Abstract
This study focuses on developing machine learning models to detect subtle alterations in hepatocyte chromatin organization due to Iron (II, III) oxide nanoparticle exposure, hypothesizing that exposure will significantly alter chromatin texture. A total of 2000 hepatocyte nuclear regions of interest (ROIs) from mouse liver tissue were analyzed, and for each ROI, 5 different parameters were calculated: Long Run Emphasis, Short Run Emphasis, Run Length Nonuniformity, and 2 wavelet coefficient energies obtained after the discrete wavelet transform. These parameters served as input for supervised machine learning models, specifically random forest and gradient boosting classifiers. The models demonstrated relatively robust performance in distinguishing hepatocyte chromatin structures belonging to the group exposed to IONPs from the controls. The study's findings suggest that iron oxide nanoparticles induce substantial changes in hepatocyte chromatin distribution and underscore the potential of AI techniques in advancing hepatocyte evaluation in physiological and pathological conditions.
Similar content being viewed by others
Introduction
The rapid development of new technologies based on machine learning (ML) has introduced radical changes in contemporary biomedical research. Machine learning (ML) models can now be trained and tested to evaluate a variety of medical signals and use them to classify and predict both physiological and pathological phenomena1,2. These signals can be one-dimensional, such as the electrocardiogram recording, or two-dimensional, such as the radiological image, nuclear magnetic resonance image, ultrasound image or a tissue micrograph3,4,5,6. In recent years, particular attention has been given to integrating supervised machine learning strategies with the analysis of tissue structure, visualized using conventional light microscopy methods7,8.
Supervised machine learning models utilize a labeled dataset, where the input data is paired with the correct output. During the training process, these methods learn to associate inputs to outputs based on the provided labels, making predictions on new, unseen data by generalizing from the training examples. Conversely, unsupervised learning models are provided with the data without explicit instructions on what to learn, and they are then required to identify patterns and relationships. The examples of supervised learning models include various forms of decision trees, random forests and the models based on gradient boosting.
Random forest, when used for classification purposes, is an ensemble learning method that creates a variety of decision trees during training and produces the mode of the classes through majority voting. It uses bagging (bootstrap aggregating) to make different subsets of the training data for each tree, which significantly reduces the risk of overfitting and increases the robustness of the model9,10. The random forest incorporates elements of randomness by including an arbitrary subset of features for each split. Gradient boosting is also an ensemble technique, with the difference that the trees are constructed sequentially, meaning that a subsequent tree will try to correct the errors of the previous ones11,12. This approach is based on the reduction of the residual errors by optimizing a loss function. The optimization is done through gradient descent and, if properly tuned, it can achieve substantial classification accuracy comparable to that of the random forest. Textural features of digital micrographs can be used for training both models, as demonstrated in previously published research13,14. Textural analysis of digital micrographs and the use of textural features for ML model training can indeed be conducted in multiple ways. The conventional Gray-level Co-occurrence Matrix (GLCM) technique8,15,16 is widely used to quantify cell and tissue parameters such as textural homogeneity based on inverse difference moment or textural uniformity based on angular second moment. One relatively unexplored approach to textural analysis of cell structure is Run Length Matrix (RLM) analysis, which can provide valuable information about the intensity and spatial relationships of micrograph components. Run Length Matrix analysis, also known as Gray-Level Run Length Matrix (GLRLM) analysis, can extract measures of cell texture such as Short Run Emphasis (ShrtREemph), Long Run Emphasis (LngREmph), and Run Length Nonuniformity (RLNonUni), quantifiers that are closely related to two-dimensional heterogeneity and the level of textural disorder17,18. Short Run Emphasis quantifies the occurrence of short runs of pixels with similar intensity levels in a two-dimensional signal and is closely related to changes in finer texture details and uniformity. Long Run Emphasis measures the frequency of longer runs of similar pixel intensities and depends on the incidence of extended homogeneous areas in the texture. Gray Level Non-Uniformity, on the other hand, relates to the variability of gray levels in the texture. A vector of these run length matrix features can later be used as valuable input for supervised ML training in classification and regression tasks. In these models, the desired output may be a class label, such as the status of the cell or tissue (i.e., damaged or intact), or a continuous variable, such as the value of a biochemical or immunological parameter. Some successfully developed ML models encompassing GLRLM or GLCM data include random forests and gradient boosting regressor algorithms, as well as multilayer perceptron artificial neural networks13,19,20,21.
In conventional pathology and toxicology research, there is a need for developing powerful AI/ML models capable of distinguishing normal tissues from those pathologically changed, as these models can greatly contribute to modern histopathological evaluation and diagnostics22. This is especially relevant in assessing liver tissue architecture, given its unique complexity, variability, and the diverse structural features that are often challenging to interpret. Hepatocytes exhibit significant variability in their morphology after exposure to various toxins, and the specific morphological patterns related to their damage can be subtle and hard to detect23.
In previously published studies, it has been demonstrated that gray level co-occurrence matrix (GLCM) analysis is adept at quantifying subtle changes in hepatocyte chromatin after exposure to iron oxide nanoparticles24. Iron and its oxides can indeed alter hepatocyte chromatin organization and distribution, and contemporary textural analysis can be used to quantify these subtle alterations. Specifically, parameters such as angular second moment, inverse difference moment, textural contrast and textural correlation can significantly change as the result of IONP treatment. Since co-occurrence matrix angular second moment is indictor of textural uniformity and inverse difference moment being an indicator of local textural homogeneity, this would indicate that cell damage resulting from IONPs leads to the increase of structural heterogeneity of the cell nucleus. This aligns with other studies suggesting that chromatin texture could be a significant biomarker for nuclear changes resulting from various physiological and pathological processes13,25,26,27. These may include malignant transformation, osmotic shock conditions, as well as the effects of various toxic chemical agents21,28,29,30. Moreover, it has been noted that wavelet coefficient energies of hepatocyte chromatin structures, which may serve as indirect measures of nuclear textural heterogeneity, may increase after IONP exposure24. This may suggest that combining gray level matrix analysis with discrete wavelet transform may further increase the sensitivity of the approach to classify exposed and intact hepatocytes.
