Abstract
The development of intracerebral hemorrhage (ICH) is a dynamic process and intervention during the acute phase of ICH is critical for subsequent recovery. Therefore, it is crucial to screen potential signature genes and therapeutic target genes in the acute phase of ICH. In this study, based on the results of mRNA sequencing in mouse ICH and mRNA sequencing of human ICH from online databases, top five potential signature genes after ICH, Tyrobp, Itgb2, Tlr2, Ptprc and Itgam, were screened. Quantitative PCR results showed higher mRNA expression of Tyrobp, Itgb2, Tlr2, Ptprc, and Itgam in the 1-, 3- and 5-day mouse ICH groups compared to the sham-operated group. Immune infiltration correlation analysis shows that the top-ranked signature gene, Tyrobp, is negatively correlated with M2 macrophages and plasma cells, and Western blot analysis shows higher expression of the Tyrobp protein in the 1-, 3-, and 5-day mouse ICH groups compared to the sham-operated group. Furthermore, immunohistochemistry revealed that TYROBP protein expression was significantly higher in human ICH tissues than in normal brain tissues. Our results suggest that Tyrobp is a signature gene in the acute phase of ICH and may be a potential target for the treatment of the acute phase of ICH.
Similar content being viewed by others
Introduction
Intracerebral hemorrhage (ICH) is a devastating form of stroke characterized by bleeding into the brain parenchyma1. ICH accounts for only 10% of all strokes, but still has a mortality rate of 50%2. ICH injuries are categorized into primary and secondary injuries3,4. Primary injuries are associated with initial hematoma, hematoma expansion, and mass effect due to hydrocephalus5. Secondary injuries include inflammation, iron and blood-related toxicity, and oxidative stress6,7. Clinical treatment of ICH mainly includes symptomatic treatments such as hemostasis, lowering blood pressure, dehydration to lower cranial pressure, and surgical treatment, which are primarily directed at the primary injury caused by ICH4,8.
During the acute phase of ICH, multiple immune cells are involved in the development of brain injury9,10. Microglia are the primary resident immune cells in the CNS and the first line in the innate immune response to ICH and play a role in acute brain injury primarily through the activation and development of pro-inflammatory or anti-inflammatory phenotypes11,12. Importantly, anti-inflammatory microglia contribute to clearance of hematoma by phagocytosis of erythrocytes and tissue debris11. Following the primary injury, damage-associated molecular patterns (DAMPs) released from compromised neural cells and the extracellular matrix engage pattern recognition receptors on microglia, resulting in the activation of microglia12,13,14. The activation of microglia leads to infiltration of various circulating immune cells. A clinical study shows that inflammation peaked 2–3 days after the onset of ICH which mainly due to activated microglia/macrophages and infiltrating leukocytes15. In addition to intrinsic brain cells such as microglia, lymphocytes also accumulate in the perihematomal domain during the acute phase of ICH, and associated subsets such as CD4 + T, CD8 + T, B, and natural killer cells are observed within a day of hematoma onset and peak at around 3 days16,17. Especially, NK cells exacerbate the disruption of the blood–brain barrier and cerebral edema after ICH through cytotoxicity to brain endothelial cells and increased neutrophil recruitment in focal inflammation18. Considering the vital role of immune cells in the pathophysiological mechanisms of brain injury after ICH. Therefore, the detection of diagnostic and therapeutic targets related to the immune system in the acute phase of ICH is meaningful.
Currently, therapeutic modalities that address the pathophysiological mechanisms of secondary ICH injuries are lacking. Therefore, many clinical and preclinical studies have focused on finding therapeutic targets for secondary injury in the acute phase of ICH4. The clinical study reveals that acute serum soluble CD163 may be a useful biomarker to predict hematoma expansion, perihematomal edema expansion, and worse short-term prognosis in ICH patients19. Up-regulation of CD163 expression to enhance microglia phagocytosis may be an effective treatment strategy for ICH. TGF-β1 is find to modulate microglia-mediated neuroinflammation and promote functional recovery after ICH, while ICH patients with high plasma concentrations of TGF-β1 has a better prognosis, suggesting that TGF-β1 may be an effective therapeutic target for ICH20. Inhibition of NLRP3 inflammasome activation can significantly reduce secondary brain injury after ICH21,22. AQP4, which is distributed mainly in astrocytes, is also a therapeutic target for alleviating brain edema after ICH23,24. Furthermore, TSPO induction after ICH could be an intrinsic mechanism to prevent an exacerbated inflammatory response and TSPO may be a viable therapeutic target and prognostic biomarker in ICH25,26,27. A related in vivo study finds that the TSPO ligand etifoxine reduces brain damage and inflammation after ICH28. Despite several studies on diagnostic and therapeutic targets for the acute phase of ICH, the search for appropriate biomarkers remains meaningful.
In this study, we screened differentially expressed genes (DEGs) during the acute phase of ICH by mRNA sequencing. After screening for co-expression DEGs (co-DEGs) in human and mouse ICH, the hub gene Tyrobp was identified by Cytoscape analysis based on the result of protein–protein interaction (PPI) of those co-DEGs. TYROBP (tyrosine kinase binding protein) is a transmembrane polypeptide that expressed in several cell types, including peripheral blood monocytes, macrophages, natural killer cells, dendritic cells, and osteoblasts29. In central nervous system (CNS), TYROBP mainly expressed in microglia and oligodendrocytes and is an important regulator of microglia activity30,31. Furthermore, immune infiltration analysis was performed using the CIBERSORT method and TYROBP expression was verified in different tissues from ICH by immunoblotting and immunohistochemical assays, respectively. Our results suggest that TYROBP has the potential to be a signature gene and even a therapeutic target in the acute phase of ICH.
