Abstract
Non-invasive prediction for KIT/PDGFRA status in GIST is a challenging problem. This study aims to evaluate whether CT based sarcopenia could differentiate KIT/PDGFRA wild-type gastrointestinal stromal tumor (wt-GIST) from the mutant-type GIST (mu-GIST), and to evaluate genetic features of GIST. A total of 174 patients with GIST (wt-GIST = 52) were retrospectively identified between January 2011 to October 2019. A sarcopenia nomogram was constructed by multivariate logistic regression. The performance of the nomogram was evaluated by discrimination, calibration curve, and decision curve. Genomic data was obtained from our own specimens and also from the open databases cBioPortal. Data was analyzed by R version 3.6.1 and clusterProfiler (http://cbioportal.org/msk-impact). There were significantly higher incidence (75.0% vs. 48.4%) and more severe sarcopenia in patients with wt-GIST than in patients with mu-GIST. Multivariate logistic regression analysis showed that sarcopenia score (fitted based on age, gender and skeletal muscle index), and muscle fat index were independent predictors for higher risk of wt-GIST (P < 0.05 for both the training and validation cohorts). Our sarcopenia nomogram achieved a promising efficiency with an AUC of 0.879 for the training cohort, and 0.9099 for the validation cohort with a satisfying consistency in the calibration curve. Favorable clinical usefulness was observed using decision curve analysis. The additional gene sequencing analysis based on both our data and the external data demonstrated aberrant signal pathways being closely associated with sarcopenia in the wt-GIST. Our study supported the use of CT-based assessment of sarcopenia in differentiating the wt-GIST from the mu-GIST preoperatively.
Similar content being viewed by others
Introduction
Gastrointestinal stromal tumor (GIST) is a common interstitial tumor occurring in gastrointestinal tract1,2. Most GISTs harbor KIT/PDGFRA mutant gene named KIT/PDGFRA mutant GIST (mu-GIST), and the remaining cases (about 10–15%) are KIT/PDGFRA wild-type GISTs (wt-GIST) which do not have mutant KIT/PDGFRA genes2. It is important to differentiate these two subtypes of GIST for selecting effective treatment3. For example, imatinib is an active multikinase inhibitor that mainly targets C-kit tyrosine-kinase receptors. Although imatinib has become the standard first-line treatment worldwide for patients with the mu-GIST, it almost ineffective for patients with the wt-GIST. Clinically, neo-adjuvant therapy with imatinib was strongly recommended for mu-GIST patients prior to surgery with the purpose of sparing important organs which may be locally invaded by tumor4,5. Currently, the only method for preoperative molecule identification of the GIST subtypes is through aspiration biopsy, which is invasive and may have potential risk of tumor rupture, perforation of gastrointestinal tract or needle track implantation6. There is an unmet need for developing a robust non-invasive tool for differentiating wt-GIST from mu-GIST before treatment.
Abdominal computed tomography (CT) is routinely used for clinic management of GIST. Sarcopenia (loss of lean muscle mass) can be evaluated on the routine abdominal CT images obtained for clinical care7,8,9. A recent study has identified the presence of sarcopenia in GIST patients and highlighted the predictive value of sarcopenia on treatment toxicity10. However, there is a paucity of literature regarding clinical value of pre-treatment sarcopenia for patients with GIST regarding their response to treatment and prognosis. Converging evidences have indicated that sarcopenia may possibly underline the pathogenesis of various cancers, because signal pathways involved in sarcopenia are usually anomalous in cancer8,11. We hypothesized that different molecular pathways for wt-GIST and mu-GIST may lead to significant differences in muscle mass of patients with different subtypes of GIST.
Here, we retrospectively analyzed the lean muscle mass on abdominal CT images of 174 patients with GIST and evaluated whether CT assessment of sarcopenia could differentiate the wt-GIST from the mu-GIST. In addition, we also performed genetic analysis on both our own data and an external dataset.
