Abstract
Several reports have presented that balanced chromosomal rearrangements (BCRs) carriers with normal phenotypes may be carriers of complex rearrangements. However, the incidence and PGT clinical outcomes of cryptic complex chromosome rearrangements (CCCRs) in individuals with BCRs is remain unknown. We recruited a cohort of 1,264 individuals with BCR carriers from 2016 to 2021 at the Reproductive and Genetic Hospital of CITIC Xiangya. Peripheral blood was collected for karyotyping and genomic DNA extraction and the PGT-SR clinical outcomes of CCCRs carriers were analyzed and compared with those of BCR carriers. Our findings revealed that 3.6% (45/1,264) of BCR carriers had CCCRs, involving 3–25 breakpoints on 1–3 chromosomes. Furthermore, when mate-pair sequencing was employed, 63.3% (19/30) of CCCR carriers were found to have chromosome rearrangements that were different from those identified by the MicroSeq technique. And the transferable embryo rate of CCCR carriers with 3 chromosomes was significantly lower than that of CCCR carriers with only 1–2 chromosomes. In this research, we revealed that some of the BCR carriers were actually CCCR carriers, and the prognosis of PGT in CCCR carriers with one or two chromosomes is better than that of CCCR carriers with three chromosomes.
Similar content being viewed by others
Introduction
Chromosomal structural variations are intricate rearrangements within the structure of chromosomes, with consequences that can range from benign to pathogenic1. These alterations can emerge during fundamental processes such as DNA replication, recombination, or repair2,3. The most important mechanisms of chromosomal structural rearrangements are non-allelic homologous recombination (NAHR), non-homologous end-joining (NHEJ), fork stalling and template switching (FoSTeS), and microhomology-mediated Break-induced replication (MMBIR). They are notably detected in the context of prenatal diagnosis, describing possible risks to the developing fetus4,5,6, in children with developmental abnormalities where they can be a cause of these issues 7,8,9, in couples facing recurrent miscarriages, where they may explain repeated pregnancy losses10, and in cases of male infertility, where they can be an underlying factor contributing to reproductive challenges11.
These chromosome structural rearrangements can be broadly classified into two principal categories: unbalanced and balanced chromosome rearrangements (BCRs). Unbalanced rearrangements, described by deletions or duplications, often manifest as discernible changes in an individual’s traits, whereas BCRs, which encompass translocations, inversions, and insertions, tend not to result in a net gain or loss of genetic material and usually do not give rise to conspicuous physical alterations in affected individuals12. While BCRs are relatively rare, it is important to note that they can be linked to abnormal phenotypes, including congenital abnormalities and reproductive issues such as infertility or miscarriages13.
Complex chromosomal rearrangements (CCR) are rare chromosome rearrangements involving more than two chromosomes or more than two breakpoints in one chromosome14. Segregation during meiosis in CCR carriers is the main driver of structural alterations, generating unbalanced gametes containing random combinations of derivative and normal homologues; the probability of producing gametes with balanced genomes decreases as the number of chromosomes involved in the rearrangements increases. The general risk for spontaneous abortions for CCR carriers is 77.6% and 9.7% for an affected child. Most unbalanced CCRs are often associated with intellectual disability15. There is a direct relationship between the number of breakpoints detected and the resolution of the analysis method used to assess CCRs because when the resolution increases, the identified number of breakpoints also increases16. CCR patients commonly seek help from assisted reproduction technology (ART) and preimplantation genetic testing (PGT), which can identify balanced euploid embryos for intrauterine transplantation and subsequent development into a healthy infant17.
Although case reports have indicated the presence of CCCR in BCR carriers18, no comprehensive reports on the prevalence, features, and PGT clinical outcomes of CCCR in BCR carriers have been documented. This study employed MicroSeq technology to investigate chromosome rearrangements in 1,264 BCR carriers and revealed that 3.6% of them had CCCRs. Furthermore, the characteristics of these CCCRs and the clinical outcomes of PGT were also analyzed.
