Abstract
Thirteen known Yr gene-associated markers pertaining to genes (Yr5, Yr10, Yr15, Yr24/Yr26) were used to identify the genes in selected wheat germplasm which were found resistant under field conditions at two locations in Punjab, India against stripe rust. In field evaluation, 38 genotypes exhibited highly resistant response, with a final rust severity (FRS) ranging from 0 to TR. Seven genotypes expressed a resistant to moderately resistant response with FRS ranging from 5MR–10S. In race-specific phenotying against most prevalent pathotypes of Puccinia striiformis tritici (46S119,110S119 &238S119) by seedling reaction test (SRT) 14 genotypes (29.2%) were found to be immune (IT = 0), 28 genotypes (58.3%) were resistant (IT = 1), and 3 genotypes (6.3%) were moderately resistant (IT = 2). Yr5 was detected in sixteen lines with the help of two markers Xwmc175 and Xgwm120 linked with Yr5. Yr10 was detected in ten lines with the marker Xpsp3000 and Yr15 was detected in fourteen lines with two linked markers; Xgwm413 and Xgwm273. Likewise, Yr24/26 was detected in 15 lines with two linked markers, namely Xbarc181 and Xbarc187. Based on the race specific phenotyping data and marker data, fourteen lines were found to carry a single gene, 16 showed the presence of two gene combinations, and seven genotypes were found to have a combination of three genes. Frequencies of Yr5, Yr15 and Yr26/Yr24 was high among test wheat germplasm in comparison to Yr10.
Similar content being viewed by others
Introduction
Wheat, scientifically known as Triticum aestivum L., is the most widely consumed food grain, with a global per capita consumption of 67.4 kg/year1. In India, wheat production was recorded 106.41 MT in 2021–22, with 3484 kg/ha average national productivity2 Uttar Pradesh, Punjab, and Haryana are the main contributors to central pool in North-western Plains zone of India3. Wheat production in North-western Plains zone of India is threatened by a variety of factors, including pests and diseases. Approximately 16 percent losses due to pathogens have been reported globally4,5. Among-est the diseases stripe rust has been identified as the most devastating wheat disease, as it prefers a cool climate and can establish its infection during the early stages of crop growth and the infection prevails till the crop matures6,7. Under favourable conditions, this disease can result in yield losses of up to 90 percent6,8. In its moderate to a severe form, stripe rust caused 68.8% yield losses in Punjab during 20089. Puccinia striiformis, the pathogen that causes stripe rust, is extremely diverse, possibly due to a combination of long-distance migration capacity10, high rates of mutation from avirulence to virulence11, adaptation to different climatic conditions12, the existence of recombinant and highly diverse populations13,14 and the potential creation of new variants through a sexual cycle15. Mutations in the pathogen's DNA can result in the emergence of new virulence races, allowing the pathogen to infect previously resistant plants16,17. In India, P. striiformis virulence in the cultivar Kalyansona was first appeared around 1971, followed by virulence in the cultivar Sonalika between 1984–199018. The first detection of virulence forYr9 was in 1996, followed by a pathotype with combined virulence for Yr9 and Yr27 in 200119,20. Finally, three new pathotypes 110S119, 238S119, and 110S84 with additional virulence have been identified on Riebesel 47/51, Suwon × Omar, Yr11 and Yr1421. The resistance genes Yr5, Yr10, Yr15 Yr24/Yr26, Yr32 and YrSp are still effective against all the prevalent pathotypes of P. striiformis tritici in India, while Yr2, Yr3, Yr4, Yr6, Yr7, Yr8, Yr9, Yr17, Yr18, Yr19, Yr21, Yr22, Yr23, Yr25, Yr27 and YrA have become ineffective to the prevailing and recently evolved pathotypes. Till date, 84 permanently designated stripe rust resistance genes, 100 temporarily designated genes, and 363 quantitative trait loci (QTLs) with different names have been reported in wheat22,23,24. Despite of so many genes have been identified which impart resistance against stripe rust throughout the crop season or at adult plant stage; we are left with only few effective Yr genes as many of them have been defeated due to the continuous and fast evolution of the pathogen in one or another part of the world. To broaden our gene pool for stripe rust resistance gene mining is a continuous process in the wheat breeding programs globally and further to achieve durability of resistance pyramiding is the need of the hour to keep pace with the ever-evolving pathogen. The known genes, namely Yr5, Yr10, Yr15, Yr36, Yr40, Yr47 etc. have been extensively exploited in our in house breeding program at Punjab Agricultural University. However, the climate change and the fungal evolution is always anticipated to create newer pathotypes/races of the pathogen. In view of this, the novel genes need to be streamlined and characterized genetically for its ready mobilization whenever required. Since, the germplasm collections from National Bureau of Plant Genetic Resources (NBPGR) constitute the diverse germplasm collected from variable sites, it holds the promise to harbour newer and unidentified resistance sources. So, in the present study efforts were made to evaluate the wheat germplasm against the most prevalent and virulent pathotypes of the stripe rust pathogen At seedling stage as well as at adult plant response against stripe rust under field conditions and further molecular markers were deployed to identify the already known stripe rust resistance genes in the resistant germplasm.