Materials and methods
Intracerebral hemorrhage (ICH) model
Eight-week-old male C57BL/6 J mice were used in this study. The experimental mice were purchased and kept at the Laboratory Animal Center of Chongqing Medical University. The mouse ICH model was induced by injection of autologous blood (25 μL) at a rate of 2 µL/min. The injection coordinates relative to 0.2 mm posterior to the bregma, 2 mm lateral to the midline, and 3.5 mm inferior to the skull. The sham-operated group underwent the same surgical trauma treatment and without injection32. The tissues around the hematoma (1–2 mm) were collected 1, 3, and 5 days after the model was constructed and stored at -80 °C, respectively. These tissues of the sham-operated and ICH model group were used for subsequent transcriptomics sequencing. The experimental protocols were approved by the Institutional Animal Care and Use of Chongqing Medical University (No. IACUC-CQMU-2023–0260), and all methods were carried out in accordance with the regulations for the management of experimental animals, and all methods are reported in accordance with ARRIVE guidelines.
Hematoxylin–eosin (HE) staining
The paraffin sections were first dewaxed and rehydrated. Subsequently, the nuclei were stained with hematoxylin for 5 min and the cytoplasm was stained with eosin for 1 min. Finally, the sections were dehydrated and sealed with neutral gum.
Data acquisition and differentially expressed genes (DEGs) analysis
The GSE24265 dataset was downloaded from the GEO database33. The GSE24265 dataset consists of microarray gene expression data obtained from the GPL570 (Affymetrix Human Genome U133 Plus 2.0 Array) platform for the analysis of 11 brain samples from four patients who died from supratentorial ICH, including the perihematomal area and the corresponding contralateral white matter and gray matter. The perihematomal area served as the experimental group, and the contralateral white and gray matter served as the control group. The differences between these two groups were analyzed based on the expression profile data through the standard difference analysis process of the limma package. |log2(FC)|> 1 and adjusted p-value < 0.05 were considered DEGs.
Subsequently, the mRNA sequencing data was compared between the sham-operated group and the groups 1, 3, and 5 days after the mouse ICH model, respectively. All mRNA sequencing data were subjected to PCA analysis and later visualized. Two-group difference analysis based on transcriptome Counts and DEGs were identified by the DESeq2 R software package. Briefly, |log2(FC)|> 1 and p-value < 0.05 were considered DEGs. All raw data from mRNA sequencing were uploaded to the Sequence Read Archive (SRA) of NCBI (BioProject ID: PRJNA1048733).
Gene ontology (GO) and kyoto encyclopedia of genes and genome (KEGG) analyzes
RNA-seq (mRNA) DEGs for mouse ICH model of different comparison groups were analyzed using GO and KEGG analyses34 which were performed using the R package “clusterProfiler” (version 4.4.4). GO analyses consisted of biological process (BP), cellular composition (CC), and molecular function (MF).
Construction and analysis of the protein–protein interaction (PPI) network
The PPI network of all the DEGs in this study were constructed in the String database35. The minimum required interaction score was set to medium confidence (0.4). The active interaction sources come from textmining, experiments, databases, co-expression, neighborhood, gene fusion, and co-occurrence. The meaning of network edges is confidence. Subsequently, we used the Degree algorithms in cytoHubba of Cytoscape (version 3.9.1) to select and visualized the hub genes based on the PPI results.
Immune infiltration prediction model
In this study, we used the CIBERSORT method, which is the most used method for analyzing immune cell infiltration, to analyze RNA-seq (mRNA) from sham and ICH models in mice. A mouse immune cell dataset set as the reference dataset36. Spearman statistical methods were used for the correlation analysis of infiltrating immune cells. The R software package “ggplot2” (version 3.3.6) was used to visualize the differences in immune cell infiltration of each group and the correlation of immune cells infiltrating.
Quantitative PCR
Tissue samples were collected from the striatum at 1, 3 and 5 after ICH. Total RNA was isolated from tissue samples using a commercial kit (FastPure Cell/Tissue Total RNA Isolation Kit, Vazyme, Nanjing, China, RC101-01). Reverse transcription was performed using 0.8 μg of total RNA and a commercial kit (ABScript III RT Master Mix for qPCR with gDNA Remover, ABclonal, Wuhan, China, RK20429). Quantitative PCR (qPCR) assays were performed using fluorescence quantitative SYBR technology (2X Universal SYBR Green Fast qPCR Mix, ABclonal, Wuhan, China, RK21203). Primers for the qPCR assay are shown in Supplementary Table S1 online.
Western blotting
Equal amounts of protein were separated by electrophoresis on 12.5% PAGE gel electrophoresis and transferred electrophoretically to 0.22 μm PVDF membranes. Blots were cut prior to hybridization with antibodies during blotting. After blocking with 5% skim milk for 2 h, membranes were incubated with anti-TYROBP (1:1000, Abcam, UK, ab280568) and β-actin (1:20,000, ABclonal, Wuhan, China, AC026) at 4 °C overnight. Subsequently, incubated with HRP-conjugated anti-rabbit IgG (1:10,000, BIOMIKY, Shanghai, China, MK103A) for 2 h at 25 °C. Finally, the signals were developed using an extra-hypersensitive ECL chemiluminescence kit (BeyoECL Star, Beyotime, Shanghai, China, P0018AM).