Materials and methods
Patients
This single-center retrospective study including all experimental protocols, was approved by the Institutional Review Board of Xiangya hospital (IRB#: 202009685). The written informed consents were waived by the Institutional Review Board of Xiangya hospital due to the retrospective nature of this study. All methods were carried out in accordance with relevant guidelines and regulations.
Patients with pathologically proved GIST and with genotyping information were retrospectively identified and their data was collected after searching our institutional medical database from January 2011 to October 2019. This cohort was divided into two groups: the wt-GIST group and the mu-GIST group. Details of the patient recruiting process and exclusion criteria were presented in Fig. 1. Next, the patients were randomly selected into a training cohort and a validation cohort at a ratio of 7:3 for future analysis.
Re-analysis of pathology specimen
For each patient, all pathological slides were re-analyzed by two pathologists specialized in GIST (HYZ and GHG with 17 and 6 years of experience in solid tumor, respectively). Consensus was reached by discussion if differences in opinions occurred. KIT and PDGFRA gene were amplified and sequenced by Sanger sequencing set kit according to the manufacturer’s instructions using primers designed for the c-KIT exon 9, 11, 13, 14, 17, 18 and PDGFRA exon 12, 18.
Body composition evaluation
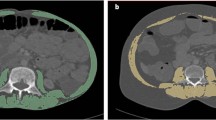
Initial pre-treatment abdominal CT images were retrieved from Picture Archiving and Communication Systems (PACS) for analysis. Two axial unenhanced images were selected at the level of the third lumbar vertebra body (for measurement of skeletal muscle) and at the navel level (for measurement of visceral fat) respectively. Total body fat area, visceral fat area (VFA), fat area in muscle (MFA), subcutaneous fat area (SFA), and skeletal muscle area (SMA) were measured manually on the selected axial image by using a medical image analysis software (SliceOmatic software; Tomovision, Quebec, Canada). Bone structures, and the central spinal canal were excluded manually in the selected axial images for accurate measurement of specific areas on a workstation (Advantage Windows workstation 4.6, GE Healthcare, Milwaukee, Wisconsin, USA). According to a prior study10, corresponding fat area (between − 50 and − 150 Hounsfield units, Hu) and the area of the abdominal wall and back muscles (psoas, paraspinal, transversus abdominis, rectus abdominis, internal oblique muscles, and external oblique muscles) (between − 29 and 150 Hu) were measured by calculating the area of pixels with corresponding attenuation in demarcated areas, respectively. L3 skeletal muscle index (SMI) was calculated as the area of total L3 skeletal muscle (cm2) divided by height square (m2). Visceral or subcutaneous fat index (VFI, MFI or SFI) was calculated as the area of visceral, intramuscular or subcutaneous fat (cm2) at the navel level divided by the square (m2) of height. The details for body composition measurement was presented in Fig. 2.
Treatment, follow-up and outcome parameters
All patients underwent surgery, and post-surgical follow-up was obtained for each patient. The primary endpoints of the study were disease free survival (DFS). DFS was defined as the time from surgery to recurrence of GIST or metastasis for expired patients or the last follow-up for surviving patients.
Genomic data analyses
Genomic DNA data for 28 patients in our cohort including 15 wt-GIST samples (sanger sequencing negative for c-KIT exon 9, 11, 13, 14, 17, 18 and PDGFRA exon 12, 18) and 13 KIT mu-GIST samples (sanger sequencing positive for c-KIT exon 11) were obtained from our archived formalin-fixed paraffin-embedded (FFPE) tissues. Ten peripheral blood lymphocyte (PBL) samples from the 15 wt-GIST patients were collected as germline controls. The comprehensive genetic alterations of these 28 GIST samples were analyzed using targeted deep sequencing of 1021 cancer related genes region. The Next Generation Sequencing was executed according to the manufacturer’s instructions (Illumina, San Diego, CA, USA). The somatic mutations of the wt-GISTs were investigated for genomics landscape description with the ComplexHeatmap R package.