In this article, we hypothesize that exposure to iron oxide nanoparticles will significantly alter the chromatin organization in hepatocytes, detectable through advanced textural analysis and machine learning models. We present our findings which suggest that iron oxide nanoparticles can affect GLRLM and other characteristics of hepatocyte nucleus, to a degree that enables the effective training of machine learning classifiers, specifically random forest and gradient boosting models. We also show that these ML models are capable of distinguishing between intact hepatocytes and those exposed to iron oxide nanoparticles with robust performance.
Materials and methods
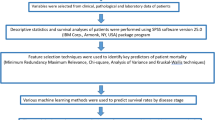
Textural analysis and ML model training were performed on a total of 2000 regions of interest of hepatocyte nuclei created on digital micrographs of mouse liver tissue. The tissue was obtained from previously conducted experiments on healthy male, 16-weeks-old C57BL/6 (C57 black 6, B6) mice in which the experimental group (N = 10) was exposed to 2 mg/kg/day i.p. of Iron (II, III) oxide nanoparticles (80–100 nm, Hongwu International Group Ltd. HWNANO materials, Guangzhou, Guangdong, CN) for 3 days while the controls (N = 10) were received saline for the same time period. The research was granted approval by the University of Belgrade's Faculty of Medicine Ethics Commission for the Protection and Welfare of Experimental Animals (approval number 229/2) as well as the Ministry of Agriculture, Forestry, and Water Management—Veterinary Division of the Republic of Serbia (approval number 323-07-07783/2020-05). The methodologies employed adhered to the standards set by the Universal Declaration of Animal Welfare (WSPA, London, 2000), the European Convention on the Protection of Vertebrates Used for Experimental and Other Scientific Purposes (1998), along with a range of national and international guidelines that advocate for the ethical and humane treatment of experimental animals.
Liver tissue samples were preserved in Carnoy's solution, a mixture of 60% ethanol, 30% chloroform, and 10% glacial acetic acid. Subsequently, these samples were embedded in Paraplast and tissue sections were created for glass slides. The sections were stained using These sections were first stained with hematoxylin to color the nuclei blue and then eosin to stain the cytoplasm and extracellular matrix pink. The staining protocol concluded with dehydration, clearing, and mounting the tissue on slides.
Digital images of the liver tissue were captured using the Pro-MicroScan DEM 200 device (Oplenic Optronics, Hangzhou, CN) connected to an OPTIC900TH Trinocular Biological Microscope (COLO LabExperts, Novo Mesto, Slovenia). These micrographs were produced at a resolution of 1200 × 1600 pixels, a 24-bit depth, and a uniform horizontal and vertical resolution of 96 dpi (Fig. 1). The micrographs were saved in BMP format for run length matrix and discrete wavelet transform analysis. The OPTIC900TH microscope features a trinocular head with an interpupillary distance of 20:80, high eye-point plan eyepiece WF 10×/20, infinity plan achromatic objective 40× (S) N.A.0.70, Abbe condenser 0.9/1.25, Kohler illuminator with field diaphragm, and a 6V 20W tungsten-halogen lamp.
A total of 1000 ROIs originating from experimental group and 1000 ROIs from controls were selected and analyzed in Mazda software (version 4.6), which was developed during the COST B11 European project titled "Quantitative Analysis of Magnetic Resonance Image Texture" and the COST B21 European project "Physiological modelling of MR Image formation." by researchers at the Institute of Electronics, Technical University of Lodz (TUL), Poland32,33,34,35. The selection of ROIs was done in a way to minimize potential bias. We did our best to ensure representative sampling of the entire tissue section by randomly choosing ROIs from different areas of the tissue. Each ROI was of uniform size to ensure consistency and the ROIs were selected to avoid regions with unclear boundaries or overlapping structures that could complicate the analysis. Observers involved in selecting ROIs were blinded to the experimental group assignments to prevent any unconscious bias in selecting ROIs from the experimental or control groups.
Mazda is a comprehensive tool designed for the calculation of texture indicators in digitized images and it enables Region of Interest (ROI) selection, techniques for feature selection and reduction (e.g. Fisher coefficients, principal component analysis, linear discriminant analysis and nonlinear discriminant analysis), preprocessing options and tools for data classification, as well as 3D data analysis. In a digital micrograph, up to 16 regions of interest can be defined and customized. For texture evaluation, the software can calculate parameters derived from histograms, co-occurrence matrices, run-length matrices, autoregressive models, and wavelet transforms32,33,34,35.