Immunohistochemistry (IHC) analysis
Paraffin sections of normal human brain and ICH tissues were obtained from the Pathology Center of Chongqing Medical University, and informed consent was obtained from the patients for the use of the sections. Those paraffin sections were used for the IHC assay according to the manufacturer’s instructions (ZSGB-BIO, Beijing, China, PV-9000). After incubation with anti-TYROBP (1:50, Sangon, Shanghai, China, D260256) and the corresponding secondary antibody for 30 min at 25 °C. The DAB Colorimetric kit (ZSGB-BIO, Beijing, China, ZLI-9018) was used for section visualization. The intensity of immunostaining was scored as 0 (no immunostaining), 1 (weak immunostaining), 2 (moderate immunostaining) or 3 (strong immunostaining). The percentage of positive cells was scored as 0 (< 5%), 1 (5—25%), 2 (26—50%), 3 (51—75%) and 4 (76—100%). The IHC score was obtained by multiplying the immunostaining intensity score by the positive percentage cell score37.
Statistical analysis
Immune infiltration correlation analysis was performed using R version 4.2.1 for Spearman correlation analysis. Hematoma volume, quantitative PCR, immunoblotting and IHC results were analyzed using GraphPad Prism 9.0.0 with Student's t-test for two groups and one-way ANOVA for multiple groups. The date was presented as mean ± SEM and statistical significance was established at p < 0.05.
Results
Identification of ICH in mice
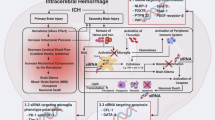
Figure 1 shows a schematic diagram of the research methods and results. As shown in Fig. 2A, brain tissue samples from mice were collected after different autologous blood injection times. Subsequently, we characterized the mouse ICH model by HE staining. As shown in Fig. 2B–F, compared to the sham-operated group, the ICH group, especially the 1-day and 3-day groups, show hematomas of different degrees in the region of the basal ganglia. HE staining results indicated successful brain hematoma formation in ICH group of mice.
Flow diagram of the entire research process. Parts of the figure were drawn by using pictures from Servier Medical Art. Servier Medical Art is licensed under CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
ICH model. (A) Schematic diagram of the construction of mouse ICH model. The tissues around the hematoma were collected 1, 3 and 5 days after autologous blood injection. Representative HE staining in the sham-operated group (B) and 1 (C), 3 (D), and 5 days (E) after ICH. (F) Relative hematoma volume in the sham-operated group and each ICH group, n = 3, one-way ANOVA was used for statistical analysis. *p < 0.05, **p < 0.01, ***p < 0.001. Parts of the figure were drawn by using pictures from Servier Medical Art. Servier Medical Art is licensed under CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
DEGs screening of mouse ICH
All mRNA sequencing data were subjected to PCA analysis and the result shows that PC1 is 64% variance and PC2 is 13% variance (Fig.S1). After validating the reliability of the mouse ICH model and mRNA sequencing data, we further analyzed the data to screen for DEGs, as these genes are more likely to play a role in the pathophysiological process of ICH. Expression profiles of different days of the ICH groups compared to the sham-operated group using p < 0.05 and logFC > 2 as cut-off values, and the sham-operated group as the control. Among them, 1,228 genes upregulated, and 264 genes downregulated in the 1-day ICH group (Fig. 3A). 645 up-regulated genes and 151 down-regulated genes were verified in the 3-day ICH group (Fig. 3B). And 381 up-regulated genes and 8 down-regulated genes were identified in the 5-day ICH group (Fig. 3C). We then cross-compared the 1-, 3-, and 5-day ICH groups (see Supplementary Fig. S3 online). The 3-day group screens 497 up-regulated genes and 688 down-regulated genes compared to the 1-day group (Fig. S3A). The 5-day group identified 126 up-regulated genes and 241 down-regulated genes compared to the 3-day group (Fig. S3B). Meanwhile, the 5-day group filtered 460 up-regulated genes and 926 down-regulated genes compared to the 1-day group (Fig. S3C).
DEGs screening and functional enrichment analysis in mouse ICH34. Volcano plots showing DEGs in the sham-operated group versus the group at 1 (A), 3 (B), and 5 days (C) after ICH of C57BL/6 J mice. (D) GO and KEGG analysis of DEGs in Fig. 2A. (E) GO and KEGG analysis of DEGs in Fig. 2B. (F) GO and KEGG analysis of DEGs in Fig. 2C.
GO and KEGG pathway analyses of mouse ICH
To reveal the potential biological functions of DEGs in mouse ICH, we performed pathway analysis of these DEGs. GO analysis showed that the BP, CC, and MF components of the 1-day ICH group are predominantly present in positive regulation of response to external stimulus, collagen-containing extracellular matrix, and immune receptor activity, respectively (Fig. 3D). The 3-day ICH group was predominantly present in leukocyte cell–cell adhesion, collagen-containing extracellular matrix, and immune receptor activity, respectively (Fig. 3E). The 5-day ICH group was predominantly presented in negative regulation of immune system process, phagocytic vesicle, and immune receptor activity, respectively (Fig. 3F). KEGG analysis in the 1-day ICH group focused on cytokine-cytokine receptor interactions, the TNF signaling pathway and the NF-kappa B signaling pathway (Fig. 2D). Phagosome, viral protein interaction with cytokine and cytokine receptor, rheumatoid arthritis for 3-day ICH group (Fig. 2E). And phagosome, leishmaniasis, tuberculosis for the 5-day ICH group (Fig. 2F). Furthermore, GO and KEGG analyses of up-regulated and down-regulated genes in 1-, 3- and 5-day mouse ICH were performed respectively, and the results are demonstrated in Figure S2A-C. In addition, GO and KEGG analyses in 1-day ICH versus 3-day ICH group focused primarily on leukocyte migration, collagen-containing extracellular matrix, cytokine activity, and cytokine-cytokine receptor interactions, respectively (Fig.S3D). The 3-day ICH versus 5-day ICH group mainly focused on monoamine transport, axon terminus, peptide receptor activity, and neuroactive ligand-receptor interaction, respectively (Fig.S3E). The 1-day ICH versus 5-day ICH group mainly focused on leukocyte migration, collagen-containing extracellular matrix, cell adhesion molecule binding, and cytokine-cytokine receptor interactions, respectively (Fig.S3F).
