents an important achievement in the perspective of a genetic improvement of this crop through the production of new varieties less hazardous towards the chemistry of the atmosphere. At the same time, such varieties would be ideal candidates for the establishment of iodine-biofortification programs. As a matter of fact, since silencing of AtHOL1 resulted in a significant increase of iodine content in Arabidopsis plants53, it is as much plausible that OsHOL1 silencing may lead to increase of iodine in rice plants. The development of new rice varieties with low OsHOL1 activity, provided this does not affect their agronomic performance, will therefore represent a necessary and unavoidable strategy to effectively enrich of iodine rice plants, possibly increasing its translocation to the seeds and thus making this micronutrient more available for the human diet, without having more negative impact on the environment.
Methods
Plant material
Seeds of Oryza sativa, ssp. Japonica, cv. Nipponbare, from the Genetic Stocks Oryza (GSOR) Collection (Dale Bumpers National Rice Research Center, AR, USA), were used. Seeds were sterilized with 70% ethanol for 1 min followed by 30 min in 6% sodium hypochlorite with shaking and washed six times for 15 min in sterile water. To examine the expression of HOL genes, rice seeds were pre-germinated in water in the dark for 3 days and then sown in soil, placing three/four seeds in each pot. Plants were grown in a growth chamber at 28 °C day/24 °C night, with a 12 h photoperiod, 150/200 μmol photons m−2 s−1 and 35% relative humidity. Roots, shoots and single leaves were collected after 2 weeks and stored at − 80 °C. For callus induction, 100 sterilized seeds, 10 per Petri dish, were inoculated on modified callus induction media supplemented with vitamins and 2,4-Dichlorophenoxyacetic acid (2,4-D) and incubated in darkness at 28 °C for 2 weeks54. Seeds of Arabidopsis thaliana, Columbia-0 ecotype, were used. For protoplast isolation, seeds were sown in a hydroponic system55 and plants were cultured in a growth chamber for 3 weeks at 23 °C/19 °C night with a 12 h photoperiod at 120 μmol photons m–2 s–1.
Expression analysis of OsHOL1 and OsHOL2 genes
Total RNA was extracted from rice leaves and roots as previously described56. Quality and integrity of RNA were checked by electrophoresis analysis on 1% agarose gel. RNA concentration was measured with a Multiskan Microplate Spectrophotometer (Thermo Fisher Scientific, USA). RNA was reverse transcribed using the “iScript TM cDNA synthesis kit” (Bio-Rad Laboratories, USA). RT-qPCR analysis was performed with technical duplicates in an ABI Prism 7300 Sequence Detection System (Thermo Fisher Scientific), using the “PowerUP SYBR Green Mastermix” (Thermo Fisher Scientific). Three biological replicates were analyzed. The reference gene OsGAPDH and the target genes OsHOL1 and OsHOL2 were amplified using the oligonucleotide primers listed in Table S2. Relative gene expression was calculated using the geometric averaging method57.
Sequence analyses
DNA and protein sequences were analyzed through online-available sequence analysis software. Multiple Sequence Alignments were carried out by using the CLUSTALW (Version 1.83) tool (www.genome.jp/tools-bin/clustalw). Protein Domains and Macromolecular Structures analyses were performed through the tools available on the NCBI website (www.ncbi.nlm.nih.gov/Structure/index.shtml). Sanger sequencing of PCR products were performed at Eurofins Genomics (Eurofins Scientific, Germany).