In addition, mutational landscapes and corresponding clinical information were downloaded from cBioPortal as external data (http://cbioportal.org/msk-impact). Data was analyzed by R version 3.6.1 and KEGG enrichment analysis was performed by clusterProfiler. The pathway with p-value less than 0.05 were kept for the following comparison analysis.
Statistical analysis
Statistical analysis was implemented with SPSS 22.0, SAS9.4, and R software (http://www.Rproject.org). Multivariate binary logistic regression, nomograms, calibration plots, decision curve and correlation matrix plots were done with the “Caret”, “PROC”, “rmda”, “corrplot”, “forestplot”, “rms” and “corrplot” package. Decision curve was performed with the “rmda” package. And “survival” package was used for the survival analysis. KEGG analysis12 was performed by R version 3.6.1 through application of clusterProfiler. The statistical significance levels were all two-sided with statistical significance set at 0.05.
Ethical approval
Institutional Review Board approval was obtained (IRB#: 202009685).
Contribution to the field statement
We assessed body composition features and sarcopenia on the pretreatment abdominal CT images in patients with Gastrointestinal stromal tumor (GIST). Our prediction model combining sarcopenia parameters and clinical variables showed robust performance, indicating the potential for using such a non-invasive method for predicting KIT/PDGFRA status.
Results
Patients’ clinicopathological information
In total, 174 consecutive patients with GIST were included into our study. The clinicopathological characteristics of the patients in the wt-GIST group and mu-GIST group were summarized in Table 1. The medium Neutrophil lymphocyte ratio (NLR) value of the patients with wt-GIST was slightly higher than that of the patients with mu-GIST. There were no significant differences in the clinical characteristics between the wt-GIST group and the mu-GIST group, either in the training cohort or in the validation cohort.
Body composition status for GIST subtypes
The VFA, VFI, MFI and SFI of patients in the wt-GIST group were all significantly larger than those of patients in the mu-GIST Group. The SMI and VFI/SFI Ratio of the wt-GIST Group were significantly lower than those in the mu-GIST Group (Table 1).
Taking into considerations of the influence of age and gender on skeletal muscle index (SMI), we developed a new index termed sarcopenia score based on the three features through multivariate logistic regression. Not surprisingly, the sarcopenia score for the wt-GIST group (− 0.597, [− 8.940 to 1.620]) was significantly higher than that of the mu-GIST group (− 1.060, [− 2.470 to 0.900]) (P < 0.001).
There was no significant difference on obesity (calculated based on BMI) between the wt-GIST group and the mu-GIST Group (23.1% vs 30.3%) (P = 0.33). The incidence of sarcopenia and visceral obesity in the wt-GIST group was much higher than that of the mu-GIST group (75.0% vs 48.4%, 76.9% vs 36.9%, respectively) (P < 0.001). Interestingly, the inconsistent diagnosis of adiposity, including obesity diagnosed based on BMI and visceral obesity based on VFA, occurred in 32.18% (56/174) of patients with GIST. BMI was normal in forty-six GIST patients with visceral obesity, while ten obese patients did not actually have visceral obesity.
Identification of risk factors associated with wt-GIST
Single factor logistic regression analysis showed that the patients with wt-GIST were associated with NLR, VFA, MFA, SFA, VFA/SFA ratio, SMI, VFI, MFI, SFI, sarcopenia score, sarcopenia and visceral obesity. Further multivariate logistic regression analysis showed that, only sarcopenia score (P = 0.000298, OR 10.216 [95% CI 3.207–41.151]) and MFI (P = 0.005, OR 1.141 [95% CI 1.079–1.380]) were identified as independent risk factors for wt-GIST.
