Abstract
To evaluate the prognostic significance of the maximum standardized uptake value (SUVmax) in nasopharyngeal carcinoma (NPC), establish a gene signature that correlates with SUVmax, and explore the underlying biological behaviors associated with these correlations for the prediction of clinical outcomes. A cohort of 726 patients with NPC was examined to identify correlations between SUVmax and various clinical variables. RNA sequencing was performed to identify genes related to SUVmax, and these genes were used to develop an SUV signature. Additionally, transcriptome enrichment analysis was conducted to investigate the potential biological behaviors underlying the observed correlations. Higher SUVmax was associated with an increased tumor burden and worse prognosis. The SUV signature, which consisted of 10 genes, was positively correlated with SUVmax, and it predicted worse survival outcomes. This signature was highly expressed in malignant epithelial cells and associated with hypoxia and resistance to radiotherapy. Additionally, the signature was negatively correlated with immune function. SUVmax is a valuable prognostic indicator in NPC, with higher values predicting worse outcomes. The SUV signature offers further prognostic insights, linking glucose metabolism to tumor aggressiveness, treatment resistance, and immune function, and it could represent a potential biomarker for NPC.
Similar content being viewed by others
Introduction
Nasopharyngeal carcinoma (NPC), arising from the mucosal epithelium of the nasopharynx, poses a significant clinical challenge because of its aggressiveness and distinctive anatomical location1. The primary treatment modality for NPC is radiotherapy, particularly with the advancements provided by intensity-modulated radiation therapy (IMRT)2. Although IMRT has significantly improved the 5-year overall survival (OS) rate, recurrent and distant metastatic NPC remains a therapeutic challenge3. Identifying effective biomarkers is crucial for predicting clinical outcomes and tailoring individualized treatments to enhance overall prognosis.
F18-FDG PET-CT, a functional imaging modality allowing quantitative assessment of glucose uptake, plays a crucial role in NPC diagnosis and staging4. Notably, the maximum standard uptake value (SUVmax) derived from F18-FDG PET-CT has displayed predictive capabilities for survival outcomes in various malignancies5,6. Studies consistently highlight SUVmax as a significant predictor of clinical prognosis in NPC4,7. Recently, Lee et al.8 revealed that high tumor FDG uptake in hepatocellular carcinoma (HCC) is associated with a hypoxic tumor microenvironment. Hypoxia is a significant impediment to the efficacy of radiotherapy. In the 1950s, Gray et al.9 revealed that the radiation dose needed to kill hypoxic cells was three-fold larger than that needed to kill normoxic cells. As radiotherapy is the predominant treatment for NPC, the resistance to radiotherapy attributable to hypoxia could be linked to the poor prognosis of NPC. Because SUVmax is correlated with hypoxia, it is speculated that the utility of SUVmax for predicting poor prognosis in NPC might be mediated through the hypoxic pathway. Further investigation is needed to confirm this hypothesis. Additionally, research suggests a negative correlation between SUVmax and the expression of fructose-1,6-diphosphatase 1, indicating the potential role of SUVmax as a prognostic indicator through mechanisms associated with glucose metabolism5. However, the detailed exploration of glucose metabolism genes at the transcriptome level remains limited, and the association of SUVmax with hypoxia remains unknown.
This study aimed to identify significant glucose metabolism genes associated with SUVmax using RNA sequencing datasets and formulate a gene marker, termed the SUV signature. Furthermore, the study investigated the correlations of SUVmax with prognosis and hypoxia through enrichment analysis.
Materials and methods
Study design and patients
A brief overview of the study workflow is presented in Fig. S1. This retrospective study included 726 individuals with newly diagnosed NPC who underwent complete treatment at our center between January 2012 and December 2018. The inclusion criteria were as follows: confirmation of primary NPC through biopsy; availability of comprehensive baseline clinical data; pre-treatment whole-body 18F-FDG PET-CT; and receipt of radical treatment. The exclusion criteria were as follows: SUVmax of the primary tumor lower than 2.5; presence of distant metastasis; other concurrent malignancies; coexistence of a fatal disease; history of cancer treatment; and insufficient follow-up data and imaging information. All patients underwent restaging according to the 8th edition of the AJCC/TNM staging system.