For each nuclear region of interest, we conducted run-Length matrix (RLM) analysis and discrete wavelet transform (DWT). In the RLM analysis, we assessed the variations in the intensities of resolution units within the ROIs which is crucial for understanding the texture of the two-dimensional signal. This is achieved by measuring “runs”—sequences of consecutive resolution units with the same gray-level intensities. During the RLM analysis, a specialized matrix that reflects texture anisotropy and other relational aspects within the ROI is created. In our study, particularly for machine learning applications, we focused on quantifying specific values such as Long Run Emphasis (LngREmph), Short Run Emphasis (ShrtREemph), and Run Length Nonuniformity (RLNonUni). We also focused on nuclear structures that despite being morphologically similar, have distinct values of textural features (Fig. 1). In MaZda software, the number of bits per pixel used for image quantization can be selected. Typically, 5 bits per pixel are used for RLM parameters. We used the default setting for normalization referring to the intensity range of the image under analysis, without applying “± 3 Sigma” option and “1–99%” option. The details on the mathematical and computational algorithm for quantification of textural parameters can be found in previous publications32,33,34,35.
Long Run Emphasis quantifies the prominence of longer sequences (runs) of resolution unit gray intensities in an image, and it is significantly related with the texture's coarseness. The formula for Long Run Emphasis is defined as follows:
In this formula, p(i,j) represents the frequency of runs with length j that have a gray value of i. Here, Ng is the total number of gray values, and Nr is the count of specific run lengths analyzed in the study. The coefficient C in the equation is derived from these parameters as:
Short Run Emphasis, in contrast, highlights the occurrence of shorter runs of resolution unit gray intensities, which is influenced by uniform and finer texture aspects of a two-dimensional signal. The calculation method for Short Run Emphasis is distinct and can be presented as:
Run Length Nonuniformity is related to distributional aspects of run lengths within the ROI. It is related to the textural heterogeneity in the signal. The quantification approach for Run Length Nonuniformity is as follows:
When compared to the short run emphasis, run length nonuniformity is generally of greater magnitude since it sums the squares of run lengths. This results in larger numerical values, especially in images with varied textures and longer runs. The coefficient C is consistently used across the calculations of Long Run Emphasis, Short Run Emphasis, and Run Length Nonuniformity.
During the discrete wavelet transform, after the creation of subband images at various decomposition levels, we calculated the value of a nuclear ROI wavelet coefficient energy. These energies are determined after multi-level breakdowns of two-dimensional signal data, processing them via wavelet filters. In this research, we calculated the energies (EnHL and EnLH) of a wavelet coefficient (d) using the combination of high (H) and low-pass (L) filters as follows:
where x and y are subband coordinates and n represents a number of resolution units. In the MaZda software, the transform was performed using a cascade of filters followed by factor-2 subsampling, and the wavelet features were calculated for multiple scales, with the maximum of 7 scales.
The above described RLM features (short run emphasis, long run emphasis and run length nonuniformity) and two wavelet coefficient energies were used to train and test ML models. In this study we focused on creating two decision tree—based supervised machine learning models. The first model was built on the random forest classifier10, while the second employed the gradient boosting classifier algorithm11. The random forest as a powerful and versatile ensemble method, operates by assembling a multitude (“forest”) of decision trees from data subsets after which it applies majority voting for prediction. Sample bootstrapping and other features makes it relatively resilient to overfitting and in some cases can lead to significant classification accuracy9,10,36. Potential limitations of this model include issues related to data imbalance, complexity, resource utilization and interpretability10. The second model was based on the gradient boosting classifier, also a versatile and potentially powerful ensemble technique that in a multitude of decision trees systematically corrects errors from preceding trees11,12,37. Like random forest, the potential strength of this type of supervised ML model is reduced probability of overfitting which is in this case partially achieved by stochastic gradient boosting and tree constraints. The potential weaknesses are also similar to RF model and include the relative complexity and lack of interpretability. In our study, both models were developed using Python 3, scikit-learn library38 in the Google Colaboratory environment, the hosted Jupyter notebook service, and a CPU hardware accelerator in Google Cloud's Hosted Runtime. The code for the models was previously partially developed as a part of the project SensoFracTW of the Science Fund of the Republic of Serbia based on the Saccharomyces cerevisiae cell model for evaluation of sublethal cell damage. The models were validated using a fivefold cross-validation approach by implementing through the “GridSearchCV” module from the “sklearn.model_selection” library. This required partitioning the dataset into five equal parts with four parts having been used for training the model, and the remaining part for testing. The training and testing were repeated five times and the he resulting performance metrics were averaged over the five iterations.
A comprehensive grid search was conducted to optimize hyperparameters. In the case of random forest, these were number of trees (n_estimators), the number of features considered for splitting (max_features), the maximum depth of the trees (max_depth), the minimum number of samples required to split a node (min_samples_split), the minimum number of samples required to be at a leaf node (min_samples_leaf), and the use of bootstrap samples (bootstrap). For the gradient boosting classifier, we optimized hyperparameters including the number of boosting stages (n_estimators), the learning rate, the maximum depth of the trees (max_depth), the minimum number of samples required to split a node (min_samples_split), the minimum number of samples required to be at a leaf node (min_samples_leaf), the fraction of samples used for fitting the individual base learners (subsample), and the number of features considered for splitting (max_features). For the random forest model, the best hyperparameters were n_estimators = 100; max_features = 'Auto', max_depth = 20, min_samples_split = 2, min_samples_leaf = 2, and bootstrap = True. For the gradient boosting classifier, the best-performing hyperparameters were: n_estimators = 100, learning_rate = 0.1, max_depth = 5, min_samples_split = 10, min_samples_leaf = 1, subsample = 0.8, and max_features = 'None'.
