Abstract
Allium mongolicum Regel is a wild and sandy vegetable with unique flavours. In this study, a complete chloroplast (cp) genome of A. mongolicum was obtained (Genbank accession number: OM630416), and contained 153,609 base pairs with the GC ratio as 36.8%. 130 genes were annotated including 84 protein-coding genes, 38 tRNA, and 8 rRNA genes. The large single-copy (LSC) region was 82,644 bp, and a small single-copy (SSC) region was 18,049 bp, which were separated by two inverted repeats (IRs, including IRa and IRb) of 26,458 bp. Comparative genome analyses of 55 Allium species suggested that genomic structure of genus Allium was conserved, and LSC and SSC regions were outstanding with high variability. Among them, more divergent loci were in the SSC region covering ycf1-rrn4.5 and ndhF-ccsA. Phylogenetic analysis on cp genomes of 55 Allium determined that all members were clustered into 13 clades, and A. mongolicum had close relationship with A. senescens. Corresponding analyses of four protein-coding genes (ycf1, ndhF, rpl32, and ccsA) in aforementioned divergent loci confirmed that ycf1 was finally chosen as the candidate gene for species identification and evolutionary classification of genus Allium. These data provide valuable genetic resources for future research on Allium.
Similar content being viewed by others
Introduction
Allium mongolicum Regel is famous for its unique flavour belonging to the genus Allium L. of Amaryllidaceae family. Combining the growth habitat and the same shape as green onions, it is called sandy onion. This species is naturally distributed in desert areas such as sandy land, gobi, arid hillside and dry river bed of China, Mongolia, Russia, Kazakhstan1. With strong drought-resistance, it is distributed from the northwest to northeastern of China (located between 36° 28′–46° 14′ N and 88° 39′–116° 05′ E) including Qinghai Province, Gansu Province, Shaanxi Province, Ningxia Hui Autonomous Region, Liaoning Province and Inner Mongolia Autonomous Region. In recent years, A. mongolicum has been widely planted in greenhouses after artificial domestication. Significantly, the A. mongolicum has greatly ornamental and ecological value2. Most studies have focused on nutrition and medicinal value3,4, and there has been no in-depth report of phylogenetic relationship with other Allium Species.
Allium is one of the largest and more important taxa in monocots, and covers multitudinous economical crops with unique nutritional and medicinal value, such as shallot, garlic, leek and onion5. However, the classification or phylogeny studies of Allium were controversial. Previous studies on classification of Allium had been mainly focused on the basis of subtle morphological difference, and some recent researches further classified the members by one or two loci such as internal transcribed spacer (ITS), rps16, and so on6. These results suggested Allium species can divide into 15 subgenera7. However, the phylogenetic studies of Allium have to be settled urgently.
Chloroplasts, originating from photosynthetic autotrophs of cyanobacteria, are unique independent organelles with a semi-autonomous genetic system, which can provide nutrients for plant growth and development. The complete chloroplast (cp) genomes are reliable elements for deducing phylogeny and evolutionary history deriving from highly conserved gene structure and gene content, and the cp genomes are short of recombination with low mutation rate8. Recently, many cp genomes have been extensively applied for phylogenetic reconstruction, and especially in Allium species9,10,11,12,13,14,15,16,17,18. However, these researches were carried out according to few members of Allium species. For example, sixteen cp genomes from Cepa were assessed the genes arrangement and variation, and a phylogeny of Cepa was constructed to explain the domestication of the common onion19. Xie et al. performed analyses of six cp genome structure, and uncovered the selective pressure in Daghestanica20. In addition, Xie et al. collected 39 Allium cp genomes and revealed that the divergence time of three traditional evolutionary lineages was presumably in the early Eocene to the middle Miocene21. However, excellent genes selected for reflecting classification and evolution of species in the genus Allium are scarce. Hence, the phylogenetic relationships from the point of total Allium species need to be further studied.
Here, high-throughput sequencing and bioinformatics technology were used to assemble a cp genome sequence of A. mongolicum (Genbank accession number: OM630416). 54 cp genomes from genus Allium were compared to explore the structure and evolution of the A. mongolicum chloroplast genome. Subsequently, phylogenomic analyses were performed based on these 55 chloroplast genomes to obtain a gene of choice for identifying and classifying of members from genus Allium. Our study will provide references for studying the genetic diversity and phylogenetic relationship in genus Allium.