Co-differentially expressed genes (co-DEGs) screening and functional enrichment analysis in mouse ICH after different days
To screen for signature genes in the acute phase of ICH, it is necessary to specify that these genes have significant expression differences from the sham-operated group on different days in the acute phase after mouse ICH. As shown in Fig. 4A, the Venn plots demonstrated 202 co-DEGs in the 1-, 3-, and 5-day ICH groups. The heatmap of these 202 co-DEGs in each sample is shown in Fig. 4B, and most of the genes showed up-regulated levels. Then, we performed GO and KEGG analyses to verify the function of these genes. GO analysis showed that the BP, CC, and MF components of these 202 co-DEGs mainly presented in positive regulation of immune effector process, endocytic vesicle, and cell adhesion molecule binding, respectively (Fig. 4C–E). KEGG analyzes in these genes focused mainly on phagosome, myeloid leukocyte activation, and tumor necrosis factor superfamily cytokine production (Fig. 4F).
Co-DEGs screening and functional enrichment analysis in mouse ICH34. (A) Venn plot showing DEGs intersection in the 1-, 3-, and 5-day groups after ICH. (B) Heatmap shows the expression of 202 genes in each group that are overlapping genes in DEGs at 1, 3, and 5 days after ICH. GO analysis, including BP (C), CC (D) and MF (E), and KEGG analysis (F) of 202 overlapped genes.
Co-differentially expressed genes (co-DEGs) screening in human and mouse ICH
In order to make the screened signature genes for the acute phase of ICH work in either human or mouse ICH. It is necessary to further screen the signature genes for the acute phase of ICH from the DEGs screened for co-expression in both human and mouse models of ICH. Firstly, adjust p < 0.05 and logFC > 2 as critical values, 441 up-regulated genes and 327 down-regulated genes were selected from the GSE24265 database (Fig. 5A). Then, GO and KEGG analysis on the up-regulated and down-regulated genes were performed (Fig.S4A). To screen for DEGs that play an important role in both human and mouse ICH. Subsequently, the intersection of 202 co-DEGs from mouse ICH and 768 DEGs from human ICH showed that a total of 42 overlapped genes were screened (Fig. 5B). We considered these 42 overlapped genes as potential signature genes. The heatmap of those 42 potential signature genes in mouse ICH was shown in Fig. 5C. GO and KEGG analyses of these 42 genes were performed in humans and mice, respectively, and the results were demonstrated in Figure S4B-C.
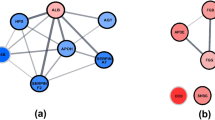
Co-DEGs screening and signature genes identification in human and mouse ICH. (A) Volcano plot showing human ICH DEGs in GSE24265 dataset. (B) 42 overlapped genes of human and mouse ICH DEGs. (C) Heatmap of 42 overlapped genes in mouse ICH. PPI analysis of 42 genes in human (D) and mouse (E). Ten signature genes of human (F) and mouse (G) screened by cytoHubba plugin of Cytoscape based on the results of PPI analysis for 42 overlapped genes. (H) Heatmap of 184 genes related to Tyrobp in mouse ICH. (I) Heatmap of GO and KEGG analyses for184 genes related to Tyrobp in mouse ICH.
Protein–protein interaction (PPI) analysis and identification of signature genes
To identify marker genes, the possible PPI of potential hub genes was constructed in the String database. The interactions between the genes were indicated by confidence and homo sapiens and mus musculus organisms were selected and analyzed separately (Fig. 5D, E). Then, we used the Degree algorithms in cytoHubba of Cytoscape to select and visualize hub genes based on the results of Fig. 5D and Fig. 5E, respectively (Fig. 5F, G). Interestingly, these results showed that Tyrobp, Itgb2, Tlr2, Ptprc, and Itgam were among the top 5 genes in both human and mice. Interestingly, all of these genes are expressed exclusively in myeloid cells, which is consistent with the significant increase in myeloid cell numbers after ICH reported in previous literature16,17. Meanwhile, Tyrobp is the top ranked gene in both human and mouse PPI analysis results, so we chose Tyrobp as the signature gene. Interestingly, Friend analysis of these potential hub genes also showed that TYROBP is also the top ranked gene (see Supplementary Fig. S5 online). After identifying the hub gene, the 184 genes associated with the hub gene Tyrobp were screened using spearman correlation analysis with spearman's rank correlation coefficient greater than 0.9 and an adjusted p-value less than 0.05 (Fig. 5H). When spearman rank correlation coefficient is greater than 0.9, it is usually considered highly correlated. GO and KEGG analyzes of these 184 genes and the associated top 66 genes were shown in Fig. 5I. The BP components of these 184 genes mainly presented myeloid leukocyte activation, leukocyte activation involved in immune response, and cell activation involved in immune response. The CC component mainly included focal adhesion, cell-substrate junction, and phagocytic vesicle. The MF component mainly involved in cell adhesion, molecule binding, and immune receptor activity. Furthermore, the KEGG analysis of these genes was presented mainly in osteoclast differentiation, leishmaniasis, and tuberculosis.