For both models we determined the values of Mattheus correlation coefficient and F1 score using the scikit-learn library 'metrics' module. The Matthews correlation coefficient (MCC) in this context can be interpreted as a measure of the quality of binary classifications. Since it is essentially a coefficient of correlation between observed and predicted binary classifications its values can range between − 1 and + 1, the + 1 indicating the perfect prediction, 0 indicating that the model is no better than random prediction, and − 1 suggesting total disagreement between prediction and observation. The advantages of MCC as a performance indicator are its balance in measurement which is useful when the data distribution in classes is imbalanced. It also incorporates symmetry in performance evaluation since it treats false positives and false negatives equally. The F1 score can be regarded as a measure of a test's accuracy in binary classification problems and it is the harmonic mean of precision and recall. Tn the context of our model performance, the precision represented the number of true positive results divided by the number of all positive results, while the recall represented the number of true positive results divided by the number of positives that should have been identified. As an addition to MCC and F1 calculation, we also determined the classification accuracy of both models,alsoalso using the scikit-learn library 'metrics' module. The same module was also used for the Receiver Operating Characteristic (ROC) analysis and the calculation of the area under the ROC curve.
Results
The average value of hepatocyte nuclear long run emphasis was 8.618 ± 0.718 in ROIs belonging to the IONP-treated group and 9.660 ± 0.883 (p < 0.01, Fig. 2) in controls, indicating that IONPs lead to a statistically highly significant reduction of this parameter (p < 0.01). This result is likely associated with reduced dominance of long, uniform intensity runs (in run length matrix), suggesting an increase in heterogeneity. Conversely, the mean values of nuclear chromatin short run emphasis were significantly increased (p < 0.01), from 0.523 ± 0.021 in controls to 0.549 ± 0.017 in the experimental group. Since the SRE is related to prevalence of short runs of similar intensities, this result would be consistent with more heterogeneous, fragmented and irregularly distributed texture. The average values of run length nonuniformity behaved similarly to the values of chromatin short run emphasis, equaling 29,138.7 ± 11,974.8 in ROIs belonging to the experimental group and 26,162.4 ± 10,694.5 in controls, with highly significant differences observed (p < 0.01). Since RLN is an indirect quantifier of the variability of run lengths this result would suggest that the distribution of run lengths in the experimental is less uniform, indicating higher textural heterogeneity.
Wavelet coefficient energies were significantly higher in hepatocyte nuclear structures belonging to the IONP-treated group compared to controls (p < 0.01). The mean value of the energy where a combination low-pass/high-pass filter (EnLH) was applied was 1.780 ± 0.199 in experimental ROIs and 1.526 ± 0.199 in controls. Similarly, the average wavelet coefficient energy for a high-pass/low-pass filter combination (EnHL) was 1.290 ± 0.157 in experimental ROIs and 1.104 ± 0.155 in controls, indicating significant differences (p < 0.01). These changes in wavelet indicators would suggest greater frequency of changes in intensity over the nuclear area which may be associated with an increased complexity in the texture of the hepatocyte nuclei.
The Gradient Boosting Tree classifier model demonstrated acceptable performance in differentiating hepatocyte nuclear ROIs belonging to the IONP-treated group from controls. The classification accuracy of this model was 74.75% on the test set and 85.06% on the training set. This indicated a moderately high level of effectiveness in correctly classifying the two groups. After receiver operating characteristics analysis, the area under the ROC curve was found to be 0.84 (Fig. 3), suggesting very good discriminatory power. The Matthews correlation coefficient for this model was 0.50 on the test set and 0.70 on the training set. The F1 score was 0.74 on the test set and 0.85 on the training set. Feature importance analysis revealed that nuclear short run emphasis was the most significant contributor to model performance, with a score of 0.37, followed by long run emphasis with a score of 0.36.
The Random Forest model, also created using the Scikit-learn library demonstrated somewhat better performance in terms of discriminatory power. The accuracy of the Random Forest model was 81.35%, and the area under the ROC curve was 0.90 (Fig. 4). Accuracy of the Random Forest model on the training set was 86.01%. The Matthews correlation coefficient for this model was 0.63 on the test set and 0.72 on the training set. The F1 score was also higher at 0.80 on the testing set and 0.86 on the training set. Feature importance analysis showed that the most critical textural parameter was nuclear long run emphasis, with a score of 0.29, closely followed by nuclear short run emphasis with a score of 0.28. Confusion matrices and classification reports for the models are provided in Figs. 5 and 6, respectively. The training performance of the models surpassed that of the test performance, although the difference was not very large. The overfitting to some degree was expected given the complexity of the models and the dataset size. This suggests that although the models captured some noise from the training data, they still generalized reasonably well to new data. This observation is particularly evident for the Random Forest model which outperformed the Gradient Boosting in other areas as well.
Discussion
In this study, we present findings indicating the feasibility of creating Gradient Boosting Classifier (GBC) and Random Forest (RF) models that can effectively differentiate between nuclear hepatocytes exposed to Iron Oxide Nanoparticles (IONPs) and their unexposed counterparts. To the best of our knowledge, this is the first study to employ Run Length Matrix (RLM) textural quantifiers as inputs for training RF and GBC models under these experimental conditions. Additionally, the unique nature of this approach is highlighted by the fact that RLM and Discrete Wavelet Transform (DWT) techniques were used to quantify nuanced alterations in hepatocyte chromatin distribution and patterns associated with IONP effects, alterations that are typically not discernible in standard histopathological evaluation. The outcomes of the study provide a strong foundation for the continued development and enhancement of artificial intelligence-based technologies for investigating the toxic effects of iron and its oxides in liver tissue.