Results
Basic characteristics of the Allium mongolicum chloroplast genome
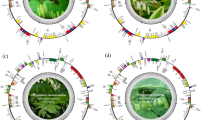
The extracted DNA was assessed, and the initial raw data with 8.98 Gbp was used to subsequent analysis. The finished cp genome was submitted to GenBank with the accession number OM630416. The length of the cp genome of the A. mongolicum was 153,609 bp, composed of a large single-copy (LSC) region of 82,644 bp and a small single-copy (SSC) region of 18,049 bp separated by two inverted repeats (IRs, including IRa and IRb) of 26,458 bp (Fig. 1). The overall GC (guanine and cytosine) content of the whole chloroplast genome was 36.8%. A total of 130 genes were annotated, including 84 protein-coding genes, 38 tRNA, and eight rRNA genes (Table 1). There are 21 chloroplast genes harbored introns, among which 19 genes contained single introns, and two genes (ycf3, clpP) contained two introns.
Characteristics and comparison of Allium cp genomes
To further explore the differences among the cp genome sequences from genus Allium, 54 reported cp genomes of Allium were compared to the cp genome sequence of A. mongolicum. The detailed information of all cp genomes was listed in the Table 2. Results showed that these genomes sequenced were similar with size and structure. The length of these genomes was generally 145,819–154,804 bp (A. paradoxum and A. monanthum, respectively) including LSC region (72,386–83,835 bp), SSC region (13,504–21,706 bp) and two IR regions (24,561–26,924 bp). A. monanthum possessed the longest cp genome in length, and this may be on account of the longest LSC region with 83,835 bp. Similarly, ungrouped A. paradoxum had the shortest cp genome in length with the shortest SSC region (13,504 bp). However, A. trifurcatum contained the longest SSC region and the shortest IR regions, A. tenuissimum accompanied by the shortest SSC region, A. cernuum was together with the longest IR region, and the three ones in length were not extreme.
In order to elucidate characteristics of sequence divergence among the 55 Allium cp genomes, nine representative species were selected to estimate Allium genome comparability using mVISTA program. The genome of A. altaicum was taken as the reference to conduct this program. Results revealed that cp genomes were relatively conserved in all Allium genomes (Fig. 2). No notably differences in gene order were detected when comparing the A. mongolicum cp genome to those of related Allium species. Generally, the highly divergent regions among representative Allium cp genomes mainly occurred in the LSC and SSC regions, and the coding regions exhibited less variations than the non-coding regions. In terms of species, high similarity and low divergence were detected in the cp genomes among A. altaicum, A.cepa and A. mongolicum, and the same things were tested among the group covering A. caeruleum and A. ampeloprasum, as well as group including A. trifurcatum, A. fetisowii, A. nerinifolium and A. nanodes.
To further exploit available polymorphic genes for identifying novel species, we further calculate the nucleotide diversity (Pi) values of overall sequences within the 600 bp window (Fig. 3). We found that Pi values ranged from 0 to 0.06769, and relatively high Pi values were determined in the SSC region, followed closely by those in the LSC region. The average Pi values of SSC and LSC regions were 0.0456 and 0.0219, respectively, but that of IR regions was 0.0052 (Table S1). These differences showed that two IR regions were more conserved than LSC and SSC regions. Here, most of cp genomes variations of Allium species existed in the SSC region, and two hypervariable regions (Pi > 0.06) were highlighted including variable loci ycf1-rrn4.5 (0.06052–0.06769) and ndhF-ccsA (0.06195–0.06516). The divergent regions called ycf1-rrn4.5 contained ycf1, tRNA-Asn, tRNA-Arg, rrn5 and rrn4.5, and ndhF-ccsA loci comprised ndhF, rpl32, tRNA-Leu and ccsA. Among these genes, four coding genes (ycf1, ndhF, rpl32, and ccsA) were understanding in terms of general conservation. Thus, these polymorphic regions might be novel candidate fragments for population genetic studies of Allium species.