FDG PET-CT
The Gemini TF 64 PET/CT scanner (Philips, The Netherlands) was employed for the study. Before the injection of 18F-FDG, all patients fasted for more than 6 h, and their serum glucose levels were maintained between 3.9 and 6.5 mmol/L. Then, 18F-FDG was intravenously administered at a dose of 148–296 MBq. After this injection, patients rested for 60 min in a dimly lit room. CT scans were acquired for all patients from the head to the proximal thigh (140 kV; 2.5 mA; matrix, 512 × 512; scan slice thickness, 4 mm). The recorded parameters included SUVmax and the metabolic tumor volume (MTV) of the primary lesion. MTV was defined as the region with SUVmax > 2.5.
Treatment and follow-up
In total, 682 patients (93.9% of the initial cohort) underwent platinum-based chemotherapy. Within the entire cohort, 624 (86%), 532 (73.3%), and 162 (22.3%) patients received neoadjuvant, concurrent, and adjuvant chemotherapy, respectively. The predominant regimen employed was platinum in combination with paclitaxel or gemcitabine. Radiotherapy was performed using intensity-modulated radiotherapy (IMRT), and the target volume and radiotherapy dose adhered to a previously established protocol10. The prescribed radiotherapy doses were as follows: GTV, 70–72.6 Gy/31–33 fractions; CTV1, 62–62.7 Gy/31–33 fractions; and CTV2, 54.4–56.2 Gy/31–33 fractions.
Local recurrence-free survival (LRFS) was defined as the duration from pathological diagnosis to local relapse or the conclusion of follow-up. Distant metastasis-free survival (DMFS) was defined as the duration of survival without cancer spreading from the primary tumor to distant organs after pathological diagnosis. Progression-free survival (PFS) was characterized as the interval from pathological diagnosis to tumor progression or death. OS was defined as the period from the pathological diagnosis to death of any cause. All patients adhered to a regular follow-up schedule, with assessments conducted every 3 months for the first 2 years, every 6 months for years 3–5, and subsequently on an annual basis.
Acquisition and processing of RNA sequencing data
An additional 29 fresh tissue samples were obtained from patients with NPC before treatment (Supplementary information, Table S1). RNA sequencing (RNA-seq) was performed at Geneplus-Beijing (Beijing, China). Total RNA was isolated from tissues using the Qiagen RNeasy Mini Kit. Following whole-transcriptome amplification, library construction was performed. Library quality analysis was conducted using the Agilent 2100 Bioanalyzer DNA 1000 Kit. The samples were sequenced using the DNBSEQ-T7 platform.
We used the NPC gene expression profile GSE102349, which includes prognostic data, to verify the prognostic ability of the SUV signature. Because there was no external NPC transcriptome dataset with available SUV data, we utilized the GSE135565 dataset for breast cancer to verify whether the SUV signature represents SUVmax. Additionally, the NPC single-cell dataset GSE150430 was used. These datasets were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/).
Construction of the SUV signature
Relevant genes encoding glucose metabolism proteins were selected from the Molecular Signatures Database. In total, 326 genes were manually curated into the glucose metabolism gene sets for further transcriptomic analysis (ementary information, Table S2). To make the results more representative, among the 29 patients who underwent transcriptome sequencing, the transcriptome data from the eight patients with the lowest SUVmax and the eight patients with the highest SUVmax were analyzed. Through Pearson correlation analysis of the glucose metabolism gene set and SUVmax, all related genes (P < 0.05) were defined as SUVmax-related glucose metabolism genes, and 55 genes were selected for subsequent analysis (ementary information, Table S3). The selected SUVmax-related glucose metabolism genes were modeled using LASSO logistic, and tenfold cross-validation was performed. The lambda of the model with the lowest average cross-validation error was taken, and the coefficients were extracted and added to obtain the transcriptome tag of SUVmax, which was defined as the SUV signature.