The decision to use both random forest and gradient boosting models was made to enhance the robustness of our study, as the models, despite their similarities, have distinct characteristics. Both models use a decision tree-based approach; however, random forest averages multiple decision trees, while gradient boosting sequentially corrects errors made by previous trees. The fact that both models yielded similar results strengthened the validity of our findings. Therefore, employing both models allowed us to utilise their complementary strengths and provided a comprehensive evaluation of our machine learning approach.Iron oxide nanoparticles can affect hepatocyte chromatin in multiple ways. Direct interaction with DNA and histones may influence various transcription factors and enzyme which can further affect chromatin structure and distribution31. Epigenetic changes related to DNA methylation patterns and the expression of non-coding RNAs are also a potential effect of both iron and iron oxides. According to some authors, IONPs can also induce generation of reactive oxygen species (ROS) and subsequent oxidative damage to DNA and chromatin structure39,40,41. In our study, it is well possible that discrete alterations in chromatin distribution detected through the changes in run length matrix and wavelet parameters were the result of early chromatin condensation or changes in euchromatin/heterochromatin ratio. It is also possible that the subtle changes in nuclear structure associated with early programmed cell death may have also contributed to the observed differences in GLRLM quantifiers.
In this research, EnHL and EnLH features represented energy metrics derived from the discrete wavelet transform and we calculated them calculated from the horizontal-low/vertical-high (HL) and horizontal-high/vertical-low (LH) subbands after nuclear ROI decomposition. These energies in essence describe the distribution and intensity of vertical and horizontal variations within the hepatocyte nuclear regions. These variations can be indicative of various pathological processes such as chromatin condensation, chromatin marginalization and increased heterochromatin/euchromatin ration. For example, chromatin condensation may create more distinct and abrupt changes in intensity within the nucleus which may lead to higher wavelet frequency components being more pronounced. It may be expected that tightly packed chromatin with dense regions and sharp boundaries will have higher values of EnLH and EnHL parameters compared to the less condensed chromatin. Also, it is possible that the condensed chromatin regions can have more uniform and higher intensity compared to the surrounding areas which also affects not only RLM but also wavelet indicators. Unfortunately, these assumptions will need to be confirmed in future studies since both DWT and RLM analyses in this context remain a relatively unexplored areas of research.
In our previously published research, we employed the Gray Level Co-occurrence Matrix (GLCM) technique to describe unique and intricate changes in hepatocyte chromatin following in vivo treatment with iron oxide nanoparticles31. We observed a dose-dependent reduction in the values of GLCM inverse difference moment, indicating a substantial decrease in nuclear local textural homogeneity. Additionally, a significant reduction in angular second moment as a measure of textural uniformity was also detected. These changes were accompanied by an increase in wavelet coefficient energies, quantified by the discrete wavelet transform technique. While there are clear similarities with the current study, it is important to carefully connect the results due to methodological differences. These differences include the approaches used in textural analysis (GLCM versus GLRLM), the total doses of IONPs utilized, and the specific staining techniques employed. Nevertheless, it should be noted that both studies observed changes that could be attributed to the IONP-related increase in textural heterogeneity of hepatocyte chromatin.
In our recent, yet unpublished study on Saccharomyces cerevisiae cells, we employed GLRLM (Gray Level Run Length Matrix) analysis to detect changes in nuclear structure induced by the toxic chemical agent oxidopamine, known for its ability to induce oxidative stress under certain experimental conditions13. We observed significant changes in Short Run Emphasis (SRE), Long Run Emphasis (LRE), and Run Length Nonuniformity (RLN) indicating that these GLRLM metrics might be useful indicators of discrete changes in nuclear textural patterns caused by external toxic chemical agents. In a manner akin to our current research, measures derived from GLRLM were effectively employed in the development of classifiers using both Random Forest and Gradient Boosting Tree methods. These classifiers demonstrated comparable performance in aspects such as classification accuracy and the area under the Receiver Operating Characteristic (ROC) curve. In future research, it may be crucial to investigate IONP-related GLRLM (Gray Level Run Length Matrix) and DWT (Discrete Wavelet Transform) changes separately for perivenous and periportal hepatocytes. These two types of hepatocytes, while having almost identical morphological parameters, play distinct physiological roles and could vary in the values of textural indicators. Additionally, these cell populations differ in their susceptibility to various types of liver damage, with some toxins preferentially affecting periportal hepatocytes due to their first contact with incoming blood. Furthermore, future studies might also focus on investigating GLRLM and DWT changes in the entire hepatic tissue cytoarchitecture, rather than in individual cells and nuclei. Previous research that attempted to develop Random Forest and Classification and Regression Tree (CART) algorithms based on GLCM indicators for differentiation of hepatic tissue exposed to iron oxide nanoparticles yielded mixed results14. However, it is still unknown whether such models, including Gradient Boosting Trees (GBT), can be used to detect wider structural changes in liver tissues associated with the toxic effects of iron and iron oxides. In our current study machine learning classifiers were created solely for classifying nuclear ROIs, and additional training or revision may be required to apply them to tissue micrographs and animal samples.
Also, it will be necessary to further broaden these approaches to better prevent, manage, and mitigate potential overfitting of the random forest and gradient boosting models. In addition to a comprehensive analysis comparing the training and testing performance metrics, external validation with an independent dataset should be conducted to demonstrate the robustness of the machine learning models. Our current study represents the first instance of using Run Length Matrix and Discrete Wavelet Transform analysis in this specific setting, and consequently, there is no available external independent dataset for comparison. For future work, we plan to collect additional data on the discrete changes in hepatocyte chromatin distribution to facilitate external validation and further demonstrate the potential applicability of our models. Several limitations were present in our research that require attention. First, the fact that random forest and gradient boosting algorithms are two contemporary ML techniques does not inherently guarantee their excellence. These methods have been chosen due to their proven effectiveness in handling textural features as input data as demonstrated in previous works36. Nevertheless, it should be acknowledged that other supervised machine learning methods not only exist but may also surpass decision tree—based approaches in certain situations. As mentioned earlier, probably a prime example would be the versatile family artificial neural networks encompassing multilayer perceptrons and convolutional neural networks which are especially adept in computer vision applications42,43,44.