Immune infiltration prediction model
Immune cells play an important role in the acute phase of ICH. Then, we predicted immuno-infiltration in mouse ICH. Firstly, we compared the ICH group on different days with the sham-operated group, and as shown in Fig. 6A, there was no statistically significant difference in the composition of the same immune cells between the different groups in the box plot. However, the composition of the various types of immune cells was different between each group (Fig. 6B). Subsequently, we compared all ICH groups on different days after merging them with the sham-operated group, the boxplot of differences in immune cell infiltration revealed that plasma cells (p < 0.001), M2 macrophages (p < 0.001), T cells CD4 naïve (p = 0.004), and Th17 cells (p = 0.013) infiltrate less in ICH compared to the sham-operated group (Fig. 6C). Additionally, we also analyzed the immune correlation between different immune cells in the mouse ICH model. DC actived cells with neutrophil cells, M2 macrophages with plasma cells, DC immature cells with Th2 cells were positively correlated (p < 0.001) (Fig. 6D). In contrast, Th17 cells with M0 macrophages, NK actived cells with M0 macrophages, NK actived cells with NK resting cells were negatively correlated (p < 0.001) (Fig. 6D). Then we performed a correlation analysis of the Tyrobp hub gene and various immune cells (Fig. 6E). The results reveale that Tyrobp is negatively correlated with M2 macrophage and plasma cells (R = -0.753, p = 0.005) (Fig. 6F, G).
Immune infiltration prediction and signature gene immune correlation analysis. (A) Box plot of immune cells infiltration in the sham-operated group, and the 1-day, 3-day, and 5-day groups after ICH in C57BL/6 J mice. (B) Distribution of infiltration immune cells in different subgroups. (C) Box plot of immune cells infiltration in the sham-operated group and ICH group. (D) Correlation analysis between infiltration immune cells. (E) Correlation of Tyrobp with immune-infiltrating cells. (F) Correlation of Tyrobp with M2 macrophage Cells. (G) Correlation of Tyrobp with plasma Cells. *p < 0.05, **p < 0.01, ***p < 0.001.
Verification of signature gene expression
Validation of mRNA and protein expression after screening potential signature genes is necessary after ICH. The mRNA expression of Tyrobp, Itgb2, Tlr2, Ptprc, and Itgam in mouse ICH were detected by qPCR assay using perihematomal brain tissues (Fig. 7A–E). The qPCR results showed that the mRNA levels of these genes were significantly higher in the 1-, 3-, and 5-day ICH groups than in the sham-operated group. It is basically consistent with the results of transcriptome sequencing. Furthermore, the protein level of the Tyrobp hub gene was examined using Western blotting experiments (Fig. 8A). Statistical results showed that Tyrobp protein expression was significantly higher in the 1-, 3-, and 5-day ICH groups than in the sham-operated group (Fig. 8B). Meanwhile, we performed immunohistochemical analysis of Tyrobp expression in normal human brain tissues and ICH tissues (Fig. 8C). IHC scores showed that Tyrobp expression was significantly higher in human ICH tissues than in normal brain tissues (Fig. 8D).
The mRNA expression of signature genes. Tyrobp (A), Itgb2 (B), Tlr2 (C), Ptprc (D) and Itgam (E) mRNA expression in mouse sham-operated and ICH groups, Student's t-test was used for statistical analysis, * represents the statistical results of the comparison with the sham group, n = 4, *p < 0.05, **p < 0.01, ***p < 0.001.
TYROBP protein expression after ICH. (A) Western blotting analysis of Tyrobp in mouse sham-operated and ICH groups, blots were cut prior to hybridization with antibodies during blotting and original blots are presented in Fig.S6. (B) Statistical analysis of Tyrobp protein expression in mouse sham-operated and ICH groups, compared with sham-operated group, n = 3. (C) Immunohistochemical analysis of TYROBP in normal brain and ICH tissues of human. (D) IHC score of TYROBP in normal brain (n = 8) and ICH (n = 6) tissues of human. Student's t-test was used for statistical analysis, *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
The DEGs analysis showed a significantly greater number of DEGs in 1- and 3-day ICH group than in the 5-day group, which may be related to hematoma resorption and reduction of secondary injury. Consistently, HE staining showed larger hematomas in the 1-day and 3-day groups than in the 5-day group. GO and KEGG analyzes showed that DEGs in the 1-day group were mainly involved in the acute inflammatory response through the cytokine regulation, TNF, and NF-kappa B signaling pathways. While the DEGs in the 3- and 5-day groups were mainly involved in immune-related regulatory processes.
When the ICH group, which consists of 1-, 3- and 5-day groups, compared to the sham-operated group, there was a statistical difference in plasma cells, M2 macrophages, CD4 naïve T cells, and Th17 cells. Previous studies have shown that M2 macrophages exist mainly as anti-inflammatory microglia in the brain that contribute to hematoma clearance and anti-inflammatory effects38. Plasma cells, also known as effector B cells, can synthesize and store immunoglobulin to participate in humoral immunity39, beyond that recent studies have found that plasma cells are also involved in the regulation of neuroinflammation40. In addition, CD4 naïve T cells and Th17 cells are both CD4 T cell subtypes, the reduction of these CD4 T cell subtypes will lead to the decrease of immune capacity and trigger the macrophage stimulation and cytokine storm events. These reports suggest that these immune cells may play a role in ICH, but evidence based on predictive modeling results is still limited, and more experimental results are needed to validate the role of these cells after ICH.