Single-cell RNA-seq
Single-cell mRNA-seq data were reported by Chen et al.11. The Python package Scanpy (version 1.4.6) was used to analyze this dataset. Clustering was performed using the UMAP algorithm. The SUV signature was calculated for each single cell. We then annotated the clusters with the following major cell types: B cells, epithelial cells, T cells, myeloid cells, NK cells, and plasma cells. The SUV signature of each class of cells was compared. T cells were divided into high and low score groups according to the median value of the SUV signature, and the expression of HAVCR2, PDCD1, LAG3, and TIGIT was compared between the groups.
Differential gene analysis and functional enrichment analysis
We conducted differential expression analysis in the high and low SUV groups based on the median SUVmax using the limma package. Characteristics meeting the criteria of |log2FC|> 2 and adj.P < 0.05 were designated as differential expression features. Subsequently, hub genes were used for Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis facilitated by the clusterProfiler package12. Furthermore, gene set enrichment analysis (GSEA) was executed on the gene expression matrix using the clusterProfiler package13.
Hypoxia score and immune function analysis
We selected a 15-gene expression signature (ACOT7, ADM, ALDOA, CDKN3, ENO1, LDHA, MIF, MRPS17, NDRG1, P4HA1, PGAM1, SLC2A1, TPI1, TUBB6, and VEGFA) that has been proven to perform optimally for classifying the hypoxia status14. The hypoxia score for each tumor sample was calculated using gene set variation analysis based on the 15 mRNA-based hypoxia signatures. The immune, stromal, and ESTIMATE scores and tumor purity in the study were calculated using the ESTIMATE algorithm. Then, Pearson’s correlation analysis was applied to examine the relationship between these indicators and the SUV signature in our center’s data and the GSE102349 dataset. The Pearson correlation coefficient (R) was calculated.
Statistical analysis
R software (version 3.6.1) and Python software (version 3.10) were used to organize, analyze, and visualize relevant data. We used Kaplan–Meier curves and the log rank test to compare survival data between different risk groups. The two-sample t-test was used for comparison between groups, and correlations between groups were assessed using Spearman’s rank correlation or Pearson’s correlation. Statistical significance was indicated by P < 0.05.
Results
Higher SUVmax suggests a larger tumor burden and poor prognosis
In this study, the median follow-up duration was 49.0 months (range: 3.0–118.0 months). The median patient age was 47 years (range: 11.0–78.0 years), and the majority of patients presented with AJCC stages III (43.4%) and IV (37.1%). Patient characteristics are summarized in Table 1. Throughout the follow-up period, 63 patients (8.7%) experienced local failure, 88 (12.1%) experienced distant metastasis, 161 (22.2%) encountered disease progression, and 75 (10.3%) died. The 5-year LRFS, DMFS, PFS, and OS rates were 89.7%, 87.7%, 76.7%, and 87.8%, respectively.
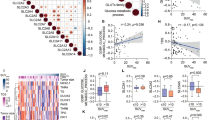
The correlations between SUVmax and clinical variables were examined. SUVmax was associated with T stage (P < 0.01, Fig. 1A), MTV (r = 0.691, P < 0.001, Fig. 1C), and lactate dehydrogenase (LDH) levels (r = 0.178, P < 0.001, Fig. 1D). SUVmax displayed a weak correlation with N stage (Fig. 1B). Subsequently, survival analysis was conducted. The ROC curve indicated the optimal cutoff of SUVmax for predicting OS, maximizing Youden’s index. Patients were then categorized into high and low SUVmax groups. The results revealed that LRFS (hazard ratio [HR] = 0.42, P = 0.003, Fig. 1E), PFS (HR = 0.71, P = 0.035, Fig. 1G), and OS (HR = 0.50, P = 0.007, Fig. 1H) were significantly worse in the high SUVmax group than in the low SUVmax group. Unexpectedly, there was no significant difference in DMFS between these groups (P = 0.318, Fig. 1F).
Associations of SUVmax with clinical variables in NPC. (A and B) Boxplots illustrate the distribution of SUVmax across different T and N stages in patients with NPC. (C and D) Scatter plots demonstrate the correlation of SUVmax with key parameters such as MTV and LDH levels. (E–H) The figure presents survival curves for LRFS, DMFS, PFS, and OS.