Materials and methods
Plant materials
Total of 45 wheat accessions from NBPGR which showed highly resistant to resistant reaction in the preliminary screening of ~ 1500 wheat germplasm accessions from national gene bank of India, ICAR-National Bureau of Plant Genetic Resources (Table 1) were selected for the present study i.e. to identify the stripe rust resistance gene(s) present in them. This included 22 indigenous accessions collected or derived in India and 23 exotic collections augmented from USA, Mexico and Australia. These lines were evaluated against the three most prevalent races (238S119, 110S119, and 46S119) of stripe rust pathogen both at seedling and adult plant stages. The highly susceptible cultivars PBW343, HD2967, and Agra Local, as well as the Avocet background near-isogenic lines (NILs), i.e., Avocet/Yr5, Avocet/Yr10, Avocet/Yr15, Avocet/Yr24, and Avocet/Yr26, were used as susceptible and resistant checks respectively in the study for comparison purpose.
Seedling reaction test
Wheat seedlings were raised in plastic germination trays (14 × 7 cups) filled with a mixture of soil having sandy loam soil cocopeet + Farm Yard Mannure and vermi compost. All the germplasm were sown in three sets for evaluation against stripe rust pathotypes 46S119, 238S119, and 110S119 separately. The leaves of 10-day-old seedlings were inoculated with uredospores of three different pathotypes (46S119, 110S119 & 238S119) separately and kept in separate poly-chambers. For a successful infection, the inoculated material was placed in a dew chamber for 48 h in the dark before being transported to a greenhouse and maintained with a photo period of 16 h light and 8 h of darkness. Regular irrigation was given to maintain the humidity. The host response for seedling infection types was recorded 14 days after inoculation using 0–4 scale by25 (0 = immune = no uredinia or other macroscopic sign of infection, = nearly immune = no uredinia, but hypersensitive necrotic or chlorotic flecks present, 1 = highly resistant = small uredinia surrounded by necrosis, 2 = moderately resistant = small to medium uredinia surrounded by chlorosis or necrosis, green island may be surrounded by chlorotic or necrotic border, 3 = moderately susceptible = medium-sized uredinia that may be associated with chlorosis, 3+, 4 = susceptible = large uredinia without chlorosis). When the susceptible checks showed the highest infection score of 33+ or 4, the inoculations were considered successful.
Field evaluation
The germplasm was evaluated for three years (2018–2021) at PAU, Ludhiana, and at RRS Gurdaspur by following the standard protocol and relevant guidelines. Wheat germplasm lines were sown at both locations during the 2nd week of November, in a 1-m long pair rows of each genotype at a row-to-row distance of 20–22 cm. To ensure the uniform spread of rust inoculum, susceptible varieties like Agra Local, HD2967, A-9-30-1 and PBW343 were planted after every 20 rows of the test material, and infector rows were sown on the periphery of the experimental material. For comparison purposes and gene postulation, along with genotypes, NILs carrying known genes for stripe rust, susceptible and resistant checks were also sown in the field (Table 2). The stripe rust epidemic was created under field conditions by repeated spray inoculations of experimental material with uredospores of Puccinia striiformis fsp tritici (Pst). Infected leaves of stripe rust susceptible varieties PBW343, Agra local, A-9-30-1, and HD2967 were immersed in water for extracting uredospores. The inoculum was prepared by suspending rust uredospores in 10 l of water using a few drops of Tween-20. The spray inoculations were done in the evening with an ultralow volume sprayer on alternate days beginning from the end of December till stripe rust appeared on the susceptible checks. Stripe rust was monitored regularly at weekly intervals starting from the second week of January to the first week of March using the modified Cobb’s Scale26. The host responses were graded: S = susceptible, large uredia surrounded by necrotic tissues; MS = moderately susceptible, small uredia surrounded by necrotic tissues; MR = moderately resistant, small uredia surrounded by necrotic tissues; M = Moderately resistant to moderately susceptible, small to medium sized pustules surrounded by necrosis and chlorosis; R = resistant, very small uredia surrounded by necrotic tissues27. The area under disease progress curve (AUDPC), and coefficient of infection (CI) were calculated by using the formula’s given below.
where, Xi is the rust intensity on date i, ti is the time in days between i and date i + 1 and n is the number of dates on which disease was recorded. Coefficient of infection (CI) was calculated by multiplying disease severity (DS) and constant values of infection type (IT). The constant values for infection types were used based on immune = 0, R = 0.2, MR = 0.4, M = 0.6, MS = 0.8, S = 128.
Identification of stripe rust resistance genes by using tagged molecular markers
Under North Indian conditions against the prevalent pathotype of the stripe rust pathogen five genes namely Yr5, Yr10, Yr15, Yr24, and Yr26 are highly effective so the tagged markers for these genes were used to identify the presence or absence of these genes in the tested germplasm. In total thirteen markers (SSR, EST-SSR, and STS) were used to profile the wheat genotypes. The Graingenes database (http://wheat.pw.usda.gov/) was employed to retrieve the sequences of available markers (Table 3) along with their previously determined chromosomal positions. Integrated DNA Technologies, Inc., synthesized the primers.
DNA extraction
Genomic DNA was extracted from 100 mg of fresh leaves collected from individual lines using the modified CTAB extraction method29,30,31. To remove the contaminant RNA from the DNA extracted from fresh leaves, 10 μl/ml RNase was added and then incubated at 37 ºC for 45 min.