The second limitation of our study concerns the relatively limited size of the sample used for training and testing the random forest and gradient boosting models. The use of only 20 mice (10 treated and 10 controls) is a significant factor that may reduce the ability of the model to capture the variability in biological responses, as well as to generalize the findings to wider and more diverse experimental settings.The 2000 ROIs were carefully selected to capture a wide range of variability in hepatocyte chromatin structures, ensuring that the models are exposed to diverse patterns during training. When combined with techniques like cross-validation, a sample of this size should be sufficient for training robust machine learning models based on random forest and gradient boosting algorithms. Nevertheless, future studies should aim to acquire a larger ROI sample to enhance the classification accuracy of RF and GBT classifiers, as well as to successfully train and test more complex models like those based on neural networks. Despite the applied strategies to minimize bias, as explained in the Methodology section of this article, it must be acknowledged that manual selection of ROIs can introduce subjective bias. In the future, it may be important to consider the introduction of automated image analysis software to identify and select ROIs based on predefined criteria.
Also, it should be noted that the RF and GBT classifiers in our study were developed solely to classify nuclear ROIs and for these models to be applicable on tissue micrograph and animal samples, additional training and validation, or even the complete revision of the entire computational approach might be required. Additionally, both random forest and gradient boosting models can be seen as somewhat "black box" in nature, as their inner workings, despite their demonstrated performance, are difficult to describe and interpret. Future work could apply advanced model interpretability techniques such as SHAP (SHapley Additive exPlanations) or LIME (Local Interpretable Model-agnostic Explanations). These approaches could contribute to the identification of the factors that are most influential in the decision-making process of the models, thereby improving the transparency of the machine learning and making the future models more understandable to the researchers in the fields of pathology and histology29.
Lastly, the Run Length Matrix (RLM) and the Discrete Wavelet Transform (DWT) methods themselves may suffer from inherent weaknesses that limit their reproducibility and application in machine learning. Similar to other texture analysis techniques, such as the Gray Level Co-occurrence Matrix (GLCM), their results can vary significantly based on the software platform used for signal analysis. The use of different histological stains, or even variances in a single staining protocol, may radically change the values of GLRLM indicators such as Short Run Emphasis (SRE), Long Run Emphasis (LRE), and Run Length Nonuniformity (RLN). Going forward, it will be crucial to test the validity of both GLRLM and DWT methods across various pathohistological protocols and experimental conditions before drawing definitive conclusions about their utility and value in current research practices.
In conclusion, our findings indicate that measures of hepatocyte chromatin texture can be used to train decision tree-based supervised machine learning models capable of distinguishing nuclear regions of interest of exposed and intact cells. Quantifiers of Gray-Level Run Length Matrix such as Short Run Emphasis, Long Run Emphasis, and Run Length Nonuniformity are useful input data for training these models, each with varying degrees of feature importance. The combination of GLRLM parameters with Discrete Wavelet Transform parameters, such as wavelet coefficient energies, is a potentially effective approach in the development of Random Forest and Gradient Boosting Tree classifiers. The results and computer code obtained during the creation of RF and GBT models represent a valuable foundation for further development of artificial intelligence-based biosensors capable of detecting discrete nuclear changes in hepatocyte chromatin following exposure to toxic chemical agents. The outcomes and computer code generated in the development of the RF and GBT models provide a potentially valuable basis for the future advancement of artificial intelligence-based biosensors aimed at detecting subtle nuclear changes in hepatocyte chromatin after exposure to toxic agents.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Dimitriadis, I., Zaninovic, N., Badiola, A. C. & Bormann, C. L. Artificial intelligence in the embryology laboratory: A review. Reprod. Biomed. Online https://doi.org/10.1016/j.rbmo.2021.11.003 (2021).
Itto-Nakama, K. et al. Prediction of ethanol fermentation under stressed conditions using yeast morphological data. J. Biosci. Bioeng. 135, 210–216. https://doi.org/10.1016/j.jbiosc.2022.12.008 (2023).
Idowu, S. O. & Fatokun, A. A. Artificial intelligence (AI) to the rescue: Deploying machine learning to bridge the biorelevance gap in antioxidant assays. SLAS Technol. 26, 16–25. https://doi.org/10.1177/2472630320962716 (2021).
Kim, D., Min, Y., Oh, J. M. & Cho, Y. K. AI-powered transmitted light microscopy for functional analysis of live cells. Sci. Rep. 9, 18428. https://doi.org/10.1038/s41598-019-54961-x (2019).
Corridon, P. R., Wang, X., Shakeel, A. & Chan, V. Digital technologies: Advancing individualized treatments through gene and cell therapies, pharmacogenetics, and disease detection and diagnostics. Biomedicines. https://doi.org/10.3390/biomedicines10102445 (2022).
Pantic, I. V., Shakeel, A., Petroianu, G. A. & Corridon, P. R. Analysis of vascular architecture and parenchymal damage generated by reduced blood perfusion in decellularized porcine kidneys using a gray level co-occurrence matrix. Front. Cardiovasc. Med. 9, 797283. https://doi.org/10.3389/fcvm.2022.797283 (2022).