Development and validation of individualized prediction model
A parsimonious model separating the patients with wt-GIST from the patients with mu-GIST was developed by incorporating the two independent predictors (sarcopenia score and MFI), and a nomogram was obtained (Fig. 3A). The ROC test provided an AUC of 0.879 (95% CI 0.816–0.943) with a sensitivity of 0.8780 (95% CI 0.7561–0.9756) and a specificity of 0.9167 (95% CI 0.8611–0.9722) in the training cohort (Fig. 3B), and a AUC of 0.9099 (95% CI 0.8324–0.9873) with a sensitivity of 0. 9412 (95% CI 0.7647–1.0000) and a specificity of 0.8871 (95% CI 0.7742–0.9839) in the validation cohort (Fig. 3C).
(A) Sarcopenic nomogram. The nomogram was developed in the primary cohort, with the sarcopenia score and MFI incorporated. The classification efficiency of the nomogram was shown by ROC curves for training (B) and validation (C) cohorts respectively. Calibration curve of the nomogram in the primary cohort (E) and the validation cohort (F).
The calibration curve of the nomogram for the probability of wt-GIST showed good agreement between prediction and observation in both the training and validation cohorts (Fig. 3E,F). The Hosmer–Lemeshow test yielded a non-significant statistic in both the training (P = 0.819) and the validation cohort (P = 0.751), which suggested no departure from perfect fit.
The decision curve analysis for the nomogram was presented in Fig. 3D. The decision curve showed that if the threshold probability of a patient or doctor was 10%, using the nomogram to predict wt-GIST added more benefit than either the diagnose-all-patients scheme or the diagnose-none scheme.
Survival analysis
Seven (13.5%, 7/52) patients were lost at the median follow-up of 31.0 months (range 1.0–73.0 months). During the follow-up period, only 3 (6.67%, 3/45) and 4 (8.89%, 4/45) deaths were observed, respectively. The relevant prognostic analysis was not completed because the majority of patients did not have an endpoint event. However, it should be noted that all endpoints (recurrence or death) occurred in patients with sarcopenia and visceral obesity. The incidence of recurrence was higher in patients who were sarcopenic compared with those who were not (11.4% vs. 0%), although the difference was not statistically significant due to the relatively short follow-up period (P = 0.561). Furthermore, two patients with visceral obesity could not be diagnosed as obesity based on their BMI values. The impact of sarcopenia and visceral obesity rather than obesity on the survival of wt patients with -GIST (DFS, OS) were observed in the log-rank test, although there was no statistical significance (Supplementary Fig. 1).
Abnormal gene pathways in GIST subtypes
As shown in Fig. 4A, in our center, the frequently mutated genes in the wt-GIST (occurred in at least 2 cases) were neurofibromatosis type 1(NF1) (5/10), B1 v-raf murine sarcoma viral oncogene homolog B1 (BRAF) (3/10), Epidermal growth factor receptor (EGFR) (3/10), AT-Rich Interaction Domain 1B(ARID1B) (2/10), FAT Atypical Cadherin 1(FAT1) (2/10), interleukin 6 signal transducer(IL6ST) (2/10) and tuberous sclerosis2(TSC2) (2/10).
Hot map and KEGG analysis in wt-GISTs in our center as internal data and in Memorial Sloan Kettering Cancer Center (MSKCC) database as external data. (A,C) Hot map of wt-GISTs (A: internal data, C: external data). Patients were arranged along the x-axis. Mutation per Mb region was shown in the upper panel. Genes with somatic mutations (The gene mutation > = 2 patients were included) were shown in the middle panel. Mutation frequencies of each gene were shown on the left. The type of alternations was shown in the right. (B,D) Enriched KEGG pathway comparison between wt-GISTs and mu-GIST (B: internal data, D: external data). Y-axis representing the enriched KEGG pathway; X-axis representing the value of − log10 (p-value).
Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis indicated that the pathway enriched in osteoclast differentiation and longevity regulating pathway was noted in the wt-GISTs. When compared to the mu-GISTs, higher forkhead box O (FoxO) signaling pathway and its upstream of phosphatidylinositol-3-kinase-Akt (PI3K-Akt) pathway were found in the wt-GISTs (Fig. 4B). Further analysis of the external dataset from Memorial Sloan Kettering Cancer Center (MSKCC) database including 137 GIST patients with 29 of them being wt-GIST confirmed that most frequently altered genes were NF1, EGFR, tumor protein p53(TP53), Succinate Dehydrogenase Complex Flavoprotein Subunit A(SDHA), Retinoblastomal 1(RB1), BRAF, Succinate Dehydrogenase Complex Flavoprotein Subunit B(SDHB), and TSC2, as shown in Fig. 4C. The analysis of KEGG pathways suggested that actin cytoskeleton, longevity regulating pathway and FoxO pathway indeed more frequently existed in the wt-GISTs. In addition, the Alzheimer disease (AD) pathway, Parkinson disease (PD) pathway, and insulin signaling pathway were present only in wt-GISTs in comparison to mu-GISTs (Fig. 4D).
Discussion
In this study, we observed the incidence and severity of skeletal muscle atrophy being significantly higher in patients with wt-GIST when compared to the patients with mu-GIST. The sarcopenic nomogram constructed in our study showed the potential to be useful as a tool in predicting patients with wt-GIST before surgery. Our gene sequencing analysis of both our own data and the external data demonstrated that those aberrant gene signal pathways being closely associated with sarcopenia in the wt-GISTs more often than that in mu-GISTs. Our results indicated the potential application of CT-based sarcopenic assessment and gene analysis in pretreatment evaluation of GIST subtypes.
The prevalence of sarcopenia varied among different type of cancers13, and the incidence and clinical impact of sarcopenia on GIST are largely unknown. To date, the only published study on sarcopenia in GIST patients reported an incidence of 38.7% in a French cohort of GISTs (n = 31)10. However, our study showed a higher incidence of 56.32%. The reason for the discrepancy of incidence of sarcopenia between our study and their study10 was not clear. We speculate the difference in incidence might be due to different racial/ethnic background affecting body composition and muscle loss in patients with GISTs. Potential case selection bias should also be considered for both the French study and our study.
The sarcopenic nomogram constructed in our study, with the sarcopenic score (fitted by using age, gender and SMI) and with the MFI incorporated, showed potential value in differentiating wt-GIST from mu-GIST. The atrophy score represented the severity of a patient’s skeletal muscle atrophy after considering the effects of age and gender. The higher the score, the more severe the muscle atrophy. MFI represented the severity of septocutaneous adipose tissue. The higher the MFI, the more serious the adipose infiltration of skeletal muscle and the worse the muscle quality. Our higher incidence of sarcopenia and more severe muscle atrophy accompanied by adipose degeneration in patients with wt-GIST suggested that patients with wild-type tumors may have a significant decrease in muscle quantity and quality when compared to patient with mu-GIST7,13. Although sarcopenia could also occur in mu-GIST patients, our study showed that sarcopenia was an independent predictor for wt-GIST. In other word, sarcopenia could be a potential useful predictor for identifying wt-GIST before treatment.
The underlying genetic mechanism responsible for the different incidence and severity of sarcopenia between wt-GIST and mu-GIST is largely unknown. In search of the underlying molecule mechanism of sarcopenia in the wt-GIST, we analyzed the available gene data in our cohort, and extended this analysis to the external data in the Cancer Genome Atlas (TCGA). Regarding the most frequently observed genes in the wt-GIST, we identified five common genes abnormality including NF1, EGFR, BRAF, FAT1, IL-6ST and TSC2 in the wt-GIST. Patients with NF1gene deficiency have shown to have enhanced resting energy expenditure and respiratory quotient, indicating premature sarcopenia14. EGFR may be correlated with slow-twitch muscle phenotype and EGFR inhibitor may impede the loss of muscle slow-twitch fiber loss15. Fat1 is a protocadherin gene. It affects muscle patterning, regionalized muscle wasting and adult muscle fiber functions. The shape of muscle defects has been observed in adult fat1-knockout animal16. IL-6ST, also name gp130, is one of the common cytokine receptor subunits. It is shared by a series of IL-6 family which are well-known inflammatory cytokines, and function as prompting muscle wasting, stimulating protein catabolism and suppressing muscle synthesis17. Other studies have shown that TSC 2 phosphorylation activated by mTOR could be the response to the resistance training for the prevention and treatment of sarcopenia18. Collectively, these gene abnormalities involved in the total energy expenditure are correlated to food consumption and body composition, mitochondrial dysfunction, essential energy-producing processes, skeletal muscle function and cytokine-induced muscle catabolism, thus accelerating loss of muscle mass and function leading to sarcopenia19.