PCR amplification and electrophoretic separation of PCR products
After quality and quantity check the amplification using Polymerase Chain Reaction (PCR) was performed in an Eppendorf Master cycler to study the genetic polymorphism among the genotypes carrying known Yr genes. PCR analysis was carried out in the reaction volume of 11 μl containing the 3 μl template genomic DNA, 1.0 μl forward and reverse primers, 5 μl of 10× PCR buffer and 0.5 μl of BSA and PVP. Specific PCR amplification protocols were followed for each primer linked to different Yr genes. Protocol for Yr5 markers (Wmc175 and Xgwm120) was given by32,33, Yr10 (Xpsp3000)34, Yr15 (Xgwm413 and Xgwm273)35,36, Yr24/26 (Barc181)37 and Barc18738. PCR products were separated through 6% PAGE using PAGE apparatus (Mega-Gel System) of CBS, Scientific, USA (C-DASG-400-50) and visualized under UV light in gel documentation (SYNGENE, G: Box, USA).
Results
Seedling reaction test
Disease data in terms of infection types and host response were recorded on selected wheat genotypes and four susceptible checks (PBW343, HD2967, A-9-30-1 and Agra Local) at the seedling stage, as shown in Table 4. Fourteen genotypes were found to be immune (ITs 0), accounting for 29.2% of all genotypes. 28 genotypes (58.3%) were resistant (ITs1), 3 genotypes (6.3%) were moderately resistant (ITs 2), and 4 genotypes (HD2967, Agra local, PBW 343& A-9-30-1) were susceptible (ITs 4) (Table 4). The Near isogenic lines for Yr5, Yr10, Yr15, Yr24/26 all showed nearly immune (0) to resistant (1) reaction.
Field evaluation
Data on adult-plant stage were collected in the field based on disease severity, host response, and AUDPC (Table 5). On pooling data collected at both locations over the three crop seasons it was found that 19 lines (30.6% of total genotypes) showed no disease i.e. final rust severity (FRS) = zero and 0 AUDPC, 19 genotype lines (30.6%) showed resistance with FRS = TR-5MR (AUDPC = 100); and 7 genotype lines (14.6%) expressed a moderately resistant response with 5MS-10S final rust severity (AUDPC = 101–200). Susceptible checks; HD2967, Agra local, PBW 343, A-9-30-1) expressed 80% disease severity (80S; AUDPC = 800). NILs carrying Yr5, Yr10, Yr15, Yr24/26 genes exhibited 0–5 MS final rust severity under artificial inoculated conditions in field.
Identification of Yr genes
Gene postulation in selected genotypes was performed using 13 Yr gene-tagged markers. The presence of one, two, or more than two gene combinations was observed. Table 6 shows the total number of alleles of each microsatellite marker recorded for all genotypes tested, for the detection of known Yr genes (Yr5, Yr10, Yr15, Yr24/ Yr26), with the amplified allele number being given as 0 for the absence and 1 for presence.
Postulation of Yr5
The dominant SSR marker Xwmc175 is 1.4 cM away from the gene35. To confirm and evaluate the diagnostic potential of the markers in wheat genotypes, two microsatellite markers Wmc175, Xgwm120, and one STS marker, STS9/10, linked to the stripe rust resistance gene Yr5 were used. The presence of the Yr5 gene was associated with a product size of 253 bp in 16 lines (31.3 percent), while the other 33 wheat genotypes (68.8 percent) failed to amplify the gene (Fig. 1). Another SSR marker, Xgwm 120, was found in 13 lines and is closely linked to Yr5 amplified alleles of 156 bp (Fig. 2). Based on Yr5 linked markers (Wmc175 and Xgwm120), 16 genotypes (IC535470, EC693621, EC636264, EC693322, EC692262, EC635577, EC664198, IC529052, EC0582305, EC0612509, EC0612504, EC0635659, IC47034, EC0635685, IC0598571, IC0594933, IC0078959-A, EC530087, Avocet/Yr5) were found to have the Yr5 gene.
Postulation of Yr10
The microsatellite marker Xpsp3000 is co-dominantly inherited and can be used to identify genotypes of individuals at any growth stage39. The fragment 260 bp was amplified in ten tested entries i.e. IC529094, IC111691, EC635701, IC47034, IC530078, IC445516, IC553914, IC469420, EC0610944, and IC0594933, which shows the presence of Yr10 gene, in rest of the 35 accessions tested it was absent (Fig. 3). In addition to the SSR Xpsp3000 marker, the E10 marker with a genetic distance of 0.5 cM from Yr1040 was used for detecting Yr10 gene in test accessions but that did not work.