Formulation of the SUV signature using glucose metabolism genes
In a cohort of 29 patients undergoing transcriptome sequencing, individuals were categorized into two groups based on the median SUVmax. Differential gene analysis (Fig. 2A), KEGG analysis (Fig. 2B), and GSEA (Fig. 2C) revealed the enrichment of differential genes in pathways related to glucose metabolism. Consequently, we constructed the SUV signature using glucose metabolism genes.
Construction of the SUV signature. (A) Heatmap depicting differential gene expression between the high and low SUVmax groups using the R package “pheatmap” version 1.0.12. (B and C) KEGG enrichment analysis and GSEA conducted on the differentially expressed genes within the high and low SUVmax groups. (D, E) Genes selected through LASSO regression and logistic analysis. (F) Contribution of the selected genes in constructing the SUV signature along with their corresponding regression coefficients.
Through correlation analysis between genes related to glucose metabolism and SUVmax, we identified 55 genes associated with glucose metabolism. LASSO logistics modeling (Fig. 2D and E) was conducted with ten-fold cross-validation, and the lambda value (0.00392) corresponding to the model with the smallest cross-validation mean error was selected. The coefficients were then extracted and combined to derive the SUV signature (Fig. 2F) using the following formula:
Verification of the ability of the SUV signature to predict SUVmax and prognosis
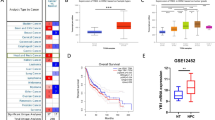
To verify whether the SUV signature can predict SUVmax, we first conducted a correlation analysis between these variables. This analysis was performed using our datasets comprising 16 cases and 29 cases (Figs. 3A and S2), as well as the GSE135565 dataset (Fig. 3B). The SUV signature was positively correlated with SUVmax, supporting supports the notion that the SUV signature could represent SUVmax. Additionally, we conducted a prognosis validation using the GSE102349 dataset, stratifying the cohorts based on the median SUV signature. Our findings revealed a significantly worse prognosis in the high SUV signature group than in the low SUV signature group (P = 0.006, Fig. 3C).
SUV signature predicts SUVmax and prognosis. (A, B) Association between the SUV signature and SUVmax in our 16-patient cohort and GSE135565. (C) Prognostic prediction using the SUV signature in GSE102349. (D) UMAP plot visualizing the clusters of each cell type in NPC. (E) Violin plot depicting the SUV signature score in each cell type. (F, G) UMAP plot illustrating the SUV signature in epithelial cells. (H, I) Density and violin plot presenting the SUV signature score in malignant epithelial cells and normal epithelial cells.
Subsequently, we utilized the single-cell RNA-seq dataset (GSE150430) for NPC to investigate the intrinsic biological characteristics associated with SUVmax. Employing UMAP, we categorized the cells into six clusters, including B cells, T cells, NK cells, myeloid cells, plasma cells, and epithelial cells (Fig. 3D). The SUV signature was then computed across these distinct clusters, revealing the highest expression in epithelial cells (Fig. 3E). Further delineating epithelial cells into malignant and normal subtypes, the SUV signature was significantly higher in malignant epithelial cells than in their non-malignant counterparts (Fig. 3F–I).
The SUV signature was associated with hypoxia and radiotherapy resistance
A correlation analysis was conducted using our center’s dataset (Fig. 4A) and the GSE102349 dataset (Fig. 4D) to examine the relationship between the SUV signature and hypoxia score. The results revealed significant correlations between these variables, as indicated by R of 0.76 in our center’s dataset and 0.62 in the GSE102349 dataset. The outcomes of GSEA further validated the enrichment of the SUV signature within hypoxia-associated hallmarks, as illustrated in Fig. 4B and E. This observation supports the notion that the SUV signature could potentially serve as an indicator of hypoxia.
Additionally, our investigation revealed a notable enrichment score of the SUV signature within the radiotherapy resistance gene set, as depicted in Fig. 4C and F. Given that radiotherapy is the primary treatment for NPC and insensitivity to radiotherapy implies a poor prognosis, the connection between the SUV signature and radiotherapy resistance is critical. Notably, hypoxia is a known mechanism contributing to radiotherapy resistance. This suggests that SUVmax could potentially predict an unfavorable prognosis by influencing sensitivity to radiotherapy through oxygen deficiency.