Targeted mutagenesis of OsHOL1 and OsHOL2 genes using the CRISPR-Cas9 system
The sequences of the genes Os03g62670 (OsHOL1) and Os06g06040 (OsHOL2) were downloaded from the Rice Genome Annotation Project website (http://rice.plantbiology.msu.edu/). Suitable DNA target sequences for guide RNA (gRNA) design were selected using the “CRISPR-P 2.0 design tool” (http://crispr.hzau.edu.cn/CRISPR2/). The gRNAs were synthesized, annealed, and inserted at the BsaI sites in the pOs-sgRNA entry vector29. Binary T-DNA pH-Ubi-Cas9-7 vectors29 for co-expression of Cas9 and the selected gRNAs were produced through Gateway™ recombination technology using the “LR Clonase II™” enzyme mix (Thermo Fisher Scientific). The sequence of each gRNA cassette after recombination in the expression vector was analyzed by PCR, using the primers listed in Table S2.
Generation of OsHOL1 and OsHOL2 knock-out plants and analysis of mutations
The expression vectors containing the CRISPR/Cas9 genetic cassettes containing the gRNAs targeted the genes of interest were introduced into Agrobacterium tumefaciens strain EHA105 by the freeze–thaw method. Rice transformation was performed starting from embryogenic calli according to the protocol described by58, as modified by54. Genomic DNA was extracted from T0 transgenic plants grown in soil using the “CTAB extraction method” and amplified for the hygromycin resistance gene using the primers listed in Table S2. Genomic fragments of 567 or 624 bp containing the target site were amplified from the genomic DNA of all the hygromycin PCR-positive plants using the specific OsHOL1 and OsHOL2 primers listed in Table S2. The PCR products were purified using the “Wizard® Genomic DNA Purification Kit” (Promega, USA) and then sequenced using the same PCR primers. The relative chromatograms were analyzed by Synthego “ICE CRISPR analysis tool” (https://ice.synthego.com/#/).
Vector construction and production of OsHOL1 overexpressing lines
The expressed cds of OsHOL1 was amplified from total RNA extracted from leaf tissue. First-strand cDNA was synthesized from RNA using the “Superscript IV® reverse transcriptase” (Thermo Fisher Scientific), according to the manufacturer’s instructions, and then amplified by PCR using the “Phusion® High-Fidelity DNA Polymerase” enzyme (Thermo Fisher Scientific) and the primers listed in Table S2. The purified full-length cds was cloned into the pENTR™/D-TOPO vectors (Thermo Fisher Scientific) and then recombined with the pIPKb003 destination vector59 using the “LR Clonase II™” enzyme mix (Thermo Fisher Scientific). The expression vector was mobilized into Agrobacterium strain EHA105 for transformation of rice calli and transgenic plants were produced as above described. Total RNA was extracted from leaf tissues of T0 plants using the “TRIzol™ Reagent” (Sigma-Aldrich, Merck, Germany) and the expression level of OsHOL1 was measured by RT-qPCR, as above described.
Measurement of methyl iodide emissions
Rice leaves (0.2 g per sample) were collected from control and transgenic plants, cut in small pieces, and incubated in a 22-ml sealed vial containing 0.5X Murashige & Skoog (MS) solution and 0.5 mM potassium iodide (KI) for 24 h at 23 °C under gentle shaking. Methyl iodide emissions were measured using a gas chromatography-tandem mass spectrometry (GC–MS/MS) CP-3800 gas chromatograph coupled to Saturn 2200 quadrupole ion trap mass spectrometer (Varian Analytical Instruments, Walnut Creek, CA, USA). 1 mL of the head space gas from the sealed vials was injected into a Mega PS264 capillary column (30 m × 0.25 mm i.d., 3.00 μm film thickness) (Mega, Milano, Italy). The carrier gas was helium, which was dried and air free, with a linear speed of 60 cm s−1. The oven temperature was maintained at 80 °C for 5 min. Injector and transfer line were set at 150 °C and the ion source temperature at 200 °C. Full scan mass spectra were obtained in EI + mode with an emission current of 10 µA and an axial modulation of 4 V. Data acquisitions was from 10 to 250 Da at a speed of 1.4 scan s−1. CH3I was identified by comparison with the retention time and spectrum ions (m/z) of a methyl iodide standard (Merck, Germany). The quantification was performed using the calibration curve with this compound. The minimum level of quantification (LOQ) and the minimum level of detection (LOD) were monitored daily with standard and with the signal/noise ratio, respectively. The same experimental setup was used to quantify methyl iodide emissions produced by rice roots (0.55 g per sample).