Postulation of Yr15
Yr15 gene diagnostics markers have been identified as Xbarc8, Xgwm273, and Xgwm413. These three markers were used in the current study to detect the presence of Yr15 gene in wheat genotypes under study. The 3.5 cM proximal SSR locus Xgwm413 produced three types of alleles (90 bp, 95 bp, and 100 bp) among the wheat genotypes used in this study. The allelic profile of Xgwm413 showed variation. Fourteen genotypes amplified specific alleles of 90 bp, 18 genotypes amplified alleles of 95 bp, and 16 genotypes did not amplify any allele. (Fig. 4). The Xgwm273 marker is located 2.1 cM from Yr15 and amplified 5 different types of alleles (156 bp, 165 bp, 180, 200 bp, and 220 bp). 14 genotypes (29.2%) produced 156 bp bands specific for Yr15 (Fig. 5). Based on confirmation of Yr15 with both Xgwm413 and Xgwm273 markers, fifteen genotypes (31.3%) (IC534662, IC336645, IC530087, EC693621, EC635701, EC693322, IC445516, IC469420, EC0582305, EC0612509, IC0078981-B, EC0610955, EC0635659, IC0591082, IC0078959-A, Avocet/Yr15) showed the presence of Yr15 gene.
Postulation of Yr24/26
The presence/absence of Yr24/Yr26 genes was detected using two markers, Barc181 and Barc187. Individual results for SSR marker Barc181 linked with Yr24 at 6.7 cM37 amplified two types of alleles; 180 bp (presence of Yr24) and 200 bp (absence of Yr24), respectively. In twelve genotypes, IC111691, IC53547, EC217835, EC33961, EC635750, IC529052, IC529094, EC0612506, EC0105966, EC0610955, ICO598571, and EC0635701 the presence of Yr24 gene was observed as the amplified products showed the presence of 180 bp specific allele, indicating the presence of Yr24. SSR marker Xbarc187, which is 2.3 cM away from Yr2637, produces three types of alleles, 200 bp, 225 bp, and 240 bp. Fourteen genotypes i.e. IC111691, IC53547, EC217835, EC33961, EC635750, IC529052, IC529094, EC0612506, EC0105966, EC0610955, ICO598571, EC693322, EC692262 and EC0635659 were detected with the Yr26 gene due to presence of 200 bp specific allele. Combined results with both the markers exhibited the presence of Yr24/Yr26 in 11 genotypes (22.9%) (Figs. 6, 7).
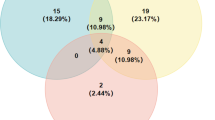
The distribution of these five Yr resistance genes among 45 wheat accessions is illustrated in Fig. 8 and Table 7. Fourteen lines were found to carry a single gene, 16 lines showed the presence of two gene combinations, and seven accessions were found to have a combination of three genes. These 45 accessions exhibited strong resistance at seedling and adult plant stage.
Discussion
Many sources of wheat yellow rust genetic resistance have proven ineffective after only a short period of deployment41. Monoculture of wheat cultivar PBW343 in Punjab, India resulted in the emergence of a new Pst race, 78S84, and a decrease in the predominance of Pst race 46S119. Because the new race 78S84 was virulent on Yr27 thus PBW343 became vulnerable. During 2008–2010, the race 78S84 was found in more than 80% of infected samples collected from North Western Plains Zone of India. However, as PBW343 was replaced by cultivars with R genes other than Yr27, 46S119 became prevalent once more, while 78S84 became the least common one (5%). Three new races, 238S119, 110S119 and 46S117, have also emerged in Punjab and their prevalence’s are increasing with every passing year42. The knockdown of race-specific single genes is always a concern for plant breeders. The pyramiding of more than one R gene into a single cultivar by MAS is the best way to develop cultivars with durable resistance and slows down the process of pathogen evolution. against current prevalent and new emerging Pst races43. In fact, continuous search for novel source of donor genotype is warranted for identification of resistance genes and their deployment in new cultivars or cultivars with defeated genes and unexploited genebank material is the best option for such kind of study. Forty-five genotypes including indigenous (IC-) and exotic collections (EC-) were selected out of ~ 1500 diverse set of genebank germplasm accessions screened for yellow rust disease during 2014–15 and 2015–16. Subsequently, this study was conducted to postulate the Yr genes in selected and pre-screened germplasm accessions for yellow rust resistance. Therefore, this study was conducted to postulate the Yr genes in selected genotypes (45) using Yr gene-tagged markers (13) and to assess the contribution of Yr genes to the current status of Pst resistance. In the current study, we have found three types (single, two, and three genes) combinations among the germplasm under study. Fourteen lines of wheat germplasm were found to carry a single gene, 16 lines showed the presence of two gene combinations, and seven genotypes were found to have a combination of three genes. These 45 genotypes lines exhibited strong resistance at seedling as well as at adult plant stage. Out of 45 germplasm lines, eight germplasm lines showed no amplification with any of the markers used, which means some other already known gene (s) or some new gene (s) may be present which confers a good level of resistance. In the rest of the 38 lines, the amplification was achieved with one/two/more of the tagged Yr gene markers, indicating the presence of single or multiple genes. Rani et al.44 found two genotypes (VL 3002 and VL 1009) with 15 Yr genes, followed by 14 genes in HI8759 and VL3010. Sobia et al.43 and Yuan et al.45 studied the frequency of Yr genes among wheat genotypes by using tagged markers. Zheng et al.46 also did the