The causes of sarcopenia are likely multifactorial and molecular alterations may be present in complex signaling pathways20. Our results showed patients with wt-GIST had a higher incidence of mutant pathways associated with sarcopenia than patients with mu-GIST. For example, Longevity regulating pathways consisted of a series of genes and pathways for aging process have a high rate of sarcopenia21,22,23. FoxO proteins such as FOXO1 and FOXO3) are transcription factors which regulate the expressions of various atrophy-related genes atrogin-1 and MuRF-1 muscle, contributing to the decrease in muscle size and number24,25. Intriguingly, our study showed that the FoxO signal pathway was present in patients with mu-GIST in our own genetic data but this pathway was only present in the wt-GISTs in the external data. The exact reason for this discrepancy is not clear. We again speculate that the racial and ethnic differences between the cohorts may play a role.
Our study suggested that wt-GIST patients with sarcopenia had a shorter overall survival, as well as recurrence-free survival than wt-GIST patients without sarcopenia. Patients with recurrence all had sarcopenia. Our results were consistent with the literature indicating sarcopenia being a prognostic factor in cancer patients26. In advanced GIST, sarcopenia was correlated with shorter PFS and OS27,28. Further study with long-term follow-up is our on-going research.
There were limitations to this study. First, the retrospective nature of the study precluded the measurement of muscle strength which ought to be more important than muscle mass in terms of adverse health outcomes. Second, the absence of national cut-off points for muscle mass may hinder standardization of data and generalizability of results from sarcopenia research. Third, it was a single center study and the results may not be generalizable to other centers. Future study with a large-scale prospective multicenter approach is needed to validate our results and to expand this line of research.
Conclusion
Our study identified higher incidence of sarcopenia, more severe muscle atrophy accompanied with more fat degeneration and more gene abnormalities in wt-GIST, than mu-GIST. Our study indicated that CT-based assessment of sarcopenia may be useful for differentiating wt-GIST from mu-GIST non-invasively before treatment. The molecule abnormalities underlying sarcopenia in wt-GIST may be potentially useful as alternative targets for detecting, mitigating and reversing sarcopenia.
Data availability
The original data will be made available to qualified the corresponding author of this paper upon request.
Abbreviations
- GIST:
-
Gastrointestinal stromal tumor
- mu-GIST:
-
Mutant GIST
- wt-GIST:
-
Wild-type GISTs
- CT:
-
Computed tomography
- PACS:
-
Picture archiving and communication systems
- VFA:
-
Visceral fat area
- MFA:
-
Fat area in muscle
- SFA:
-
Subcutaneous fat
- SMA:
-
Skeletal muscle area
- Hu:
-
Hounsfield units
- SMI:
-
Skeletal muscle index
- DFS:
-
Disease free survival
- PBL:
-
Peripheral blood lymphocyte
- NLR:
-
Neutrophil lymphocyte ratio
- NF1:
-
Neurofibromatosis type 1
- BRAF:
-
B1 v-raf murine sarcoma viral oncogene homolog B1
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- EGFR:
-
Epidermal growth factor receptor
- ARID1B:
-
T-Rich Interaction Domain 1B
- FAT1:
-
FAT Atypical Cadherin 1
- IL6ST:
-
Interleukin 6 signal transducer
- TSC2:
-
Tuberous sclerosis 2
- FoxO:
-
Forkhead box O
- PI3K-Akt:
-
Phosphatidylinositol-3-kinase-Akt
- MSKCC:
-
Memorial Sloan Kettering Cancer Center
- Tp53:
-
Tumor protein p53
- SDHA:
-
Succinate Dehydrogenase Complex Flavoprotein Subunit A
- RB1:
-
Retinoblastomal 1
- SDHB:
-
Succinate Dehydrogenase Complex Flavoprotein Subunit B
- AD:
-
Alzheimer disease
- PD:
-
Parkinson disease
References
Søreide, K. et al. Global epidemiology of gastrointestinal stromal tumours (GIST): A systematic review of population-based cohort studies. Cancer Epidemiol. 40, 39–46 (2016).