Codon usage
Codon usage is the correlation to mRNA and protein, which is an essential characteristic for gene expression in plants genomes12. We estimated in detail codon usage frequency associated with all protein-coding sequences in A. mongolicum. Results showed that leucine (L), arginine (R) and serine (S) were the highest frequent codons, and methionine (M) and tryptophan (W) were least frequent (Fig. 4). Next, 51 grouped Allium species cp genomes were used to calculated in detail codon usage frequency, and results showed that these protein-coding genes were encoded by 19,832 (A. przewalskianum) to 26,802 (A. fistulosum) codons (Table S2). Like to other monocots plants, UGA, UAG, and UAA were known as the termination codons. For these Allium species, we found that the UUA encoded leucine (Leu) had the highest relative synonymous codon usage (RSCU) value in A. przewalskianum at approximately 2.23, and the in A. przewalskianum CUG encoded leucine had the lowest RSCU value at approximately 0.29. Same to A. mongolicum, methionine and tryptophan are used the least, but only leucine is used the most. In addition, 30 codons of Allium species were > 1, and 32 codons were < 1. Similar to other monocots chloroplast genomes, the nucleotide frequency of G or C from Allium species was lower than those of A or T at the whole codon, as well as the third codon position. The preference was considered to be a comprehensive result of gene expression, natural selection, and speciation mechanisms in species19.
Contraction and expansion of IR regions
It is well known that the cp genomes are usually well conserved with typical quadripartite structure and specific order. Through diversities of cp genomes mainly depend on highly divergent and lowly variable LSC and SSC regions, the size variations of cp genomes are strongly linked to the contraction and expansion of two IR regions, which can reflect species evolution. Therefore, IR boundaries of 9 representative members were detected to explain the differences of Allium cp genome size, including the boundaries of LSC/IRb regions (JLB), boundaries of SSC/IRb regions (JSB), boundaries of SSC/IRa regions (JSA), and boundaries of LSC/IRa regions (JLA). Results exhibited that the boundaries of different regions in various Allium species were different (Fig. 5). Generally, JLB were found to be located within the rpl22 gene up to a distance of 359–366 bp except for A. caerulrum (none) and A. fetisowii (rps19), and this partial expansion of the IRb region of A. fetisowii may result in the longest LSC regions (83,657 bp). JSB from three species (A. altaicum, A. cepa, A. mongolicum) was located in the ndhF gene, and those of four species was ycf1 gene. JSA in the seven cp genomes was located in the ycf1 gene. JLA were found between rps19 gene and psbA gene. Interesting, A. mongolicum cp genome has lost the ycf1 gene on the JSB. Overall, four connections of A. mongolicum had nearly identical relative positions with A. nanodes, A. ampeloprasum, and A. cepa.
Phylogenetic analysis
To know the evolutionary location of A. mongolicum and genetic clusters of genus Allium, phylogenetic tree of 55 Allium members was constructed. Phylogenetic analysis of the cp genome suggested that all members were clustered into 13 clades, and three outgroup ones derived from one clade (Fig. 6). Among them, A. mongolicum found in this study had close relationship with A. senescens in the genus Allium. To further obtain a gene of choice on assessing phylogeny of Allium species based on aforementioned Pi values, we constructed the phylogenetic trees according to the most variable four protein-coding genes (ycf1, ndhF, rpl32, and ccsA). Results showed that these phylogenetic trees were less similar with various major clades, and the cladogram for ycf1 was accordant to those of the whole-length cp genomes (Fig. 7).
Discussion
Allium mongolicum is distributed in desert areas with strong drought-resistance, and is an infrequent Allium species of Amaryllidaceae family. Unlike other Allium plants, A. mongolicum mainly located in the barren land with territorial restrictions, and related researches were lacking. As an essential member of genus Allium, it plays a critical role in explaining the evolutionary relationship of genus Allium. In this study, we sequenced a cp genome of A. mongolicum (153,609 bp), and exhibited the typical quadripartite structure including 130 unigenes. These genes were composed of 84 coding genes, 38 tRNAs and 8 rRNAs. Compared to cp genomes from 55 genus Allium, all genomes had a high conservation in the genome structure, length and organization. It is obvious that the LSC and SSC regions were highly divergent regions than two IR regions, and analysis of nucleotide diversity also confirmed this view. We further found that the more variable loci were in the SSC region comprising ycf1-rrn4.5 and ndhF-ccsA. The traditional taxonomy of Allium is mostly based on plant morphology such as tillering characteristics and pseudo-stem morphology. For a long time, the taxonomic status of Allium genus has changed frequently. In this study, phylogenetic analysis of full-length cp genome revealed that 55 Allium members were separated into 13 clades. In addition, members from Group VI including A. mongolicum shared closer relationship and more similar gene contents with members from Group VII. The 13 clades obtaining from the phylogenetic analyses of cp genomes was in accord with the modern taxonomic classification. In addition, the detailed evolutionary analyses of four protein-coding genes from two above-mentioned key loci were performed, and ycf1 was finally chosen as the candidate gene for species identification and evolutionary classification of genus Allium.