Subcellular localization studies in Arabidopsis thaliana protoplasts
The entry vector containing the cds of OsHOL1 was recombined with the p2FGW7 destination vector60 using the Gateway recombination protocol, as previously described, to produce a 35S:GFP-OsHOL1 expression plasmid. Arabidopsis mesophyll protoplasts were isolated and transfected with 5 μg of plasmid, as described in61. An mCherry-NLS–RFP-containing vector62 was used to visualize cell nuclei. For GFP and RFP imaging, Arabidopsis protoplasts were analyzed using a Leica THUNDER Imager Model Organism fluorescence microscope (Leica Microsystems, Germany).
Study of protein–protein interactions through Split-Luciferase Complementation assay
OsHOL1 cds, from the entry vector, was recombined into the Gateway-compatible split-luciferase system composed of pDuExAn6 and pDuExDn6 vectors63. The same vectors containing AtRAP2.1231 were gently provided by Dr. Alicja Kunkowska, Institute of Life Sciences, Scuola Superiore Sant'Anna, Pisa (Italy). Arabidopsis protoplasts, isolated as above described, were transfected with 5 μg of each plasmid. After an overnight incubation, protoplasts were pelleted and sea pansy (Renilla reniformis) luciferase activity was measured using the “Luciferase Reporter Assay System” (Promega). Luminescence intensity, measured with a Lumat LB 9507 Tube Luminometer (Berthold Technologies GmbH & Co., Germany), was normalized to the protein content of each sample determined through the Bradford protein assay (Bio-Rad).
Study of protein–protein interactions through Bimolecular Fluorescence Complementation (BiFC) assay
Vectors for the expression of OsHOL1 proteins fused to the N-terminal or C-terminal YFP fragments were generated by Gateway recombination of the OsHOL1 cds entry vector with the destination vectors pE-SPYNE-GW and pE-SPYCE-GW64,65. Arabidopsis protoplasts, isolated as above described, were transformed with 5 μg of each BiFC vector. An mCherry-NLS–RFP-containing vector62 was used to visualize the cell nuclei. YFP and RFP fluorescence was visualized using a Leica THUNDER Imager Model Organism fluorescence microscope (Leica Microsystems).
References
International Rice Research Institute, AfricaRice & International Center for Tropical Agriculture. Rice as a crop. Ricepedia. http://ricepedia.org/rice-as-a-crop (2021).
Food and Agriculture Organization of the United Nations. Food and Agriculture Data. FAOSTAT. http://www.fao.org/faostat/en/#home (2020).
Hoseinzade, H. et al. Rice (Oryza sativa L.) nutrient management using mycorrhizal fungi and endophytic Herbaspirillum seropedicae. J. Int. Agric. 15, 1385–1394 (2016).
Fraiture, M.-A. et al. Biotech rice: Current developments and future detection challenges in food and feed chain. Trends Food Sci. Technol. 52, 66–79 (2016).
Zimmermann, M. B. & Boelaert, K. Iodine deficiency and thyroid disorders. Lancet Diabetes Endocrinol. 3, 286–295 (2015).
World Health Organization. Statistical annex. In The World health report: 2004: changing history (WHO Library Cataloguing-in-Publication Data, 2004).
Fuge, R. & Johnson, C. C. Iodine and human health, the role of environmental geochemistry and diet, a review. Appl. Geochem. 63, 282–302 (2015).
Gonzali, S., Kiferle, C. & Perata, P. Iodine biofortification of crops: agronomic biofortification, metabolic engineering and iodine bioavailability. Curr. Opin. Biotechnol. 44, 16–26. https://doi.org/10.1016/j.copbio.2016.10.004 (2017).