molecular characterization of 330 leading wheat cultivars and 164 advanced breeding lines in China and identified Yr9, Yr17, Yr18, and Yr26 in 134 (29.4%), 45 (9.1%), 10 (2%) and 15 (3%) entries, respectively. Yan et al. 47 observed Yr 1, Yr 13, Yr 18, Yr 14a, Yr 26, Yr 34 and Yr 46 either singly or in combination in twenty-five cultivars by testing them in the field during 2014–2018 crop seasons against stripe rust and also by using eleven molecular markers associated with known Yr genes. Yr5 has been shown to be effective against all rust virulent races in North America 6,48,49, China, Iran 6,50, Turkey, and India 51. The stripe rust resistance gene Yr5, which was derived from Triticum spelta var album, is a race-specific R-gene effective at both seedling and adult plant growth stages and is located on chromosome 2BL 52. In our study Yr5 presence was detected in eighteen (40%) lines with two linked markers, i.e., Xwmc175 and Xgwm120. Naruoka et al. 32 found six lines out of 13 carrying lines carrying the resistance-linked allele using the Xwmc175 marker. STS-9/10, developed by Chen et al. 49, co-segregates with the Yr5 locus and amplified fragments of 439 or 433 bp for resistant or susceptible plants, respectively. Using the S19M93 molecular marker, Ullah et al.53 discovered an 89% polymorphism rate of the Yr5 gene in 99 Pakistan wheat lines. The Yr5 gene was not amplified in any of the 45 genotypes using STS9/10. In India, to date, the Yr5 gene is effective against all prevalent races and can be an effective source for stripe rust resistance when used singly or in combination with other resistant genes. The Yr10 gene was also found effective against all the Pst races prevalent in India, Iran, China, Pakistan, and the United States54. The marker Xpsp3000, located at the end of chromosome 1BS, is 1.2 cM away from the stripe rust-resistant gene, Yr10 55 which is co-dominantly inherited and can be used to identify genotypes of individuals at any growth stage56 and is linked with brown glume color (0.2 cM) in wheat genotypes PI 178,383 and Moro57, while T. spleta 415 and T. vavilovii AUS22498 have white glumes34,58. Resistant genotypes with brown glumes during the phenotypic evaluation were observed in nine genotypes, which was further confirmed with the tagged marker Xpsp3000 yielded a specific allele of 260 bp. Bariana et al.34 described two Yr10 alleles, Yr10 and Yrvav, and stated Yr10-containing varieties amplified 258-260bps fragments, whereas Yrvav-containing varieties amplified 285 bp and 240 bp for varieties lacking the Yr10 gene. Similarly, Yr15 is currently the most common all-stage resistance gene used in breeding programs it is effective against all identified races in the United States59. Yr15, a dominant gene derived from Triticum dicoccoides, is found on chromosome 1BS 60. Sun et al. 61, Peng et al.62, and Murphy et al. 35 mapped the Yr15 gene to a 6.4 cM interval flanked by markers Xbarc8, located 3.9 cM to the distal side, Xgwm413 located 2.5 cM to the proximal side, and Xgwm273 located at 2.5 cM and 2.1 cM to the proximal side. On the basis of confirmation with two linked markers, i.e., Xgwm413 and Xgwm273, Yr15 was detected in fourteen lines accounting for (31%) of the total genotypes. Kokhmetova et al. 63 used Xbarc8 and Xgwm413 markers to confirm the presence of Yr15 in seven genotypes (10%) that carried the Yr15 gene. Barc8 did not work well under our conditions. Yr15 and its flanking SSR markers Xbarc8 and Xgwm413 are separated by 3.9 and 2.5 cM, respectively. The two markers that have been most frequently used to incorporate Yr15 into wheat cultivars are Xbarc8 and Xgwm413 35,64, but in our conditions, Xbarc8 did not work. Yr24, discovered from the K733 accession of T. turgidum var. durum, confers stripe rust resistance at all stages, and the Yr26 stripe rust resistant gene was discovered on chromosome 1B in the T. turgidum durum line65. Due to their similar infection types against stripe rust isolates, Yr24 and Yr26 are thought to be identical genes66 and further demonstrated by Clemence et al. 39. Combined results with both the markers exhibited the presence of Yr24/Yr26 in 11 genotypes (22.9%), namely IC111691, IC535470, EC217835, EC339610, EC635750, IC529052, IC529094, EC0612506, EC0105966, EC0610955, and ICO598571. In EC0635701 Yr24/26 were detected by Barc181 whereas in EC693322, EC692262, and EC0635659 genotypes they were detected by XBarc187. The Yr5, Yr10, Yr15, Yr24/Yr26 genes, which are specific to races, are still effective against the present dominant races prevalent in north western plains zone of India. To increase their durability, they have to be use in combination with other genes.
Data availability
The datasets generated during the current study will be available from the corresponding author on reasonable request.
Change history
22 June 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41598-023-37367-8
References
Djanaguiraman, M. et al. Alien chromosome segment from Aegilops speltoides and Dasypyrum villosum increases drought tolerance in wheat via profuse and deep root system. BMC Plant Biol. 19, 1–15 (2019).
ICAR-IIWBR. Director’s Report of AICRP on Wheat and Barley 2021–22 (Singh, G.P. Ed.) (ICAR-Indian Institute of Wheat and Barley Research, 2022).
Singh, S., Kumar, A., Sendhil, R., Khippal, A.K. & Singh, G.P. All India coordinated research project on wheat and barley. in Progress Report 2019–20. Social Sciences. Vol. 50. 1–2 (ICAR-Indian Institute of Wheat and Barley Research Institute, 2021).