Contrastive analysis of the Allium chloroplast genomes showed that the size was wide range from 145 to 154 kb, and the difference was mainly due to JSB and JSA regions. Two ycf1 genes on the boundaries of IR regions were crucial, which concatenated the single copy region and the reverse repeat region. Specifically speaking, the length of A. monanthum was longest, and two ycf1 genes of those were longer with 1130 bp and 5309 bp, respectively. On the side, two ycf1 genes of A. paradoxum were severally 521 bp and 5249 bp, resulting in the shortest cp size. This phenomenon was coincident with the studies of comparative chloroplast genomics of the genus Taxodium22, and the changes in length of the two genes brought out the reduction and expansion of two IR regions, which directly impacted the size of the chloroplast genome.
Further characteristics of Allium sequence divergence and diversity were analyzed through multiple ways in this study, and all results referred to the single copy regions were more variable than two IR regions. Interestingly, ycf1-rrn4.5 and ndhF-ccsA in the SSC region were the most clustered diversity sites in the whole chloroplast genome of genus Allium. These two sites contained 4 protein coding-genes (ycf1, ndhF, rpl32, and ccsA) and 5 non-coding genes (tRNA-Leu, tRNA-Asn, tRNA-Arg, rrn5 and rrn4.5). Researcher had reported that rpoC2 can be taken as a gene of choice in Allium phylogeo-graphical studies, but the results were pointed out by the phylogenetic tree of 17 members of genus Allium. Hence, digging the best candidate gene for genus Allium evolution analysis was extremely urgent. The phylogenetic tree of 55 Allium species was constructed from chloroplast genome sequences, and further evolution analyses of four variable protein-coding genes in all species were performed. Results suggested that the evolutionary relationship of ycf1 was consistent in the tree of the whole-length cp genomes. Hence, ycf1 was the best choice for assessing classification of Allium species.
Conclusions
In this study, we successfully provided a complete chloroplast genome of A. mongolicum with 130 genes. Compared to genome structure with other members from Allium genus, the size, structure, gene contents of A. mongolicum chloroplast genome was conserved. Fifty-five Allium species in total were used to comparative analysis, and the phylogenetic analysis using full-length genome sequence were in accordance with the results using highly diverse ycf1 gene. Hence, ycf1 gene can be employed to evaluate phylogenetic relationships in Allium. The molecular data in this study provide a valuable resource for the study of evolution in Allium genus.
Materials and methods
Sampling, DNA extraction
Allium mongolicum seeds in this study were obtained from National Wild Plant Germplasm Resource Center, Kunming, Yunnan, China (Number: 178291), and was preserved at Horqin Plant Stress Biology Research Institute of Inner Mongolia Minzu University (Tongliao, China). The study protocol complied with relevant institutional, national, and international guidelines and legislation. The genomic DNA was extracted by the CTAB method23 assessing by the values of OD260/280.
Chloroplast genome sequencing, assembly and annotation
The qualified DNA was fragmented and used to construct the library, then sequenced on an Illumina Hiseq platform by two-terminal sequencing strategy. Firstly, the initial raw data was controlled and plotted by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), seqtk (1.3-r106), fastp (0.19.5) and R (3.6.1). Then, software SPAdes v3.61 under different K-mer parameters were used for contig assembly24. Finally, software CPGAVAS2, Dual Organellar GenoMe Annotator (DOGMA, http://dogma.ccbb.utexas.edu/) and blast + (2.9.0) were used for genes annotation. The finished cp genome was submitted to GenBank (OM630416).