Results
The incidence and classification of CCCR in BCR carriers
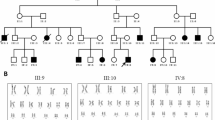
In this study, MicroSeq was used to analyze 1,264 BCR carriers previously identified through G-banding karyotyping. Interestingly, it was observed that 3.6% (45 out of 1,264) of BCR carriers had CCCR. Among them, 3 inversion carriers (8.1%, 3/37) were found to have cryptic intrachromosomal CCCRs. These CCCRs do not affect other chromosomes, but there are microdeletions or microduplications at the breakpoints. The other 42 BCR carriers (3.4%, 42/1,225) had interchromosomal CCCRs. Among them, 11 cases (26.2%, 11/42) had CCCRs that involved chromosomes other than those involved in balanced translocations. The remaining 31 cases (73.8%, 31/42) had CCCRs that exclusively involved translocation chromosomes (Supplementary Table S1) (Fig. 1). Our results suggest that the presence of CCCR is not uncommon in G-banding-characterized BCR carriers.
CCCR can often cause copy number variation of more than 100 kb
Our MicroSeq data revealed that one out of the three inversion carriers with CCCR had a 480 kb gap, suggesting the occurrence of a microdeletion. Additionally, overlaps of 300 kb were observed in the breakpoint regions of one inversion carrier with CCCR, indicating the presence of a microduplication. Out of the other 42 CCCR carriers, 16 cases exhibited microdeletions ranging from 100 kb to 8 Mb within the breakpoint regions. Additionally, 5 cases showed microduplications ranging from 100 to 400 kb within the breakpoint regions. No copy number variations (CNVs) were detected in the breakpoint regions of the remaining 21 CCCR carriers (Fig. 2 and Supplementary Table S2). Taken together, 23 out of 45 CCCR carriers (51.1%) were found to have CNVs larger than 100 kb in their breakpoint regions. We reviewed these CNVs based on the ACMG guidelines (Riggs et al. 2020) and found that all of them were CNVs of uncertain significance. Out of the 17 CCCR carriers with microdeletions, 13 cases (76.5%) showed microdeletion regions containing at least one disease-related gene as documented by OMIM (Supplementary Table S2). This finding indicates that microdeletions resulting from CCCR may increase the vulnerability to genetic diseases for CCCR carriers and their offspring.
CCCR increases the genetic risk by interrupting more genes in carriers
A total of 2689 breakpoints were identified in 1264 cases of BCR, with 226 breakpoints interrupting disease-related genes (AR, AD, and others) recorded in OMIM. To assess the genetic risks associated with gene interruption in carriers of BCR (1219 cases) and CCCR (45 cases), we conducted an analysis of the average number of interrupted disease-related genes per carrier. Our findings revealed that 1219 BCR carriers exhibited interruptions in 205 disease-related genes (0.17 genes per carrier), whereas 45 CCCR carriers showed interruptions in 18 disease-related genes (0.40 genes per carrier) (Table 1).
Structural variation spectrum of CCCRs
In the 45 CCCR carriers, we used mate-pair sequencing to characterize the CCCR in 30 of them. The remaining 15 cases were not subjected to mate-pair sequencing due to the lack of DNA samples. We identified a total of 192 rearrangement breakpoints, with an average of 6.4 (3–25) breakpoints and a maximum of 25 breakpoints. Fourteen cases showed “extreme” complex rearrangements. Among them, three cases were found to have the most breakpoints: MD19029 (25 breakpoints) (Fig. 3), MD19079 (18 breakpoints), and MD19237 (16 breakpoints). On the other hand, our results demonstrated that four cases exhibited fewer breakpoints when analyzed using mate-pair sequencing compared to characterization by MicroSeq. This may be due to the breakpoints being in close proximity to the centromere regions (Supplementary Table S3).
CCCR within 1 or 2 chromosomes does not affect the PGT clinical outcomes
The study evaluated the clinical outcome of PGT-SR by calculating the transferable embryo rate, as BCR typically leads to chromosomal disorders due to meiotic impairment. The cohort was categorized into three groups based on the complexity of the rearrangements: BCR carriers with 2 breakpoints, CCCR carriers involving only 1–2 chromosomes, and CCCR carriers involving more than 2 chromosomes. Interestingly, there is no significant difference in the transferable embryo rate between CCCR carriers involving only 1–2 chromosomes and BCR carriers with 2 breakpoints (31.0% vs. 32.2%, p > 0.05). However, the transferable embryo rate of CCCR carriers involving more than 2 chromosomes was significantly lower than that of CCCR carriers involving only 1–2 chromosomes (7.5% vs. 31.1%, p < 0.01) (Table 2).