Oerke, E. C. Crop losses to pests. J. Agric. Sci. 144, 31 (2006).
Strange, R. N. & Scott, P. R. Plant disease: A threat to global food security. Annu. Rev. Phytopathol. 43, 83–116 (2005).
Chen, X. M. Epidemiology and control of stripe rust (Puccinia striiformis f. sp. tritici) on wheat. Can. J Plant Pathol. 27(3), 314–337 (2005).
Hovmøller, M. S., Walter, S. & Justesen, A. F. Escalating threat of wheat rusts. Science 329, 369 (2010).
Sharma-Poudyal, D. et al. Virulence characterization of international collections of the wheat stripe rust pathogen, Puccinia striiformis f. sp. tritici. Pl. Dis. 97, 379–386 (2013).
Jindal, M. M., Sharma, I. & Bains, N. S. Losses due to stripe rust caused by Puccinia striiformis in different varieties of wheat. J. Wheat Res. 4, 86–88 (2012).
Brown, J. K. M. & Hovmøller, M. S. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 297, 537–541 (2002).
Hovmøller, M. S. & Justesen, A. F. Rates of evolution of avirulence phenotypes and DNA markers in a northwest European population of Puccinia striiformis f. sp. tritici. Mol. Ecol. 16, 4637–4647 (2007).
Milus, E. A., Kristensen, K. & Hovmoller, M. S. Evidence for increased aggressiveness in a recent widespread strain of Puccinia striiformis f. sp. tritici causing stripe rust of wheat. Phytopathology 99, 89–94 (2009).
Stakman, E.C., Stewart, D.M., & Loegering, W.Q. Identification of Physiologic Races of Puccinia graminis f.sp. tritici. (USDA.F, 1962).
Thach, T., Ali, S., de Vallavieille-Pope, C., Justesen, A. F. & Hovmøller, M. S. Worldwide population structure of the wheat rust fungus Puccinia striiformis in the past. Fungal Genet. Biol. 87, 1–8 (2016).
Jin, Y., Szabo, L. J. & Carson, M. Century-old mystery of Puccinia striiformis life history solved with the identification of Berberis as an alternate host. Phytopathology 100, 432–435 (2010).
Chen, X. M. Integration of cultivar resistance and fungicide application for control of wheat stripe rust. J. Plant Pathol. 36, 311–326 (2014).
Chen, X. M. et al. Wheat stripe rust epidemics and races of Puccinia striiformis f. sp. tritici in the United States in 2000. Pl. Dis. 86, 39–46 (2002).
Bhardwaj, S. C. Wheat rust pathotypes in Indian subcontinent then and now. in Wheat-Productivity Enhancement Under Changing Climate; 9–12 Feb 2011, University of Agricultural Sciences, Dharwad (Singh, S.S., Hanchinal, R.R., Singh, G., Sharma, R.K., Saharan, M.S., Sharma, I. Eds.). 227–238 (Narosa Publishing House Pvt. Ltd., 2012).
Bhardwaj, S.C. et al. Evolution of wheat rust pathogens in Indian subcontinent. in Management of Wheat and Barley Diseases (Singh, D.P. Ed.).(Apple Academic Press, 2017).
Prashar, M., Bhardwaj, S. C., Jain, S. K. & Datta, D. Pathotypic evolution in Puccinia striiformis in India during 1995–2004. Aust. J. Agric. Res. 58, 602–604 (2007).
Gangwar, O. P. et al. Characterization of three new Yr9-virulences and identification of sources of resistance among recently developed Indian bread wheat germplasm. J. Plant Pathol. 101, 955–963 (2019).
Klymiuk, V. et al. Discovery of stripe rust resistance with incomplete dominance in wild emmer wheat using bulked segregant analysis sequencing. Commun. Biol. 5, 826. https://doi.org/10.1038/s42003-022-03773-3 (2022).
McIntosh, R. A., Dubcovsky, J., Rogers, W. J., Xia, X. & Raupp, W. J. Catalogue of gene symbols for wheat: 2022 supplement. Annu. Wheat Newslett. 68, 104–113 (2022).
Wang, M. & Chen, X. Stripe rust resistance. in Stripe Rust (Chen, X., Kang, Z. Eds.). 353–378 (Springer, 2017).
Nayar, S. K., Prashar, M., & Bhardwaj, S. C. Manual of current techniques in wheat rusts. in Research Bulletin No. 2, 32P. (Regional Research Station, Directorate of Wheat Research, 1997).
Peterson, R. F., Campbell, A. B. & Hannah, A. E. A diagrammatic scale for estimating rust intensity on leaves and stems of cereals. Can. J. Res. 26, 496–500 (1948).
Akin, B., Zencirci, N. & Özseven, İ. Field resistance of wheat (Triticum aestivum L.) genotypes from different countries to leaf rust (Puccinia triticina). Turk. J. Agric. For. 32(6), 479–486 (2008).
Stubbs, R. W., Dubin, H. J., Saari, E. E. & Prescott, J. M. International Maize and Wheat Improvement Centre. Cereal Disease Methodology Manual. 46. (CIMMYT, 1986).
Murray, M. G. & Thompson, W. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326 (1980).
Saghai-Maroof, M. A., Soliman, K. M., Jorgensen, R. A. & Allard, R. Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. 81, 8014–8018 (1984).