Swanson, C. A. & Pearce, E. N. Iodine insufficiency: a global health problem?. Adv. Nutr. 4, 533–535 (2013).
Food and Agriculture Organization of the United Nations & International Life Sciences Institute. Preventing micronutrient malnutrition: A guide to food-based approaches. http://www.fao.org/3/X5244E/X5244e02.htm (1997).
Tsukada, H., Takeda, A., Tagami, K. & Uchida, S. Uptake and distribution of iodine in rice plants. J. Environ. Qual. 37, 2243–2247 (2008).
Nakamura, Y., Sumiya, M., Uchida, S. & Ohmomo, Y. Transfer of gaseous iodine to rice plants. J. Rad. Res. 27, 171–182 (1986).
Redeker, K. R. & Cicerone, R. J. Environmental controls over methyl halide emissions from rice paddies. Global Biogeochem. Cycles. https://doi.org/10.1029/2003GB002092 (2004).
Carpenter, L. J. Iodine in the marine boundary layer. Chem. Rev. 12, 4953–4962 (2003).
Redeker, K. R. et al. Emissions of methyl halides and methane from rice paddies. Science 290, 966–969 (2000).
Lee-Taylor, J. & Redeker, K. R. Reevaluation of global emissions from rice paddies of methyl iodide and other species. Geophys. Res. Lett. 32, 5 (2005).
Redeker, K. R., Manley, S. L., Walser, M. & Cicerone, R. J. Physiological and biochemical controls over methyl halide emissions from rice plants. Global Biogeochem. Cycles 18, 1007 (2004).
Wuosmaa, A. M. & Hager, L. P. Methyl chloride transferase: a carbocation route for biosynthesis of halometabolites. Science 249, 160–162 (1990).
Attieh, J. M., Hanson, A. D. & Saini, H. S. Purification and characterization of a novel methyltransferase responsible for biosynthesis of halomethanes and methanethiol in Brassica oleracea. J. Biol. Chem. 270, 9250–9257 (1995).
Attieh, J., Kleppinger-Sparace, K. F., Nunes, C., Sparace, S. A. & Saini, H. S. Evidence implicating a novel thiol methyltransferase in the detoxification of glucosinolate hydrolysis products in Brassica oleracea L. Plant Cell Environ. 23, 165–174 (2000).
Itoh, N. et al. Involvement of S-adenosylmethionine-dependent halide/thiol methyltransferase (HTMT) in methyl halide emissions from agricultural plants: isolation and characterization of an HTMT-coding gene from Raphanus sativus (daikon radish). BMC Plant Biol. 9, 116. https://doi.org/10.1186/1471-2229-9-116 (2009).
Rhew, R. C., Østergaard, L., Saltzman, E. S. & Yanofsky, M. F. Genetic control of methyl halide production in Arabidopsis. Curr. Biol. 13, 1809–1813 (2003).
Rhew, R. C., Miller, B. R. & Weiss, R. F. Natural methyl bromide and methyl chloride emissions from coastal salt marshes. Nature 403, 292–295 (2000).
Manley, S. L., Wang, N. Y., Walser, M. L. & Cicerone, R. J. Coastal salt marshes as global methyl halide sources from determinations of intrinsic production by marsh plants. Global Biogeochem. Cycles.. https://doi.org/10.1029/2005GB002578 (2006).
Saini, H. S., Attieh, J. M. & Hanson, A. D. Biosynthesis of halomethanes and methanethiol by higher plants via a novel methyltransferase reaction. Plant Cell Environ. 18, 1027–1033 (1995).
Manley, S. L. Phytogenesis of halomethanes: a product of selection or a metabolic accident?. Biogeochem. 60, 163–180 (2002).
Nagatoshi, Y. & Nakamura, T. Characterization of three halide methyltransferases in Arabidopsis thaliana. Plant Biotech. 24, 503–506 (2007).