Von, M. M. & Joensuu, H. Gastrointestinal stromal tumors. J. Clin. Oncol. 36(2), 136–143 (2018).
Bhatt, N. R., Collins, D., Crotty, P. & Ridgway, P. F. Prognosis and management of adult wild type gastrointestinal stromal tumours (GISTs): A pooled analysis and review of literature. Surg. Oncol. 25(3), 152–157 (2016).
Reichardt, P. et al. Adjuvant therapy in primary GIST: State-of-the-art. Ann. Oncol. 23(11), 2776–2781 (2012).
Casali, P. G., Fumagalli, E. & Gronchi, A. Adjuvant therapy of gastrointestinal stromal tumors (GIST). Curr. Treat. Opt. Oncol. 13(3), 277–284 (2012).
Joensuu, H. et al. Risk of recurrence of gastrointestinal stromal tumour after surgery: An analysis of pooled population-based cohorts. Lancet Oncol. 13(3), 265–274 (2012).
Chindapasirt, J. Sarcopenia in cancer patients. Asian Pac. J. Cancer Prev. 16(18), 8075–8077 (2015).
Xiaoping, Y., Jingying, Y., Liping, Z. et al. CT-based sarcopenic nomogram for predicting progressive disease in advanced non-small-cell lung cancer. Front. Oncol. (2021).
Yi, X. et al. Myosteatosis predicting risk of transition to severe COVID-19 infection. Clin. Nutr. 41, 3007–3015 (2021).
Moryoussef, F. et al. Reversible sarcopenia in patients with gastrointestinal stromal tumor treated with imatinib. J. Cachexia Sarcopenia Muscle. 6(4), 343–350 (2015).
Maurício, S. F. et al. Relationship between sarcopenia and mTOR pathway in patients with colorectal cancer: Preliminary report. Nutr. Cancer. 71(1), 172–177 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M., Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. (2022).
Ongaro, E. et al. Sarcopenia in gastric cancer: When the loss costs too much. Gastr. Cancer 20(4), 563–572 (2017).
Souza, M. L. R. et al. Increased resting metabolism in neurofibromatosis type 1. Clin. Nutr. ESPEN. 32, 44–49 (2019).
Ciano, M. et al. EGF receptor (EGFR) inhibition promotes a slow-twitch oxidative, over a fast-twitch, muscle phenotype. Sci. Rep. 9(1), 9218 (2019).
Puppo, F. et al. Identification of variants in the 4q35 gene FAT1 in patients with a facioscapulohumeral dystrophy-like phenotype. Hum. Mutat. 36(4), 443–453 (2015).
Johnson, R. W. et al. The primary function of gp130 signaling in osteoblasts is to maintain bone formation and strength, rather than promote osteoclast formation. J. Bone Miner. Res. 29(6), 1492–1505 (2014).
Ziaaldini, M. M., Marzetti, E., Picca, A. & Murlasits, Z. Biochemical pathways of sarcopenia and their modulation by physical exercise: A narrative review. Front. Med. (Lausanne). 4, 167 (2017).
Lipina, C. & Hundal, H. S. Lipid modulation of skeletal muscle mass and function. J. Cachexia Sarcopenia Muscle. 8(2), 190–201 (2017).