Liu, Q. et al. A histopathologic image analysis for the classification of endocervical adenocarcinoma silva patterns depend on weakly supervised deep learning. Am. J. Pathol. 194, 735–746. https://doi.org/10.1016/j.ajpath.2024.01.016 (2024).
Pantic, I. V. et al. Computational approaches for evaluating morphological changes in the corneal stroma associated with decellularization. Front. Bioeng. Biotechnol. 11, 1105377. https://doi.org/10.3389/fbioe.2023.1105377 (2023).
Hansch, R. Handbook of Random Forests: Theory and Applications for Remote Sensing (World Scientific Publishing Company Pte Limited, 2024).
Yifan, C., Jianfeng, S. & Jun, P. Development and validation of a random forest diagnostic model of acute myocardial infarction based on ferroptosis-related genes in circulating endothelial cells. Front. Cardiovasc. Med. 8, 663509. https://doi.org/10.3389/fcvm.2021.663509 (2021).
Ahn, J. M., Kim, J. & Kim, K. Ensemble machine learning of gradient boosting (XGBoost, LightGBM, CatBoost) and attention-based CNN-LSTM for harmful algal blooms forecasting. Toxins. https://doi.org/10.3390/toxins15100608 (2023).
Kefalas, A. et al. Estimation of combustion parameters from engine vibrations based on discrete wavelet transform and gradient boosting. Sensors (Basel). https://doi.org/10.3390/s22114235 (2022).
Pantic, I. & Paunovic, J. Artificial intelligence approaches for the detection of discrete nuclear alterations induced by oxidopamine: A comparative study of random forest and gradient boosting techniques. Chemico-biol. Interact. (2024) (under review).
Paunovic Pantic, J., Vucevic, D., Pantic, I., Valjarevic, S. & Radosavljevic, T. Development of random forest machine learning model for the detection of changes in liver tissue after exposure to iron oxide nanoparticles. Med. Investig. V57 (2024) (in press).
Valjarevic, S. et al. Gray-level co-occurrence matrix analysis of nuclear textural patterns in laryngeal squamous cell carcinoma: Focus on artificial intelligence methods. Microsc. Microanal. 29, 1220–1227. https://doi.org/10.1093/micmic/ozad042 (2023).
Chen, X. et al. Differentiation of true-progression from pseudoprogression in glioblastoma treated with radiation therapy and concomitant temozolomide by GLCM texture analysis of conventional MRI. Clin. Imaging 39, 775–780. https://doi.org/10.1016/j.clinimag.2015.04.003 (2015).
Mishra, A. et al. Role of textural analysis parameters derived from FDG PET/CT in differentiating hepatocellular carcinoma and hepatic metastases. Nucl. Med. Commun. 44, 381–389. https://doi.org/10.1097/MNM.0000000000001676 (2023).
Ouyang, Z. et al. Dynamic PET imaging using dual texture features. Front. Comput. Neurosci. 15, 819840. https://doi.org/10.3389/fncom.2021.819840 (2021).
Anand, L. et al. Diagnosis of prostate cancer using GLCM enabled KNN technique by analyzing MRI images. Biomed. Res. Int. 2023, 3913351. https://doi.org/10.1155/2023/3913351 (2023).
Pantic, I. et al. Artificial neural networks in contemporary toxicology research. Chem. Biol. Interact. 369, 110269. https://doi.org/10.1016/j.cbi.2022.110269 (2023).
Pantic, I. et al. Gray level co-occurrence matrix, fractal and wavelet analyses of discrete changes in cell nuclear structure following osmotic stress: Focus on machine learning methods. Fractal Fract. 7, 272 (2023).
Pantic, I., Cumic, J., Dugalic, S., Petroianu, G. A. & Corridon, P. R. Gray level co-occurrence matrix and wavelet analyses reveal discrete changes in proximal tubule cell nuclei after mild acute kidney injury. Sci. Rep. 13, 4025. https://doi.org/10.1038/s41598-023-31205-7 (2023).
Paunovic, J. et al. Gray-level co-occurrence matrix analysis of chromatin architecture in periportal and perivenous hepatocytes. Histochem. Cell Biol. 151, 75–83. https://doi.org/10.1007/s00418-018-1714-5 (2019).
Paunovic, J. et al. Effects of iron oxide nanoparticles on structural organization of hepatocyte chromatin: Gray level co-occurrence matrix analysis. Microsc. Microanal. 27, 889–896. https://doi.org/10.1017/S1431927621000532 (2021).
Dos Santos, J. B. et al. Nuclear morphometry and chromatin texture changes in hepatocellular carcinoma samples may predict outcomes of liver transplanted patients. BMC Gastroenterol. 22, 189. https://doi.org/10.1186/s12876-022-02262-5 (2022).
Kouzine, F. & Levens, D. The texture of chromatin. Cell 179, 579–581. https://doi.org/10.1016/j.cell.2019.09.026 (2019).
Lee, H. K. et al. A paradigm shift in nuclear chromatin interpretation: From qualitative intuitive recognition to quantitative texture analysis of breast cancer cell nuclei. Cytometry. Part A J. Int. Soc. Anal. Cytol. 99, 698–706. https://doi.org/10.1002/cyto.a.24260 (2021).
Nikolovski, D., Cumic, J. & Pantic, I. Application of gray level co-occurrence matrix algorithm for detection of discrete structural changes in cell nuclei after exposure to iron oxide nanoparticles and 6-hydroxydopamine. Microsc. Microanal. 25, 982–988. https://doi.org/10.1017/S1431927619014594 (2019).