SUV signature is associated with immune function
In the single-cell RNA dataset, T cells were stratified into high and low SUV signature groups based on the median SUV signature. Our observations revealed a significant upregulation of immunosuppressive markers, including HAVCR2, PDCD1, LAG3, and TIGIT, in T cells with high SUV signature scores (Fig. 5A–D).
Relationship between the SUV signature and immune function. (A–D) Violin plots depict the expression of HAVCR2, PDCD1, LAG3, and TIGIT in T cells with high and low SUV signature scores. (E–H) The correlations of the SUV signature with the immune score, the stromal score, the predictive score, and tumor purity.
Expanding our analysis to the GSE102349 cohort, we further explored the associations of the SUV signature with the immune, stromal, and ESTIMATE scores and tumor purity (Fig. 5E–H). Remarkably, the SUV signature was negatively correlated with the immune, stromal, and ESTIMATE scores and positively corelated with tumor purity. These findings suggest an inverse relationship between the SUV signature and immune function.
Discussion
In this study, our focus on genes associated with glucose metabolism enabled the formulation of transcriptomic signatures linked to SUVmax in patients with NPC. To the best of our knowledge, this is the first study to delineate the transcriptomic profile of glucose uptake, assess its prognostic value, and explore its correlations with hypoxia, radiotherapy resistance, and immune function in individuals with NPC. The findings underscored that patients with a high SUV signature score have an unfavorable prognosis. We established that the SUV signature could function as an indicator of hypoxia through enrichment analysis, suggesting a plausible association between elevated SUVmax and hypoxia-induced phenomena in patients with NPC, such as radiotherapy resistance and immunosuppression. These insights contribute to a better understanding of the prognostic landscape and potential therapeutic targets in NPC.
The well-established Warburg effect posits that cancer cells preferentially metabolize glucose through glycolysis rather than more efficient mitochondrial oxidative phosphorylation, even under conditions of ample oxygen. FDG PET-CT capitalizes on increased glucose uptake precisely for tumor detection15. Notably, studies have identified substantial differences in the expression of glucose uptake-dependent transporters between high and low SUVmax groups of patients with HCC5. Therefore, employing the expression of gene associated with glucose metabolism to predict NPC prognosis emerges as a potentially valuable tool for precisely stratifying risk groups. In this study, we focused on 10 glucose metabolism-related genes associated with SUVmax in NPC: PKM, SEH1L, LHPP, LDHA, PPFIA4, PFKFB3, MPI, ZBTB20, CLDN9, and PGAM4. Pyruvate kinase, encoded by the PKM gene, crucially regulates cellular metabolism by converting phosphoenolpyruvate into pyruvate in glycolysis16. SEH1L encodes a protein integral to the Nup107-160 nuclear pore complex, impacting cell growth and proliferation17. LHPP encodes proteins participating in phosphorylation metabolism with anticancer effects18. LDHA-encoded proteins, part of the LDH family, are frequently overexpressed in cancer, promoting glycolysis and cancer progression19. PPFIA4 encodes the Liprin-α4 protein, which is associated with abnormal metabolic processes. Silencing PPFIA4 has been found to attenuate glycolysis, regulate glycolysis-related genes (PFKFB3 and ENO2), and inhibit cancer cell proliferation, migration, and invasion20. Phosphomannose isomerase, encoded by MPI, plays a vital role in glycosylation reactions21. ZBTB20, a transcriptional repressor, influences neurogenesis, glucose, and lipid metabolism. ZBTB20 induces ChREBP-A promoter transcriptional activity, regulating glycolytic and lipogenic enzyme genes and affecting glycolipid metabolism22. CLDN9 is correlated with glycolytic levels in endometrial carcinoma and esophageal adenocarcinoma23. PGAM4, encoding proteins catalyzing the reversible reaction of 3-phosphoglycerate to 2-phosphoglycerate in the glycolysis pathway, regulates metabolism and energy production24.