Mankhong, S. et al. Experimental models of sarcopenia: Bridging molecular mechanism and therapeutic strategy. Cells 9(6), 1385 (2020).
Sharples, A. P. et al. Longevity and skeletal muscle mass: The role of IGF signalling, the sirtuins, dietary restriction and protein intake. Aging Cell 14(4), 511–523 (2015).
Mazucanti, C. H. et al. Longevity pathways (mTOR, SIRT, Insulin/IGF-1) as key modulatory targets on aging and neurodegeneration. Curr. Top. Med. Chem. 15(21), 2116–2138 (2015).
Salas-Pérez, F. et al. DNA methylation in genes of longevity-regulating pathways: Association with obesity and metabolic complications. Aging (Albany NY). 11(6), 1874–1899 (2019).
Park, S. S., Seo, Y. K. & Kwon, K. S. Sarcopenia targeting with autophagy mechanism by exercise. BMB Rep. 52(1), 64–69 (2019).
Raue, U., Slivka, D., Jemiolo, B., Hollon, C. & Trappe, S. Proteolytic gene expression differs at rest and after resistance exercise between young and old women. J. Gerontol. A Biol. Sci. Med. Sci. 62(12), 1407–1412 (2007).
Shachar, S. S., Williams, G. R., Muss, H. B. & Nishijima, T. F. Prognostic value of sarcopenia in adults with solid tumours: A meta-analysis and systematic review. Eur. J. Cancer. 57, 58–67 (2016).
Su, H., Ruan, J., Chen, T., Lin, E. & Shi, L. CT-assessed sarcopenia is a predictive factor for both long-term and short-term outcomes in gastrointestinal oncology patients: A systematic review and meta-analysis. Cancer Imaging 19(1), 82 (2019).
Blauwhoff-Buskermolen, S., Langius, J. A. E., Becker, A., Verheul, H. M. W. & de van der Schueren, M. A. E. The influence of different muscle mass measurements on the diagnosis of cancer cachexia. J. Cachexia Sarcopenia Muscle. 8(4), 615–622 (2017).
Acknowledgements
We thank staff members in the Departments of Radiology, General Surgery, and Pathology at Xiangya Hospital for their efforts in collecting the information used in this study. Editing assistance was provided by our scientific writer Nancy Linford, PhD.
Funding
This work was granted by National Natural Science Foundation of China (81702278, 81974367), Key Subject Education Department of Hunan ([2012]594), Scientific Research Project of Hunan Provincial Department of Education (18K001), the Project Program of National Clinical Research Center for Geriatric Disorders (Xiangya Hospital, Grant No. 2022LNJJ09), Natural Science Foundation of Hunan Province (2019JJ40485, 2022JJ30979), and China Post-Doctoral Science Foundation (2022M713536).
Author information
Authors and Affiliations
Contributions
X.Y.: Conceptualization, methodology, writing- original draft preparation, investigation, validation, reviewing and editing. G.Z.: Data curation, writing-original draft preparation, investigation. Y.F.: Data curation, writing- original draft preparation, investigation. J.W.: Data curation, investigation. C.C.: Investigation. H.Z.: Investigation. Q.H.: Methodology, software, visualization. P.P.: Methodology, software, visualization. H.Z.: Data curation, investigation. G.G.: Data curation, investigation. T.L.: Data curation, investigation. F.T.: Data curation, investigation. H.L.: Conceptualization, investigation, supervision, validation. B.L.: Conceptualization, methodology, writing-original draft preparation, supervision, validation, reviewing and editing. B.T.C.: Methodology, supervision, reviewing and editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yi, X., Zhou, G., Fu, Y. et al. CT-based assessment of sarcopenia for differentiating wild-type from mutant-type gastrointestinal stromal tumor. Sci Rep 13, 3216 (2023). https://doi.org/10.1038/s41598-022-27213-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-27213-8