Takekawa, Y. & Nakamura, T. Rice OsHOL1 and OsHOL2 proteins have S-adenosyl-L-methionine-dependent methyltransferase activities toward iodide ions. Plant Biotechnol. 29, 103–108 (2012).
Miao, J. et al. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23, 1233–1236 (2013).
Schubert, H. L., Blumenthal, R. M. & Cheng, X. Many paths to methyltrasfer: a chronicle of convergence. Trends Biochem. Sci. 28, 329–335 (2003).
Weits, D. A. et al. Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat. Commun. 5, 3425–3427 (2014).
Kiferle, C. et al. Evidences for a nutritional role of iodine in plants. Front. Plant Sci. 12, 616868. https://doi.org/10.3389/fpls.2021.616868 (2021).
Landini, M., Gonzali, S. & Perata, P. Iodine biofortification in tomato. J. Plant Nutr. Soil Sci. 174, 480–486 (2011).
Borst Pauwels, G. W. F. H. An investigation into the effects of iodide and iodate on plant growth. Plant Soil 16, 284–292 (1962).
Zhu, Y. G., Huang, Y. Z., Hu, Y. & Liu, Y. X. Iodine uptake by spinach (Spinacia oleracea L.) plants grown in solution culture: effects of iodine species and solution concentrations. Environ. Intern. 29, 33–37 (2003).
Duborská, E., Urík, M. & EŠeda, M. Iodine biofortification of vegetables could improve iodine supplementation status. Agronomy 10, 1574. https://doi.org/10.3390/agronomy10101574 (2020).
Medrano-Macías, J., Leija-Martínez, P., González-Morales, S., Juárez-Maldonado, A. & Benavides-Mendoza, A. Use of iodine to biofortify and promote growth and stress tolerance in crops. Front. Plant Sci. 7, 146. https://doi.org/10.3389/fpls.2016.01146 (2016).
Attieh, J. et al. Cloning and functional expression of two plant thiol methyltransferases: a new class of enzymes involved in the biosynthesis of sulfur volatiles. Plant Mol. Biol. 50, 511–521 (2002).
Sato, Y. et al. RiceXPro Version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 41, D1206–D1213 (2013).
Koerner, E. Evolution, function and manipulation of methyl halide production in plants. https://ueaeprints.uea.ac.uk/id/eprint/42416/1/2012KoernerEPhD.pdf (2012).
Ascoli, F. & Kahan, F. M. Iodination of nucleic acids in organic solvents with iodine monochloride. J. Biol. Chem. 241, 428–431 (1966).
Anderson, D. M. & Folk, W. R. Iodination of DNA. Studies of the Reaction and Iodination of Papovavirus DNA. Biochem. 5, 1022–1030 (1976).
Chouhan, B. P. S., Maimaiti, S., Gade, M. & Laurino, P. Rossmann-fold methyltransferases: taking a “β-Turn” around their cofactor, S-Adenosylmethionine. Biochem. 58, 166–170 (2019).
Ohsawa, N., Tsujita, M., Morikawa, S. & Itoh, N. Purification and characterization of a monohalomethane-producing enzyme S-adenosyl-L-methionine: halide ion methyltransferase from a marine microalga, Pavlova pinguis. Biosci. Biotechnol. Biochem. 65, 2397–2404 (2001).
Schmidberge, J. W., James, A. B., Edwards, R., Naismith, J. H. & O’Hagan, D. Halomethane biosynthesis: structure of a SAM-dependent halide methyltransferase from arabidopsis thaliana. Angew. Chem. Int. Ed. 49, 3646–3648 (2010).
Noel, J. P., Dixon, R. A., Pickersky, E., Zubieta, C. & Ferrer, J. L. Structural, functional and evolutionary basis for methylation of plant small molecules. Recent Adv. Phytochem. 37, 37–58 (2003).