The prognostic model incorporating these genes, termed the SUV signature, demonstrated robust stratification of PFS. The SUV signature might hold significant implications for individualized precision therapy. Furthermore, our exploration into the cellular biology associated with the SUV signature, utilizing single-cell RNA-seq data, revealed that the SUV signature was highest in malignant epithelial tissues. This aligns with findings of Meng6, supporting the notion of increased F18-FDG uptake by tumor cells. SUVmax is relevant to various factors such as tumor metabolic activity, tumor cell counts, serum glucose levels, and fat content. In parallel with these considerations, our study uncovered associations of SUVmax with MTV, LDH levels, and T stage. The associations of SUVmax with MTV and LDH levels, reflective of the tumor burden25, suggested that SUVmax is linked to tumor burden. The T stage, indicative of the biological aggressiveness of tumors, further supported the association between SUVmax and the biological aggressiveness of NPC. Previous studies also indicated the clinical prognostic value of SUVmax in NPC7,26. For instance, in a study involving 253 patients with newly diagnosed metastatic NPC, those with higher SUVmax exhibited lower 3-year OS7. Consistent with these findings, our survival analysis demonstrated that OS, PFS, and LRFS were worse in patients with high SUVmax than in those with low SUVmax. Simultaneously, the transcriptome signature representing SUVmax, as constructed in our study, also indicated poor PFS, underscoring the prognostic significance of SUVmax in NPC. The ability of SUVmax to predict distant metastasis remains a topic of debate. Whereas Aktan et al.26 identified SUVmax as an important prognostic factor for distant metastasis and its utility in evaluating disease-free survival, Kitajima et al.27 found no significant correlation between primary tumor SUVmax and DMFS. Our study aligns with the latter findings, indicating that SUVmax could better predict prognosis regarding local treatment failure than distant metastasis.
Several studies have examined the association between SUVmax or its related genomic features and radiosensitivity in various malignancies6,28. Recently, evidence has emerged that radiomic characteristics derived from PET-CT, in combination with clinical parameters, hold promise for accurate therapeutic effect evaluation in NPC29. The potential of PET-CT to predict radiosensitivity was further demonstrated in an NPC xenograft nude mouse model30. Consistent with these findings, our enrichment analysis demonstrated an enrichment of gene sets associated with radiotherapy resistance in the high SUV signature group. Given that radiotherapy is the primary treatment for NPC, a high SUV signature in our study suggests that NPC is not sensitive to radiotherapy, implying a poor prognosis. Additionally, both correlation analysis and enrichment analysis in our study highlighted a close relationship between the SUV signature and hypoxia. This aligns with prior reports indicating a hypoxia gene signature association with FDG uptake in liver and lung cancers5,31. It is plausible that byproducts of glycolysis drive crucial biosynthetic pathways, providing a selective advantage to rapidly dividing cancer cells, and hypoxia signaling confers a survival advantage to tumor cells in a normoxic environment32. Studies also indicated that tumor hypoxia leads to radiotherapy resistance, primarily dependent on the generation of reactive oxygen species and formation of irreparable DNA damage resulting from peroxidation events33. Therefore, SUVmax represents a potential biomarker of hypoxia, influencing radiotherapy sensitivity and serving as a predictor of poor prognosis.
Moreover, in a study involving 84 patients with NPC, SUVmax was positively correlated with PD-L1 expression in tumor cells and negatively correlated with PD-L1 expression in tumor-infiltrating immune cells34. A phase 1/2 trial similarly identified T cells within neoplastic lesions as pivotal mediators of immunotherapy as assessed by whole-body PET35. Consistent with these findings, our study also revealed a significant upregulation of immunosuppressive markers in T cells with high SUV signature scores. The correlation between SUVmax and immunosuppression might be mediated through the effects of hypoxia. Hypoxia affects immune function through hypoxia-induced factor-136. Immunosuppressive cells under hypoxic conditions can utilize fatty acid oxidation for cellular energy, maintaining immunosuppressive capabilities against effector T cells37. Consequently, the SUV signature could represent a tool to characterize the tumor immune microenvironment and potentially guide the use of immunosuppressive agents.