Genome comparison
Fifty-four reported cp genomes of genus Allium were downloaded from National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov), and the detailed information was listed in the Table 2. The divergence of 9 representative genomes was operated by mVISTA (https://genome.lbl.gov/vista/index.shtml) in Shuffle-LAGAN mode25,26. MAFFT program was used to align all Allium species cp genomes27. Pi of all cp genomes was counted using Launch DnaSP628, and results were presented through a sliding window analysis with a window length of 600 bp and step size of 200 bp. Boundaries of IR regions, contraction and expansion of cp genomes were visualized by Program IRscope29.
Codon usage analysis
We selected Program codonW 1.4.4 to obtain the values of RSCU for evaluating codon preference30.
Phylogenetic analysis
A phylogenetic tree of 55 genus Allium members was constructed using MEGA-X through Maximum likelihood (ML) method31. Three related species were adopted as the outgroup containing Lilium brownie (MK493294), Ophiopogon bodinieri (NC_051508) and Polygonatum kingianum (MW373520). Phylogenetic trees of four targets genes (ndhF, rpl32, ccsA and ycf1) were constructed by the same way.
Data availability
The datasets generated during the current study are available in the NCBI repository (Genbank accession number: OM630416).
References
Chen, Y. Effects of Allium mongolicum Regel and its flavonoids on constipation. Biomolecules 10, 14. https://doi.org/10.3390/biom10010014 (2019).
Wang, W. et al. Phenolic compounds and bioactivity evaluation of aqueous and methanol extracts of Allium mongolicum Regel. Food Sci. Nutr. 7, 779–787. https://doi.org/10.1002/fsn3.926 (2019).
Liu, W. & Ao, C. Effect of dietary supplementation with Allium mongolicum Regel extracts on growth performance, carcass characteristics, and the fat color and flavor-related branched-chain fatty acids concentration in ram lambs. Anim. Biosci. 34, 1134–1145. https://doi.org/10.5713/ajas.20.0246 (2021).
Zhao, L. et al. Effects of Allium mongolicum Regel and its extracts on the quality of fermented mutton sausages. Food Sci. Nutr. 10, 169–178. https://doi.org/10.1002/fsn3.2657 (2021).
Herden, T., Hanelt, P. & Friesen, N. Phylogeny of Allium L. subgenus Anguinum (G. Don. ex W.D.J. Koch) N. Friesen (Amaryllidaceae). Mol. Phylogenet. Evol. 95, 79–93. https://doi.org/10.1016/j.ympev.2015.11.004 (2016).
Li, Q. Q. et al. Phylogeny and biogeography of Allium (Amaryllidaceae: Allieae) based on nuclear ribosomal internal transcribed spacer and chloroplast rps16 sequences, focusing on the inclusion of species endemic to China. Ann. Bot. 106, 709–733. https://doi.org/10.1093/aob/mcq177 (2016).
Friesen, N., Fritsch, R. M. & Blattner, F. R. Phylogeny and intrageneric classification of Allium (Alliaceae) based on nuclear ribosomal DNA ITS sequences. Aliso A J. Syst. Evol. Bot. 22, 372–395. https://doi.org/10.5642/aliso.20062201.31 (2006).
Raubeson, L. A. & Jansen, R. K. Chloroplast genomes of plants. In Diversity and Evolution of Plants–Genotypic and Phenotypic Variation in Higher Plants (ed. Henry, R. J.) 45–46 (CABI Publishing, 2005).
Kim, S. & Yoon, M. K. Comparison of mitochondrial and chloroplast genome segments from three onion (Allium cepa L.) cytoplasm types and identification of a trans-splicing intron of cox2. Curr. Genet. 56, 177–188. https://doi.org/10.1007/s00294-010-0290-6 (2010).
Lee, J., Chon, J. K., Lim, J. S., Kim, E. K. & Nah, G. Characterization of complete chloroplast genome of Allium victorialis and its application for barcode markers. Plant Breed Biotechnol. 5, 221–227. https://doi.org/10.9787/PBB.2017.5.3.221 (2017).
Filyushin, M. A., Beletsky, A. V., Mazur, A. M. & Kochieva, E. Z. Characterization of the complete plastid genome of lop-sided onion Allium obliquum L. (Amaryllidaceae). Mitochondrial DNA B Resour. 3, 393–394. https://doi.org/10.1080/23802359.2018.1456369 (2018).
Huo, Y. M. et al. Complete chloroplast genome sequences of four Allium species: Comparative and phylogenetic analyses. Sci. Rep. 9, 12250. https://doi.org/10.1038/s41598-019-48708-x (2019).