Zubieta, C., He, X. Z., Dixon, R. A. & Noel, J. P. Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat. Struct. Mol. Biol. 8, 271–279 (2001).
Zhang, X. H., Tee, L. Y., Wang, X. G., Huang, Q. S. & Yang, S. H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids 4, e264. https://doi.org/10.1038/mtna.2015.37 (2015).
Haeussler, M. et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148. https://doi.org/10.1186/s13059-016-1012-2 (2016).
Lee, H. & Kim, J. S. Unexpected CRISPR on-target effects. Nat. Biotechnol. 36, 703–704. https://doi.org/10.1038/nbt.4207 (2018).
Zhang, H. et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12, 797–807 (2014).
Feng, Z. et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. P. Natl. Acad. Sci. USA 111, 4632–4637 (2014).
Landini, M. et al. Metabolic engineering of the iodine content in Arabidopsis. Sci. Rep. 2, 338. https://doi.org/10.1038/srep003382 (2012).
Sahoo, K. K. et al. An improved protocol for efficient transformation and regeneration of diverse indica rice cultivars. Plant Methods 7, 49. https://doi.org/10.1186/1746-4811-7-49 (2011).
Kamachi, K., Yamaya, T., Mae, T. & Ojima, K. Hydroponic culture growing system for rice and arabidopsis (shoot branching inhibition test for strigolactones). Plant Physiol. 96, 411–417 (1991).
Loreti, E., Valeri, M. C., Novi, G. & Perata, P. Gene regulation and survival under hypoxia requires starch availability and metabolism. Plant Physiol. 176, 1286–1298 (2018).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. https://doi.org/10.1186/gb-2002-3-7-research0034 (2002).
Hiei, Y. & Komari, T. Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat. Protoc. 3, 824–834 (2008).
Himmelbach, A. et al. Set of modular binary vectors for transformation of cereals. Plant Physiol. 145, 1192–1200 (2007).
Karimi, M., Inzé, D. & Depicker, A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 (2002).
Yoo, S. D., Cho, Y. H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007).
Shukla, V. et al. Endogenous hypoxia in lateral root primordia controls root architecture by antagonizing auxin signaling in Arabidopsis. Mol. Plant 12, 538–551 (2019).
Kato, N. & Jones, J. The split luciferase complementation assay. Method Mol. Biol. 655, 359–376 (2010).
Walter, M. et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438 (2004).
Weltmeier, F. et al. Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. EMBO J. 12, 3133–3143 (2006).
Acknowledgements
S.G., P.P. and L.M. acknowledge the support of the Center for Instrument Sharing of the University of Pisa (CISUP).
Author information
Authors and Affiliations
Contributions
M.C. designed and performed experiments, analyzed data and contributed to manuscript writing; L.M. designed and performed experiments and analyzed data; F.G. designed and performed experiments and analyzed data; F.F. designed and conceived experiments and analyzed data; P.P. designed and conceived experiments and analyzed data; S.G. designed and conceived experiments, analyzed data and contributed to manuscript writing. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Carlessi, M., Mariotti, L., Giaume, F. et al. Targeted knockout of the gene OsHOL1 removes methyl iodide emissions from rice plants. Sci Rep 11, 17010 (2021). https://doi.org/10.1038/s41598-021-95198-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-95198-x
This article is cited by
-
Fostering Nutritional Equity: Biofortification Strategies, Socioeconomic Implications, and Regulatory Policies for Developing Biofortified Staple Crop
Journal of Soil Science and Plant Nutrition (2025)
-
Agronomic Biofortification of Plants with Iodine and Selenium: A Potential Solution for Iodine and Selenium Deficiencies
Biological Trace Element Research (2024)
-
Improvement in fruit yield and tolerance to salinity of tomato plants fertigated with micronutrient amounts of iodine
Scientific Reports (2022)
-
Genome editing
Scientific Reports (2022)