Xu, G. W., Magill, C. W., Schertz, K. F. & Hart, G. E. A RFLP linkage map of Sorghum bicolor (L.) Moench. Theor. Appl. Genet. 89, 139–145 (1994).
Naruoka, Y. et al. Identification and validation of SNP markers linked to the stripe rust resistance gene Yr5 in wheat. Crop Sci. 56, 3055–3065 (2016).
Sun, Q., Wei, Y., Ni, Z., Xie, C. & Yang, T. Microsatellite marker for yellow rust resistance gene Yr5 in wheat introgressed from spelt wheat. Plant Breed. 121, 539–541 (2002).
Bariana, S. et al. Characterization of Triticum vavilovii derived stripe rust resistance using genetic, cytogenetic and molecular analyses and its marker-assisted selection. Theor. Appl. Genet. 104, 315–320 (2002).
Murphy, L. R. et al. A Linkage maps of wheat stripe rust resistance genes Yr5 and Yr15 for use in marker-assisted selection. Crop Sci. 49, 1786–1790 (2009).
Revathi, P., Tomar, S. M. S. & Singh, N. K. Marker assisted gene pyramiding of leaf rust resistance genes Lr24, Lr28 along with stripe rust resistance gene Yr15 in wheat (Triticum aestivum L.). Indian J. Genet. Plant Breed. 70, 349–354 (2010).
Wang, C. et al. SSR and STS markers for wheat stripe rust resistance gene Yr26. Euphytica 159, 359–366 (2008).
Wen, W. E. et al. Development of an STS marker tightly linked to Yr26 against wheat stripe rust using the resistance gene-analog polymorphism (RGAP) technique. Mol. Breed. 22, 507–515 (2008).
Clemence, M. et al. BED-domain-containing immune receptors confer diverse resistance spectra to yellow rust. Nat. Plants 4, 662–668 (2018).
Shao, Y. et al. Identification of an AFLP marker linked to the stripe rust resistance gene Yr10 in wheat. Chin. Sci. Bull. 46, 1466–1468 (2001).
Boyd, L. A. Can Robigus defeat an old enemy?–Yellow rust of wheat. J. Agric. Sci. 143, 233–243 (2005).
Kaur, J. et al. Current status of wheat diseases in Punjab. Agric. Res. J. 55, 113–116 (2018).
Sobia, T., Muhammad, A. & Chen, X. Evaluation of Pakistan wheat germplasms for stripe rust resistance using molecular markers. Sci. China Life Sci. 53, 1123–1134 (2010).
Rani, R., Singh, R. & Yadav, N. R. Evaluating stripe rust resistance in Indian wheat genotypes and breeding lines using molecular markers. C. R. Biol. 342, 154–174 (2019).
Yuan, C. et al. Distribution, frequency and variation of stripe rust resistance loci Yr10, Lr34/Yr18 and Yr36 in Chinese wheat cultivars. J. Genet. Genom. 39, 587–592 (2012).
Zeng, Q. D. et al. Stripe rust resistance and genes in Chinese wheat cultivars and breeding lines. Euphytica 196, 271–284 (2014).
Yan, X. et al. QTL mapping of adult plant resistance to stripe rust in the Fundulea 900× Thatcher RIL population. Czech. J. Gen. Pl. Breed. 57, 1–8 (2021).
Bux, H., Ashraf, M., Hussain, F., Rattu, A. U. R. & Fayyaz, M. Characterization of wheat germplasm for stripe rust (Puccinia striiformis f. sp. tritici) resistance. Aust. J. Crop Sci. 6, 116–120 (2012).
Chen, X., Soria, M. A., Yan, G., Sun, J. & Dubcovsky, J. Development of sequence tagged site and cleaved amplified polymorphic sequence markers for wheat stripe rust resistance gene Yr5. Crop Sci. 43, 2058–2064 (2003).
Afshari, F. Prevalent pathotypes of Puccinia striiformis f. sp. tritici in Iran. J. Agric. Sci. Technol. 10, 67–78 (2008).
Zeybek, A. & Yigit, F. Determination of virulence genes frequencies in wheat stripe rust (Puccinia striiformis f. sp. tritici) populations during natural epidemics in the regions of Southern Aegean and Western Mediterranean in Turkey. Pak. J. Biol. Sci. 7, 1967–1971 (2004).
Law, C.N. Genetic control of yellow rust resistance in Triticum spelta album, Annual Report. 108–109 (Plant Breeding Institute, 1976).
Ullah, N. et al. Markers assisted selection for multiple stripe rust resistance genes in spring bread wheat lines. Int. J. Biol. 8, 63–74 (2016).
Chatrath, R., Mishra, B., Ortiz Ferrara, G., Singh, S. K. & Joshi, A. K. Challenges to wheat production in South Asia. Euphytica 157, 447–456 (2007).
Wang, L. F., Ma, J. X., Zhou, R. H., Wang, X. M. & Jia, J. Z. Molecular tagging of the yellow rust resistance gene Yr10 in common wheat, P.I.178383 (Triticm aestivum L.). Euphytica 124, 71–73 (2002).