Discussion
BCR is not rare in the general population, with an incidence of balanced translocations occurring in approximately 1 in 500 to 1 in 625 individuals, Robertsonian translations occurring in about 1 in 1000 individuals, inversions affecting approximately 1 to 2 in 1000 people, and insertions occurring in around 0.3 in 1000 individuals19,20. However, complex chromosome rearrangements (CCRs) are extremely rare in the general population, while spontaneous miscarriages and developmental abnormalities in offspring resulting from chromosomal imbalances are prevalent17,21. Despite occasional case reports of BCR carriers being identified as cryptic complex chromosome rearrangement (CCCR) carriers18,22,23, there is currently a lack of comprehensive data on the prevalence of CCCR among BCR carriers. In this investigation, 1264 carriers of BCR were examined using molecular cytogenetic techniques. The analysis identified 45 cases (3.6%) as CCCR carriers, with 34 cases (75.6%) showing CCCR exclusively involving BCR-related chromosomes, and the remaining 11 cases (24.4%) involving other chromosomes as well as the BCR chromosomes. Furthermore, 48.9% CCCR carriers (21 translocation carriers and 1 inversion carrier, 22/45) exhibit no CNVs, indicating they are apparently balanced chromosome rearrangement (ABCR) carriers. Our study suggests that CCCR is not rare among carriers of BCR, with the majority of CCCRs being intricate rearrangements involving only BCR-related chromosomes, and maybe only half of them are ABCRs.
CCCR is very common in cancers. The mechanism of CCCR formation is very complex. It may result from chromoanagenesis, a process of chaotic shattering and restructuring of chromosomes. Chromoanagenesis may give rise to stable and heritable complex chromosomal rearrangements (CCCRs). These CCCR events often involve extensive chromosome breakage, frequently accompanied by copy number variations (CNVs). Chromosome breakage can result from mitotic failure or other stress, leading to the random reassembly of fragmented chromosomes in complex configurations24.
The characteristics of de novo CNV flanking sequences indicate potential mechanisms for CNV, including breakage-fusion-bridge cycles (BFB), non-homologous end joining (NHEJ), non-allelic homologous recombination (NAHR), microhomology-mediated end joining (MMEJ), microhomology-mediated break-induced replication (MMBIR), and fork-stalling and template switching (FoSTeS)25. Our previous study showed that 54% of rearrangement breakpoints of BCRs had 1–8 bp of microhomologous sequence at the breakpoints, and 30% of chromosome breaks of BCRs were blunt end junctions, indicating that MMBIR and NHEJ were the main mechanisms of BCR. Furthermore, we previously observed that BCR is typically not associated with larger CNVs in proximity to chromosomal breakpoints26. However, this study showed that 51.1% of CCCR carriers exhibited CNVs larger than 100 kb at the sites of chromosome breakage. Our results suggest that the mechanism of CCCR formation may be more complex than that of BCR.
Gene disruption is a common outcome of chromosomal rearrangements caused by chromosomal breaks. Our previous investigation revealed that 45.9% of balanced translocations result in gene disruption26. The study uncovered that CCCR not only induces a higher frequency of gene breaks in carriers, but also frequently coincides with chromatin loss near the chromosome break site. This occurrence may result in haploinsufficiency of these individual genes in CCCR carriers, indicating that CCCR might contribute to increased genetic risks. Several case reports have demonstrated that individuals with CCRs suffer from genetic diseases caused by chromosome breakage12,27. Therefore, when assessing the genetic risk for offspring of CCCR carriers, it is essential to comprehensively consider both the gene disruption resulting from chromosomal breakpoints and the CNV caused by CCCR. CCCR carriers should be advised to undergo testing for pathogenic mutations in the corresponding genes of their spouses to accurately evaluate the risk of their offspring developing related genetic diseases if they are also CCCR carriers.