The signature gene Tyrobp was selected by Degree algorithms in cytoHubba of Cytoscape. After the signature gene was identified, the experimental results showed that Tyrobp mRNA and protein levels were significantly higher in the ICH tissues than in the sham-operated group or in the normal brain tissues. In addition, Tyrobp is negatively correlated with immune cells such as M2 macrophages and plasma cells, which may confer the potential of Tyrobp as a therapeutic target for ICH. TYROBP (tyrosine kinase binding protein) is a transmembrane polypeptide that in the CNS predominantly found in microglia and oligodendrocytes41. Also, TYROBP is expressed in several cell types, including peripheral blood monocytes, macrophages, natural killer cells, dendritic cells, and osteoblasts29. TYROBP acts as a critical regulator of microglial activity30,31. Multiple studies have shown that TYROBP expression is increased in Alzheimer's disease (AD) patients and in mouse models42,43. Meanwhile, the expression of TYROBP is also increased after traumatic brain injury (TBI)44. Several studies had reported that TYROBP can bind to several TYROBP-related receptors in brain, such as TERM45, SIRPβ146 and MDL147. The TYROBP-associated receptor provides docking sites for various ligands after attachment to TYROBP that can initiate intracellular signaling through TYROBP and its immunoreceptor tyrosine-based activation motifs (ITAMs)48.In addition, the TREM2/TYROBP receptor complex is able to control microglia activity, thus affecting the fate of damaged neurons after neuronal injury and in neurodegenerative diseases29,49. Recent studies have identified TREM2 and TYROBP/DAP12 as components of a key molecular hub linking inflammation and microglia to the pathophysiology of Alzheimer's disease and possibly traumatic brain injury44,50. Furthermore, TYROBP gene knockout in microglia was demonstrated to reduce this interaction between damaged neurons and microglia and reduce neuronal cell apoptosis rates51. Interesting, the regulatory role of TYROBP in microglia is consistent with the results of the immune correlation analysis that Tyrobp is negatively correlated with M2 macrophages. GO analysis of Tyrobp-related genes was also found to be closely associated with the activation and regulation involved in the immune response. TYROBP may play a role in the immune response after ICH, particularly regarding its effect on microglia activity. Based on those results, we speculate that TYROBP exacerbates neuronal injury after ICH by modulating microglia activation, but its specific mechanism requires further study.
Conclusions
In conclusion, Tyrobp mRNA and protein expression were significantly higher than those of normal brain tissues in human and mouse ICH tissues. Meanwhile, Tyrobp protein and mRNA expression were elevated during the acute phase of ICH, e.g. 1, 3, and 5 days after ICH, and thus Tyrobp has the potential to be a biomarker in the acute phase of ICH. Additionally, Tyrobp is negatively correlated with immune cells such as M2 macrophages and plasma cells, and Tyrobp-related genes are also found to be closely associated with the activation and regulation involved in the immune response, which may confer the potential of Tyrobp as a therapeutic target for ICH. However, further experiments are still needed to verify the effect and molecular mechanisms of Tyrobp on ICH.
Data availability
All mRNA sequencing raw data were uploaded to Sequence Read Archive (SRA) of NCBI (BioProject ID: PRJNA1048733). The GSE24265 dataset was downloaded from GEO NCBI database.
References
Manoel, A. L. D. et al. The critical care management of spontaneous intracranial hemorrhage: A contemporary review. Crit. Care https://doi.org/10.1186/s13054-016-1432-0 (2016).
Magid-Bernstein, J. et al. Cerebral hemorrhage: pathophysiology, treatment, and future directions. Circ. Res. 130, 1204–1229. https://doi.org/10.1161/circresaha.121.319949 (2022).
Li, Z. et al. Therapeutic strategies for intracerebral hemorrhage. Front. Neurol. https://doi.org/10.3389/fneur.2022.1032343 (2022).
Kim, J. Y. & Bae, H. J. spontaneous intracerebral hemorrhage: Management. J. Stroke 19, 28–39. https://doi.org/10.5853/jos.2016.01935 (2017).
Zheng, H. P., Chen, C. L., Zhang, J. & Hu, Z. P. Mechanism and therapy of brain edema after intracerebral hemorrhage. Cerebrovasc. Dis. 42, 155–169. https://doi.org/10.1159/000445170 (2016).
Aronowski, J. & Zhao, X. R. Molecular pathophysiology of cerebral hemorrhage secondary brain injury. Stroke 42, 1781–1786. https://doi.org/10.1161/strokeaha.110.596718 (2011).
Shao, A. W., Zhu, Z. Y., Li, L. F., Zhang, S. Z. & Zhang, J. M. Emerging therapeutic targets associated with the immune system in patients with intracerebral haemorrhage (ICH): From mechanisms to translation. Ebiomedicine 45, 615–623. https://doi.org/10.1016/j.ebiom.2019.06.012 (2019).
Hemphill, J. C. III. et al. Guidelines for the management of spontaneous intracerebral hemorrhage a guideline for healthcare professionals from the American heart association/American stroke association. Stroke 46, 2032–2060. https://doi.org/10.1161/str.0000000000000069 (2015).
Greenhalgh, A. D., David, S. & Bennett, F. C. Immune cell regulation of glia during CNS injury and disease. Nat. Rev. Neurosci. 21, 139–152. https://doi.org/10.1038/s41583-020-0263-9 (2020).
Wofford, K. L., Loane, D. J. & Cullen, D. K. Acute drivers of neuroinflammation in traumatic brain injury. Neural Regen. Res. 14, 1481–1489. https://doi.org/10.4103/1673-5374.255958 (2019).
Lan, X., Han, X. N., Li, Q., Yang, Q. W. & Wang, J. Modulators of microglial activation and polarization after intracerebral haemorrhage. Nat. Rev. Neurol. 13, 420–433. https://doi.org/10.1038/nrneurol.2017.69 (2017).
Bai, Q., Xue, M. & Yong, V. W. Microglia and macrophage phenotypes in intracerebral haemorrhage injury: therapeutic opportunities. Brain 143, 1297–1314. https://doi.org/10.1093/brain/awz393 (2020).