Davidovic, L. M. et al. Gray-level co-occurrence matrix analysis for the detection of discrete, ethanol-induced, structural changes in cell nuclei: An artificial intelligence approach. Microsc. Microanal. https://doi.org/10.1017/S1431927621013878 (2021).
Dincic, M. et al. The fractal and GLCM textural parameters of chromatin may be potential biomarkers of papillary thyroid carcinoma in Hashimoto’s thyroiditis specimens. Microsc. Microanal. 26, 717–730. https://doi.org/10.1017/S1431927620001683 (2020).
Paunovic, J. et al. Effects of iron oxide nanoparticles on structural organization of hepatocyte chromatin: Gray level co-occurrence matrix analysis. Microsc. Microanal. 27, 1–8. https://doi.org/10.1017/S1431927621000532 (2021).
Szczypinski, P., Strzelecki, M. & Materka, A. In Proc. of ISITC 2007, November 23–23, 2007 245–249 (Republic of Korea, 2007).
Szczypinski, P., Strzelecki, M., Materka, A. & Klepaczko, A. MaZda-A software package for image texture analysis. Comput. Methods Progr. Biomed. 94, 66–76 (2009).
Kociołek, M., Materka, A., Strzelecki, M. & Szczypinski, P. In Proc. of Interational Conference onSignals and Electronic Systems, 18–21 September 2001. 163–168.
Strzelecki, M., Szczypinski, P., Materka, A. & Klepaczko, A. A software tool for automatic classification and segmentation of 2D/3D medical images. Nucl. Instrum. Methods Phys. Res. A 702, 137–140 (2013).
Pantic, I., Topalovic, N., Corridon, P. R. & Paunovic, J. Oxidopamine-induced nuclear alterations quantified using advanced fractal analysis: Random forest machine learning approach. Fractal Fract. 7, 771 (2023).
Qin, L. et al. Machine learning-based gray-level co-occurrence matrix (GLCM) models for predicting the depth of myometrial invasion in patients with Stage I endometrial cancer. Cancer Manag. Res. 14, 2143–2154. https://doi.org/10.2147/CMAR.S370477 (2022).
Pedregosa, F. et al. Scikit-learn: Machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Ghosh, S., Ghosh, I., Chakrabarti, M. & Mukherjee, A. Genotoxicity and biocompatibility of superparamagnetic iron oxide nanoparticles: Influence of surface modification on biodistribution, retention, DNA damage and oxidative stress. Food Chem. Toxicol. 136, 110989. https://doi.org/10.1016/j.fct.2019.110989 (2020).
Ranjbary, A. G. et al. Superparamagnetic iron oxide nanoparticles induce apoptosis in HT-29 cells by stimulating oxidative stress and damaging DNA. Biol. Trace Elem. Res. 201, 1163–1173. https://doi.org/10.1007/s12011-022-03229-z (2023).
Siddiqui, M. A. et al. Iron oxide nanoparticles induced cytotoxicity, oxidative stress, cell cycle arrest, and DNA damage in human umbilical vein endothelial cells. J. Trace Elem. Med. Biol. Organ Soc. Miner. Trace Elem. 80, 127302. https://doi.org/10.1016/j.jtemb.2023.127302 (2023).
Lopez-Sanchez, M., Hernandez-Ocana, B., Chavez-Bosquez, O. & Hernandez-Torruco, J. Supervised deep learning techniques for image description: A systematic review. Entropy. https://doi.org/10.3390/e25040553 (2023).
Salahuddin, Z., Woodruff, H. C., Chatterjee, A. & Lambin, P. Transparency of deep neural networks for medical image analysis: A review of interpretability methods. Comput. Biol. Med. 140, 105111. https://doi.org/10.1016/j.compbiomed.2021.105111 (2022).
Zhao, Y., Wang, X., Che, T., Bao, G. & Li, S. Multi-task deep learning for medical image computing and analysis: A review. Comput. Biol. Med. 153, 106496. https://doi.org/10.1016/j.compbiomed.2022.106496 (2023).
Acknowledgements
This research was supported by the Science Fund of the Republic of Serbia, Grant No. 7739645 “Automated sensing system based on fractal, textural and wavelet computational methods for detection of low-level cellular damage”, SensoFracTW. The author(s) acknowledge support of the The Ministry of Science, Technological Development and Innovation of the Republic of Serbia, Grant No. 451-03-66/2024-03/200110, subgrant entitled "Development of artificial intelligence models based on the random forest algorithm for the detection of discrete structural changes in the cell nucleus" (Head of the subgrant Prof. Igor Pantić). Prof. Peter Corridon acknowledges funding from the Khalifa University of Science and Technology under Award No. RC2-2018-022 (HEIC) and Research Fund FSU-2020-25.
Author information
Authors and Affiliations
Contributions
Conceptualization: J.P.P., I.P., P.R.C., S.V., J.C., L.B.; data curation and methodology: I.P.; analysis and model training: I.P.; software: I.P.; writing—original draft preparation: I.P., J.P.P., D.V., T.R.; supervision: I.P., D.V., T.R.; resources: S.V., J.C., I.P.; writing—review and editing: J.P.P., I.P., P.R.C., S.V., J.C., T.R., D.V, L.B.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Paunovic Pantic, J., Vucevic, D., Radosavljevic, T. et al. Machine learning approaches to detect hepatocyte chromatin alterations from iron oxide nanoparticle exposure. Sci Rep 14, 19595 (2024). https://doi.org/10.1038/s41598-024-70559-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-70559-4