This study uncovers instances where disease-associated genes disrupted by balanced chromosomal rearrangements (BCRs) do not manifest any phenotypic anomalies. When a BCR disrupts the AR gene while the other allele remains intact, the individual will display a normal phenotype. Furthermore, individuals with BCR-disrupted AD genes exhibited normal phenotypes in this investigation. Potential explanations include the degradation of truncated proteins, the lack of dosage sensitivity in certain genes, and the onset of diseases resulting from truncation mutations in specific genes showing late-onset or incomplete penetrance. It is noteworthy that case MD19051 involves an 8 Mb BCR-caused deletion at 4p15.33p15.2. According to the ACMG guidelines, the CNV score is only 0.6, indicating it is a variant of uncertain significance (VUS). Follow-up assessments identified mild learning difficulties in the patient. Nevertheless, the connection between the patient’s learning challenges and the CNV remains undetermined.
Our research suggests that MicroSeq can reliably determine the CCCRs that involve BCR-related chromosomes. However, MicroSeq’s limitation in evaluating only chromosomal breakage sites hinders the accurate assessment of CCCR patterns and the detection of CCRs that are not related to chromosome rearrangements in BCR carriers. Hence, a more precise CCCR assessment using genome-wide detection techniques is essential. Previous studies have shown that mate pair sequencing, Bionano technology, and long-read sequencing technique can accurately determine CCCR at the whole-genome level23,28,29. Our study shows that mate pair sequencing can effectively identify the presence of most CCCRs throughout the genome, along with the orientation of CCCR rearrangement fragments. However, its detection capability is diminished in regions with complex genome sequences, particularly those proximal to the centromere. Our previous study has also demonstrated the limited capacity of long-read NGS technology in detecting chromosomal rearrangements within intricate genomic sequences. This requires a careful approach when interpreting results obtained from these techniques for detecting chromosome rearrangements.
CCR carriers are not recommended for natural pregnancy due to their high tendency to produce imbalanced gametes. Reproductive solutions suitable for CCR carriers include preimplantation genetic testing. Existing literature indicates that the clinical outcomes of PGT for CCR carriers are significantly poorer compared to those of BCR carriers17. This study unexpectedly discovered the presence and frequency of CCCR in carriers of BCR. CCCR can be categorized as those exclusively involving BCR-related chromosomes and those involving chromosomes not related to BCR. Our study suggests that there is no statistically significant difference in PGT clinical outcomes between carriers of CCCR involving only BCR-related chromosomes and BCR carriers. However, carriers of CCCR involving other chromosomes exhibit significantly worse PGT clinical outcomes. These results indicate that the fertility outcomes of CCCR carriers are associated with the number of chromosomes involved in CCCR, rather than the complexity and number of breakpoints on the same chromosome.
While MicroSeq technology accurately detects the presence of CCCR near the chromosomal breakpoint of BCR carriers, it is unable to identify CCCR in other areas of the genome. Furthermore, given the low incidence of CCCR, there is a requirement for extensive multicenter data to precisely assess the prevalence of CCCR in BCR carriers. Likewise, while our study did not identify any statistically significant difference in PGT clinical outcomes between CCCR carriers with exclusively BCR-related chromosomes involved and BCR carriers, this conclusion requires validation with a larger sample size.
Nevertheless, our study indicates that CCCR is not rare in carriers of BCR identified by karyotyping, and they potentially present an increased genetic risk. The fertility outcomes of carriers with CCCRs involving only BCR-related chromosomes are similar to those of BCR carriers. However, if CCCR involves chromosomes other than BCR, the fertility outcomes of such carriers will significantly deteriorate. The accurate identification and analysis of CCCR is beneficial for evaluating genetic risk and selecting assisted reproduction techniques for CCCR carriers.
Materials and methods
Study subjects
All methods were performed in accordance with the relevant guidelines and regulations. The study was conducted after approval by the Reproductive and Genetic Hospital of CITIC Xiangya Ethics Committee (LL-SC-2019-021) and informed consent was obtained from the BCR carriers, including 1225 cases of balanced translocations, 37 cases of inversions, and 2 cases of insertions. Robertsonian translocations were excluded from this study. All BCR carriers experienced infertility, either primary or secondary, and/or spontaneous abortion after marriage, without any substantial phenotypic abnormalities being observed in their clinical records.