Sun, K. et al. The complete chloroplast genome of Allium ovalifolium var. leuconeurum, an endemic plant in Southwest China. Mitochondrial DNA B Resour. 4, 1681–1682. https://doi.org/10.1080/23802359.2019.1602009 (2019).
Wang, H. et al. Characterization of the complete chloroplast genome of Allium tuberosum. Mitochondrial DNA B Resour. 4, 2863–2864. https://doi.org/10.1080/23802359.2019.1661302 (2019).
Yang, X., Xie, D. F., Zhou, S. D. & He, X. J. Characterization of the complete chloroplast genome of Allium kingdonii. Mitochondrial DNA B Resour. 4, 868–869. https://doi.org/10.1080/23802359.2019.1573118 (2019).
Liu, L., Yusupov, Z., Suyunkulov, H. & Jiang, Z. The complete chloroplast genome of Allium ferganicum. Mitochondrial DNA B Resour. 5, 2772–2773. https://doi.org/10.1080/23802359.2020.1788454 (2020).
Ren, F. et al. Complete chloroplast genome of the rare medicinal vegetable Allium hookeri. Mitochondrial DNA B Resour. 7, 6–7. https://doi.org/10.1080/23802359.2021.2003262 (2021).
Wang, H. et al. Characterization of the complete chloroplast genome of Allium mongolicum. Mitochondrial DNA B Resour. 5, 2030–2031. https://doi.org/10.1080/23802359.2020.1756951 (2020).
Yusupov, Z. et al. Phylogenomics of Allium section Cepa (Amaryllidaceae) provides new insights on domestication of onion. Plant Divers 43, 102–110. https://doi.org/10.1016/j.pld.2020.07.008 (2021).
Xie, D. F. et al. Phylogeny of Chinese Allium species in section Daghestanica and adaptive evolution of Allium (Amaryllidaceae, Allioideae) species revealed by the chloroplast complete genome. Front. Plant Sci. 10, 460. https://doi.org/10.3389/fpls.2019.00460 (2019).
Xie, D. F. et al. Insights into phylogeny, age and evolution of Allium (Amaryllidaceae) based on the whole plastome sequences. Ann. Bot. 125, 1039–1055. https://doi.org/10.1093/aob/mcaa024 (2020).
Duan, H. et al. Comparative chloroplast genomics of the genus Taxodium. BMC Genomics 21, 114. https://doi.org/10.1186/s12864-020-6532-1 (2020).
Doyle, J. J. & Doyle, J. L. A Rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem. Bull. 19, 11–15 (1986).
Bankevich, A. et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–77. https://doi.org/10.1089/cmb.2012.0021 (2012).
Brudno, M. et al. Glocal alignment: Finding rearrangements during alignment. Bioinformatics 19(S1), i54–i62. https://doi.org/10.1093/bioinformatics/btg1005 (2003).
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M. & Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279. https://doi.org/10.1093/nar/gkh458 (2004).
Zhang, D. et al. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20, 348–355. https://doi.org/10.1111/1755-0998.13096 (2018).
Julio, R. et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. https://doi.org/10.1093/molbev/msx248 (2017).
Amiryousefi, A., Hyvöne, J. & Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34, 3030–3031. https://doi.org/10.1093/bioinformatics/bty220 (2018).
Peden, J. F. Analysis of codon usage. Dissertation, University of Nottingham (2000)
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. https://doi.org/10.1093/molbev/msy096 (2018).
Acknowledgements
We express our gratitude to the anonymous reviewers for helpful comments to improve the manuscript. This study was supported by Natural Sciences Foundation of Inner Mongolia Autonomous Region of China (No. 2020LH03009 and No. 2018LH03018), Special Fund for Scientific Research Start-up Support for Introduction of Talents in Public Institutions of Inner Mongolia Autonomous Region in 2019, and the Doctoral Research Start-up Fund of Inner Mongolia Minzu University (No. BS540).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by T.Z., B.L., C.Z. and H.H. The first draft of the manuscript was written by Y.J. and J.Z., and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jin, Y., Zhang, T., Liu, B. et al. Comparative and phylogenetic analysis of the complete chloroplast genome sequences of Allium mongolicum. Sci Rep 12, 21676 (2022). https://doi.org/10.1038/s41598-022-26354-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-022-26354-0