Elkot, M., Mohammed, H. & El-Aziz, A. Molecular identification of some stem rust and yellow rust resistance genes in Egyptian wheat and some exotic genotypes. Ass. J. Agric. Sci. 47, 124–135 (2016).
Metzger, R. J. & Silbaugh, B. A. Inheritance of resistance to stripe rust and its association with brown glume color in Triticum aestivum L., ‘PI 178383’1. Crop Sci. 10, 567–568 (1970).
Kema, G. J. & Lange, W. Resistance in spelt wheat to yellow rust. II: Monosomic analysis of the Iranian accession 415. Euphytica 63, 219–224 (1992).
Chen, X. & Kang, Z. Stripe Rust (Springer, 2017).
McIntosh, R. A. & Silk, J. Cytogenetic studies in wheat XVII. Monosomic analysis and linkage relationships of gene Yr15 for resistance to stripe rust. Euphytica 89, 395–399 (1996).
Sun, G. L., Fahima, T., Korol, A. B., Turpeinen, T. & Grama, A. Identification of molecular markers linked to the Yr15 stripe rust resistance gene of wheat originated in wild emmer wheat, Triticum dicoccoides. Theor. Appl. Genet. 95, 622–628 (1997).
Peng, J. H. et al. High density molecular map of chromosome region harboring stripe-rust resistance genes YrH52 and Yr15 derived from wild emmer wheat, Triticum dicoccoides. Genetica 109, 199–210 (2000).
Kokhmetova, A. et al. Identification of stripe rust resistance genes in common wheat cultivars and breeding lines from Kazakhstan. Plants 10, 2303. https://doi.org/10.3390/plants10112303 (2021).
Yaniv, E. et al. Evaluation of marker-assisted selection for the stripe rust resistance gene Yr15, introgressed from wild emmer wheat. Mol. Breed. 35, 1–12 (2015).
Ma, J. et al. Molecular mapping and detection of the yellow rust resistance gene Yr26 in wheat transferred from Triticum turgidum L. using microsatellite markers. Euphytica 120, 219–226. https://doi.org/10.1023/A:1017510331721 (2001).
Li, G. Q. et al. Molecular mapping of stripe rust resistance gene YrCH42 in Chinese wheat cultivar Chuanmai 42 and its allelism with Yr24 and Yr26. Theor. Appl. Genet. 112, 1434–1440. https://doi.org/10.1007/s00122-006-0245-y (2006).
Ti-Lin, F. A. N. G. et al. Molecular characterization of a stripe rust resistance gene from wheat line S2199 and its allelism with Yr5. Acta Agron. Sin. 34(3), 355–360 (2008).
Cheng, P., Xu, L. S., Wang, M. N., See, D. R. & Chen, X. Molecular mapping of genes Yr64 and Yr65 for stripe rust resistance in hexaploid derivatives of durum wheat accessions PI 331260 and PI 480016. Theor. Appl. Genet. 127(10), 2267–2277 (2014).
Zhang, X. et al. Fine mapping of wheat stripe rust resistance gene Yr26 based on collinearity of wheat with Brachypodium distachyon and rice. PLoS ONE https://doi.org/10.1371/journal.pone.0057885 (2013).
Temel, A. et al. Yr10 gene polymorphism in bread wheat varieties. Afr. J. Biotechnol. 7(14), 2328 (2008).
Acknowledgements
The USAID-funded MSU’s GRAIN Project provided funding for this study. The National Bureau of Plant Genetic Resources, New Delhi, for giving plant material; the IIWBR Regional Research Station, Flowerdale, Shimla, for the inoculum; and the Borloug Institute for South Asia, Ladowal for providing the seed of NILs (Yr genes) are gratefully acknowledged by the authors.
Author information
Authors and Affiliations
Contributions
M.H.W., S.S. and Ja.K. planned & conducted the experiments, recorded & analyzed the data and drafted the M.H.W., S.S., R.B. and P.S. helped in planning and executing the experiments. A.S. and Jy.K.: helped in maintaining the germplasm lines and also in planning. R.S.: helped in molecular work and in analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of this Article contained an error in the name of author Mohammad Waris Haider, which was incorrectly given as Mohammad Haider Waris.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Haider, M.W., Kaur, J., Bala, R. et al. Stripe rust resistance gene(s) postulation in wheat germplasm with the help of differentials and tagged molecular markers. Sci Rep 13, 9007 (2023). https://doi.org/10.1038/s41598-023-36197-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-36197-y
This article is cited by
-
Molecular and biochemical characterization of wheat resistance to yellow rust (Puccinia striiformis f. Sp. tritici) using SSR markers and antioxidant profiles
BMC Plant Biology (2025)
-
Use of Field pathogenomics approach for Puccinia striiformis f. sp. tritici race identification and phylogenomic delineation in North India
World Journal of Microbiology and Biotechnology (2025)
-
Stripe rust resistance in historical wheat cultivars at seedling and adult plant stages and tagging effective resistance genes using molecular markers
Journal of Plant Pathology (2025)
-
Appraisal of European winter wheat against Puccinia striiformis f.sp tritici pathotypes prevalent under North Indian conditions
Cereal Research Communications (2025)
-
Host resistance responses against Puccinia striiformis f. sp. tritici in wheat cultivars with different resistance levels: molecular, biochemical, and ultrastructural studies
BMC Plant Biology (2024)