Genomic DNA preparation
Genomic DNA from the patients’ peripheral blood was extracted using Qiagen Blood DNA Kit (Qiagen) according to the manufacturer’s instructions. The quality and quantity of the extracted DNA were assessed using NanoDrop 1000 Spectrophotometer and agarose gel electrophoresis.
Modified MicroSeq technique
G-banded karyotyping was conducted. The current version of the International System for Human Cytogenomic Nomenclature (2020) was used for karyotype designation. Chromosome microdissection-based next-generation sequencing (MicroSeq) was conducted as previously described with specific modifications30. Briefly, 10 μL of nucleic acid-free sterilized water was added to the split phase region before chromosome microdissection. After the chromosome microdissection, either the PicoPLEX or WGA4 kit were utilized to simplify the amplification process. For the construction of the sequencing library, the Ion Xpress™ Plus Fragment Library Kit from Thermo Fisher Scientific was employed, following the manufacturer’s provided instructions. Subsequently, the library DNA underwent amplification using the Ion PGM™ Template OT2 200 Kit (Thermo Fisher Scientific), which was conducted for a duration of 5.5 h as per the manufacturer’s recommendations.
Mate-pair sequencing
In order to obtain large fragments of 2000–5000 bp DNA, 1.0 ug genomic DNA was subjected to ultrasonic treatment in a Covaris M220 focused ultrasound instrument according to the manufacturer’s operating procedures. AMPure XP beads (Agencourt, CA, USA) were used to select DNA fragments between 2000 and 3000 bp. Then, the DNA fragments were subjected to end repair and 3’-terminal labeling with biotinylated nucleotides. The repaired DNA fragments were subsequently selected by 0.6% trillion base agarose gel electrophoresis.
To constructing a paired-end library, the 2–3 kb DNA fragments are cyclized through intramolecular ligation31. Any remaining linear molecules are removed by DNA exonuclease treatment. Circular DNA fragments were cut again using the Covaris S2 to produce fragments of 350 bp. Then, the streptavidin-coated magnetic beads were used to purify the biotinylated fragments. These fragments are subjected to end repair and A tailing to prepare for the connection to Illumina paired end oligomer adapters. Agilent Bioanalyzer DNA 2100 Kit (Agilent, California, USA) and Qubit 3.0 (Invitrogen, Carlsbad, California, USA) were used to ensure the quality of the constructed library. Finally, the Illumina NovaSeq platform is used for large-scale parallel sequencing of the libraries.
Preimplantation genetic testing—structural rearrangements (PGT-SR)
Preimplantation genetic testing—structural rearrangements (PGT-SR) was performed as previously described30. Briefly, antagonists protocol was utilized for controlled ovarian stimulation. After oocyte retrieval, all eggs were fertilized by intracytoplasmic sperm injection (ICSI). All embryos were cultured in sequential media (G1 and G2, Vitrolife, Goteborg, Sweden) to the blastocyst stage. Approximately 3–8 trophectoderm (TE) cells were aspirated using a biopsy pipette with a 30-μm internal diameter and dissected with a Zilos TK laser (Hamilton Thorne, MA, USA). Biopsied TE cells were then used for whole genome amplification (WGA) via multiple displacement amplification with a REPLI-g Single Cell Kit (Qiagen, Valencia, CA). NGS and comprehensive chromosomal screening was then performed as previously described30.
Statistical analysis
Categorical variables were presented as percentages and compared using chi-square tests. P < 0.05 was considered statistically significant. Analyses were performed using the SPSS Statistics version 26 software (IBM SPSS).
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Zhang, F. et al. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat. Genet. 41, 849–853. https://doi.org/10.1038/ng.399 (2009).
Carvalho, C. M. & Lupski, J. R. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 17, 224–238. https://doi.org/10.1038/nrg.2015.25 (2016).
Lieber, M. R. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 283, 1–5. https://doi.org/10.1074/jbc.R700039200 (2008).