Alsbrook, D. L. L. et al. Neuroinflammation in acute ischemic and hemorrhagic stroke. Curr. Neurol. Neurosci. Rep. 23, 407–431. https://doi.org/10.1007/s11910-023-01282-2 (2023).
Simpson, D. S. A. & Oliver, P. L. ROS generation in microglia: Understanding oxidative stress and inflammation in neurodegenerative disease. Antioxidants https://doi.org/10.3390/antiox9080743 (2020).
Liu, B. H. et al. CD163/hemoglobin oxygenase-1 pathway regulates inflammation in hematoma surrounding tissues after intracerebral hemorrhage. J. Stroke Cerebrovasc. Dis. 24, 2800–2809. https://doi.org/10.1016/j.jstrokecerebrovasdis.2015.08.013 (2015).
Loftspring, M. C., McDole, J., Lu, A., Clark, J. F. & Johnson, A. J. Intracerebral hemorrhage leads to infiltration of several leukocyte populations with concomitant pathophysiological changes. J. Cerebral Blood Flow Metab. 29, 137–143. https://doi.org/10.1038/jcbfm.2008.114 (2009).
Mracsko, E. et al. Leukocyte invasion of the brain after experimental intracerebral hemorrhage in mice. Stroke 45, 2107–2114. https://doi.org/10.1161/strokeaha.114.005801 (2014).
Li, Z. et al. Brain transforms natural killer cells that exacerbate brain edema after intracerebral hemorrhage. J. Exp. Med. https://doi.org/10.1084/jem.20200213 (2020).
Roy-O’Reilly, M. et al. Soluble CD163 in intracerebral hemorrhage: biomarker for perihematomal edema. Ann. Clin. Transl. Neurol. 4, 793–800. https://doi.org/10.1002/acn3.485 (2017).
Taylor, R. A. et al. TGF-β1 modulates microglial phenotype and promotes recovery after intracerebral hemorrhage. J. Clin. Investig. 127, 280–292. https://doi.org/10.1172/jci88647 (2017).
Ma, Q. et al. NLRP3 inflammasome contributes to inflammation after intracerebral hemorrhage. Ann. Neurol. 75, 209–219. https://doi.org/10.1002/ana.24070 (2014).
Ren, H. L. et al. Selective NLRP3 (pyrin domain-containing protein 3) inflammasome inhibitor reduces brain injury after intracerebral hemorrhage. Stroke 49, 184. https://doi.org/10.1161/strokeaha.117.018904 (2018).
Tang, Y. et al. Effects of aquaporin-4 on edema formation following intracerebral hemorrhage. Exp. Neurol. 223, 485–495. https://doi.org/10.1016/j.expneurol.2010.01.015 (2010).
Qing, W. G. et al. Brain edema after intracerebral hemorrhage in rats: The role of iron overload and aquaporin 4 Laboratory investigation. J. Neurosurg. 110, 462–468. https://doi.org/10.3171/2008.4.Jns17512 (2009).
Bonsack, F., Alleyne, C. H. Jr. & Sukumari-Ramesh, S. Augmented expression of TSPO after intracerebral hemorrhage: A role in inflammation?. J. Neuroinflamm. https://doi.org/10.1186/s12974-016-0619-2 (2016).
Li, M., Ren, H., Sheth, K. N., Shi, F.-D. & Liu, Q. A TSPO ligand attenuates brain injury after intracerebral hemorrhage. FASEB J. 31, 3278–3287. https://doi.org/10.1096/fj.201601377RR (2017).
Jiang, W. Q., Jin, P., Bao, Q., Wei, W. F. & Jiang, W. Prognostic significance of serum translocator protein in patients with spontaneous intracerebral hematoma:preliminary findings. Neurol. Res. 43, 412–417. https://doi.org/10.1080/01616412.2020.1866372 (2021).
Li, M. S., Ren, H. L., Sheth, K. N., Shi, F. D. & Liu, Q. A TSPO ligand attenuates brain injury after intracerebral hemorrhage. FASEB J. 31, 3278–3287. https://doi.org/10.1096/fj.201601377RR (2017).
Haure-Mirande, J.-V., Audrain, M., Ehrlich, M. E. & Gandy, S. Microglial TYROBP/DAP12 in Alzheimer’s disease: Transduction of physiological and pathological signals across TREM2. Mol. Neurodegen. https://doi.org/10.1186/s13024-022-00552-w (2022).
Tomasello, E. & Vivier, E. KARAP/DAP12/TYROBP: three names and a multiplicity of biological functions. Eur. J. Immunol. 35, 1670–1677. https://doi.org/10.1002/eji.200425932 (2005).
Kobayashi, M., Konishi, H., Takai, T. & Kiyama, H. A DAP12-dependent signal promotes pro-inflammatory polarization in microglia following nerve injury and exacerbates degeneration of injured neurons. Glia 63, 1073–1082. https://doi.org/10.1002/glia.22802 (2015).
Yan, J. et al. CCR5 activation promotes NLRP1-dependent neuronal pyroptosis via CCR5/PKA/creb pathway after intracerebral hemorrhage. Stroke 52, 4021–4032. https://doi.org/10.1161/strokeaha.120.033285 (2021).
Rosell, A. et al. Brain perihematoma genomic profile following spontaneous human intracerebral hemorrhage. Plos One https://doi.org/10.1371/journal.pone.0016750 (2011).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–D592. https://doi.org/10.1093/nar/gkac963 (2023).
Szklarczyk, D. et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646. https://doi.org/10.1093/nar/gkac1000 (2023).
Chen, Z. et al. Inference of immune cell composition on the expression profiles of mouse tissue. Sci. Rep. https://doi.org/10.1038/srep40508 (2017).