David, D. et al. Comprehensive clinically oriented workflow for nucleotide level resolution and interpretation in prenatal diagnosis of de novo apparently balanced chromosomal translocations in their genomic landscape. Hum. Genet. 139, 531–543. https://doi.org/10.1007/s00439-020-02121-x (2020).
Giardino, D. et al. De novo balanced chromosome rearrangements in prenatal diagnosis. Prenat. Diagn. 29, 257–265. https://doi.org/10.1002/pd.2215 (2009).
Halgren, C. et al. Risks and recommendations in prenatally detected de novo balanced chromosomal rearrangements from assessment of long-term outcomes. Am. J. Hum. Genet. 102, 1090–1103. https://doi.org/10.1016/j.ajhg.2018.04.005 (2018).
Feenstra, I. et al. Balanced into array: Genome-wide array analysis in 54 patients with an apparently balanced de novo chromosome rearrangement and a meta-analysis. Eur. J. Hum. Genet. 19, 1152–1160. https://doi.org/10.1038/ejhg.2011.120 (2011).
Schluth-Bolard, C. et al. Cryptic genomic imbalances in de novo and inherited apparently balanced chromosomal rearrangements: Array CGH study of 47 unrelated cases. Eur. J. Med. Genet. 52, 291–296. https://doi.org/10.1016/j.ejmg.2009.05.011 (2009).
Schluth-Bolard, C. et al. Whole genome paired-end sequencing elucidates functional and phenotypic consequences of balanced chromosomal rearrangement in patients with developmental disorders. J. Med. Genet. 56, 526–535. https://doi.org/10.1136/jmedgenet-2018-105778 (2019).
Dong, Z. et al. Genome sequencing explores complexity of chromosomal abnormalities in recurrent miscarriage. Am. J. Hum. Genet. 105, 1102–1111. https://doi.org/10.1016/j.ajhg.2019.10.003 (2019).
Chau, M. H. K. et al. Investigation of the genetic etiology in male infertility with apparently balanced chromosomal structural rearrangements by genome sequencing. Asian J. Androl. 24, 248–254. https://doi.org/10.4103/aja2021106 (2022).
Schluth-Bolard, C. et al. Breakpoint mapping by next generation sequencing reveals causative gene disruption in patients carrying apparently balanced chromosome rearrangements with intellectual deficiency and/or congenital malformations. J. Med. Genet. 50, 144–150. https://doi.org/10.1136/jmedgenet-2012-101351 (2013).
Mikelsaar, R. et al. Balanced reciprocal translocation t(5;13)(q33;q12) and 9q31.1 microduplication in a man suffering from infertility and pollinosis. J. Appl. Genet. 53, 93–97. https://doi.org/10.1007/s13353-011-0078-5 (2012).
Eisfeldt, J. et al. Comprehensive structural variation genome map of individuals carrying complex chromosomal rearrangements. PLoS Genet 15, e1007858. https://doi.org/10.1371/journal.pgen.1007858 (2019).
Campos, A. E. et al. An apparently balanced complex chromosome rearrangement involving seven breaks and four chromosomes in a healthy female and segregation/recombination in her affected son. Mol. Syndromol. 12, 312–320. https://doi.org/10.1159/000516323 (2021).
De Gregori, M. et al. Cryptic deletions are a common finding in “balanced” reciprocal and complex chromosome rearrangements: a study of 59 patients. J. Med. Genet. 44, 750–762. https://doi.org/10.1136/jmg.2007.052787 (2007).
Hu, L. et al. Clinical outcomes in carriers of complex chromosomal rearrangements: a retrospective analysis of comprehensive chromosome screening results in seven cases. Fertil. Steril. 109, 486–492. https://doi.org/10.1016/j.fertnstert.2017.11.021 (2018).
Ozer, L., Aktuna, S., Unsal, E., Baltaci, A. & Baltaci, V. An incidental detection of a cryptic complex chromosome rearrangement found during NGS based PGT-SR: a case report. J. Reprod. Infertil. 23, 303–309. https://doi.org/10.18502/jri.v23i4.10817 (2022).
Gu, S. et al. Mechanisms for complex chromosomal insertions. PLoS Genet 12, e1006446. https://doi.org/10.1371/journal.pgen.1006446 (2016).
Gardner, R. J. M., Sutherland, G. R. & Shaffer, L. G. Chromosome Abnormalities and Genetic Counseling. (Oxford University Press, 2011).
Liao, Y., Wang, L., Zhang, D. & Liu, C. Identification of a balanced complex chromosomal rearrangement involving chromosomes 3, 18 and 21 with recurrent abortion: Case report. Mol. Cytogenet. 7, 39. https://doi.org/10.1186/1755-8166-7-39 (2014).
He, P. et al. Analysis of complex chromosomal rearrangements using a combination of current molecular cytogenetic techniques. Mol. Cytogenet. 15, 20. https://doi.org/10.1186/s13039-022-00597-y (2022).
Zhang, S. et al. Detection of cryptic balanced chromosomal rearrangements using high-resolution optical genome mapping. J. Med. Genet. 60, 274–284. https://doi.org/10.1136/jmedgenet-2022-108553 (2023).
Pellestor, F. & Gatinois, V. Chromoanagenesis: a piece of the macroevolution scenario. Mol. Cytogenet. 13, 3. https://doi.org/10.1186/s13039-020-0470-0 (2020).
Burssed, B., Zamariolli, M., Bellucco, F. T. & Melaragno, M. I. Mechanisms of structural chromosomal rearrangement formation. Mol. Cytogenet. 15, 23. https://doi.org/10.1186/s13039-022-00600-6 (2022).
Xiao, Y. et al. Evaluation of genetic risk of apparently balanced chromosomal rearrangement carriers by breakpoint characterization. J. Assist. Reprod. Genet. 41, 147–159. https://doi.org/10.1007/s10815-023-02986-7 (2024).
Yamada, M., Suzuki, H., Miya, F., Takenouchi, T. & Kosaki, K. Deciphering complex rearrangements at the breakpoint of an apparently balanced reciprocal translocation t(4:18)(q31;q11.2)dn and at a cryptic deletion: Further evidence of TLL1 as a causative gene for atrial septal defect. Am. J. Med. Genet. A. 188, 2472–2478, https://doi.org/10.1002/ajmg.a.62777 (2022).
Aristidou, C. et al. Cryptic breakpoint identified by whole-genome mate-pair sequencing in a rare paternally inherited complex chromosomal rearrangement. Mol. Cytogenet. 11, 34. https://doi.org/10.1186/s13039-018-0384-2 (2018).
Mitsuhashi, S., Ohori, S., Katoh, K., Frith, M. C. & Matsumoto, N. A pipeline for complete characterization of complex germline rearrangements from long DNA reads. Genome. Med. 12, 67. https://doi.org/10.1186/s13073-020-00762-1 (2020).
Hu, L. et al. Reciprocal translocation carrier diagnosis in preimplantation human embryos. EBioMedicine 14, 139–147. https://doi.org/10.1016/j.ebiom.2016.11.007 (2016).
Mardis, E. & McCombie, W. R. Preparation of a 3-kb mate-pair library for illumina sequencing. Cold Spring Harb. Protoc. https://doi.org/10.1101/pdb.prot094656 (2017).
Acknowledgements
This study was supported by grants from the National Key R&D Program of China (Grant 2023YFC2705600), Hunan Provincial Grant for Innovative Province Construction (2019SK4012). The authors thank all the affected individuals and their families for participating in and supporting this study and all figures in this article were made by authors.
Author information
Authors and Affiliations
Contributions
H.L. and T.Y. conceived and designed the study. L.K. were involved in the patients care. G.Y. performed the IVF laboratory work. C.D., X.P., C.J. and W.X. performed the experiments and collected the data. C.D., H.I. and H.L. analyzed the data and prepared Figs. 1–3. H.I., C.D., X.Y. and H.L. wrote the manuscript. Ge Lin participated in the project supervision. T.Y. and L.G. critically reviewed the manuscript and all authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cheng, D., Ibrahim, H., Luo, K. et al. Characterization of cryptic complex chromosome rearrangements in balanced chromosomal rearrangement carriers and their PGT-SR clinical outcome assessments. Sci Rep 14, 20705 (2024). https://doi.org/10.1038/s41598-024-70566-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-70566-5