Meyerholz, D. K. & Beck, A. P. Principles and approaches for reproducible scoring of tissue stains in research. Lab. Investig. 98, 844–855. https://doi.org/10.1038/s41374-018-0057-0 (2018).
Chhor, V. et al. Characterization of phenotype markers and neuronotoxic potential of polarised primary microglia in vitro. Brain Behav. Immunity 32, 70–85. https://doi.org/10.1016/j.bbi.2013.02.005 (2013).
Lam, W. Y. & Bhattacharya, D. Metabolic links between plasma cell survival, secretion, and stress. Trends Immunol. 39, 19–27. https://doi.org/10.1016/j.it.2017.08.007 (2018).
Pioli, P. D. Plasma cells, the next generation: Beyond antibody secretion. Front. Immunol. https://doi.org/10.3389/fimmu.2019.02768 (2019).
Kiialainen, A., Hovanes, K., Paloneva, J., Kopra, O. & Peltonen, L. Dap12 and Trem2, molecules involved in innate immunity and neurodegeneration, are co-expressed in the CNS. Neurobiol. Dis. 18, 314–322. https://doi.org/10.1016/j.nbd.2004.09.007 (2005).
Audrain, M. et al. Reactive or transgenic increase in microglial TYROBP reveals a TREM2-independent TYROBP-APOE link in wild-type and Alzheimer’s-related mice. Alzheimers Dementia 17, 149–163. https://doi.org/10.1002/alz.12256 (2021).
Haure-Mirande, J.-V. et al. Deficiency of TYROBP, an adapter protein for TREM2 and CR3 receptors, is neuroprotective in a mouse model of early Alzheimer’s pathology. Acta Neuropathol. 134, 769–788. https://doi.org/10.1007/s00401-017-1737-3 (2017).
Zheng, R.-Z. et al. Integration of single-cell and bulk RNA sequencing data reveals key cell types and regulators in traumatic brain injury. Math. Biosci. Eng. 18, 1201–1214. https://doi.org/10.3934/mbe.2021065 (2021).
Takahashi, K., Rochford, C. D. P. & Neumann, H. Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J. Exp. Med. 201, 647–657. https://doi.org/10.1084/jem.20041611 (2005).
Dietrich, J., Cella, M., Seiffert, M., Bühring, H. J. & Colonna, M. Cutting edge: Signal-regulatory protein β1 is a DAP12-associated activating receptor expressed in myeloid cells. J. Immunol. 164, 9–12. https://doi.org/10.4049/jimmunol.164.1.9 (2000).
Bakker, A. B. H., Baker, E., Sutherland, G. R., Phillips, J. H. & Lanier, L. L. Myeloid DAP12-associating lectin (MDL)-1 is a cell surface receptor involved in the activation of myeloid cells. Proc. Natl. Acad. Sci. USA 96, 9792–9796. https://doi.org/10.1073/pnas.96.17.9792 (1999).
Paradowska-Gorycka, A. & Jurkowska, M. Structure, expression pattern and biological activity of molecular complex TREM-2/DAP12. Hum. Immunol. 74, 730–737. https://doi.org/10.1016/j.humimm.2013.02.003 (2013).
Konishi, H. & Kiyama, H. Microglial TREM2/DAP12 signaling: A double-edged sword in neural diseases. Front. Cellular Neurosci. https://doi.org/10.3389/fncel.2018.00206 (2018).
Elder, G. A., Ehrlich, M. E. & Gandy, S. Relationship of traumatic brain injury to chronic mental health problems and dementia in military veterans. Neurosci. Lett. https://doi.org/10.1016/j.neulet.2019.134294 (2019).
Zhou, X., Song, H., He, J., Han, W. & Li, Q. Deciphering microglial activation and neuronal apoptosis post-traumatic brain injury: The role of TYROBP in inflammation regulation networks. Mol. Med. Rep. https://doi.org/10.3892/mmr.2024.13228 (2024).
Acknowledgements
The authors would like to thank Prof. Jingxian Wu for providing the paraffin sections for IHC assay.
Funding
This research was funded by the National Natural Science Foundation of China (grant number 81971217), the National Natural Science Foundation of China (youth fund) (grant number 82301512), the project of the National Clinical Medical Research Center (grant number YBXM-2019–17), the China Postdoctoral Science Foundation (grant number 2022MD723734), the Chongqing Postdoctoral Science Foundation (grant number CST2023NSCQ-BHX0121), the Program for Youth Innovation in Future Medicine of Chongqing Medical University to Xuan Zhai, and the Chongqing Graduate Student Research Innovation Program (grant number CYB23206, CYS23338).
Author information
Authors and Affiliations
Contributions
All authors listed have made a substantial contribution to the work, agree to be accountable for all aspects of the work, and approved it for publication. Conceptualization, Y.L.; Funding acquisition, X.Z; Methodology, M.Z., Y.D., Y.L., A.P., and C. L.; Project administration, X.Z.; Resources, Y.Q.; Software, M.Z. and Y.L.; Supervision, X.Z.; Validation, M.Z., Y.D., Y.Q., and Y.H.; Writing – original draft, M.Z.; Writing – review & editing, Y.D., H.G., and N.J.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Children’s Hospital of Chongqing Medical University (No. 2021295). The animal study protocol was approved by Institutional Animal Care and Use of Chongqing Medical University (No. IACUC-CQMU-2023–0260). Informed consent was obtained from all subjects involved in the study.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, M., Duan, Y., Gan, H. et al. TYROBP serve as potential immune-related signature genes in the acute phase of intracerebral hemorrhage. Sci Rep 14, 20158 (2024). https://doi.org/10.1038/s41598-024-71132-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-71132-9