Our study had several limitations. First, the small sample size of patients undergoing transcriptomic sequencing for NPC, coupled with the absence of external transcriptomic data on SUVmax in this context, prompted us to perform external validation using breast cancer data. Consequently, further confirmation of the accuracy and reliability of the SUV signature requires larger cohort studies with dedicated NPC datasets. Second, the retrospective nature of our study introduced the possibility of bias, underscoring the need for prospective studies to mitigate potential biases and enhance the robustness of our findings. Third, although our bioinformatics analysis suggests that SUVmax could serve as a marker of hypoxia, the sensitivity and specificity of SUVmax in this role remain unclear. Rigorous studies with well-defined methodologies are essential to clarify the utility of SUVmax as a marker of hypoxia, shedding light on its potential diagnostic accuracy.
In summary, our study highlights the utility of SUVmax as a predictive indicator of poor prognosis in NPC. The development of a SUVmax-based transcriptomic signature underscores its potential as a marker of predicting tumor hypoxia. Furthermore, our findings suggest that the biological behavior associated with SUVmax could contribute to a worse prognosis by affecting radiation sensitivity and suppressing immune function. These insights offer valuable perspectives for understanding the clinical implications of SUVmax in NPC, and these findings could guide future therapeutic strategies.
Data availability
The data generated in this study are accessible to interested parties upon reasonable request, and should be directed to the corresponding author.
References
Chen, Y. P. et al. Nasopharyngeal carcinoma. Lancet 394(10192), 64–80 (2019).
Wong, K. C. W. et al. Nasopharyngeal carcinoma: An evolving paradigm. Nat. Rev. Clin. Oncol. 18(11), 679–695 (2021).
Zhang, L. et al. Gemcitabine plus cisplatin versus fluorouracil plus cisplatin in recurrent or metastatic nasopharyngeal carcinoma: A multicentre, randomised, open-label, phase 3 trial [published correction appears in Lancet. 2016 Oct 15;388(10054):1882]. Lancet 388(10054), 1883–1892 (2016).
Sanli, Y. et al. Update 2018: 18F-FDG PET/CT and PET/MRI in head and neck cancer. Clin. Nucl. Med. 43(12), e439–e452 (2018).
Xia, H. et al. Hypoxia-induced modulation of glucose transporter expression impacts 18F-fluorodeoxyglucose PET-CT imaging in hepatocellular carcinoma. Eur. J. Nucl. Med. Mol. Imaging 47(4), 787–797 (2020).
Meng, J. et al. Prognostic value of metabolic signature on 18F-FDGuptake in breast cancer patients after radiotherapy. Mol. Ther. Oncolytics. 23, 412–419 (2021).
Sun, X. S. et al. Maximal standard uptake values of 18F-fluoro-2-deoxy-D-glucose positron emission tomography compared with Epstein-Barr virus DNA as prognostic indicators in de novo metastatic nasopharyngeal carcinoma patients. BMC Cancer 19(1), 908 (2019).
Lee, H. et al. Metabolism-associated gene signatures for FDG avidity on PET/CT and prognostic validation in hepatocellular carcinoma. Front. Oncol. 12, 845900 (2022).
Gray, L. H., Conger, A. D., Ebert, M., Hornsey, S. & Scott, O. C. The concentration of oxygen dissolved in tissues at the time of irradiation as a factor in radiotherapy. Br. J. Radiol. 26(312), 638–648 (1953).
Lin, S. et al. Update report of nasopharyngeal carcinoma treated with reduced-volume intensity-modulated radiation therapy and hypothesis of the optimal margin. Radiother. Oncol. 110(3), 385–389 (2014).
Chen, Y. P. et al. Single-cell transcriptomics reveals regulators underlying immune cell diversity and immune subtypes associated with prognosis in nasopharyngeal carcinoma. Cell Res. 30(11), 1024–1042 (2020).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 16(5), 284–287 (2012).
Hänzelmann, S., Castelo, R. & Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 14, 7 (2013).
Ye, Y. et al. Characterization of hypoxia-associated molecular features to aid hypoxia-targeted therapy. Nat. Metab. 1(4), 431–444 (2019).
Hsu, P. P. & Sabatini, D. M. Cancer cell metabolism: Warburg and beyond. Cell. 134(5), 703–707 (2008).
Morita, M. et al. PKM1 confers metabolic advantages and promotes cell-autonomous tumor cell growth. Cancer Cell. 33(3), 355–367 (2018).
Platani, M., Samejima, I., Samejima, K., Kanemaki, M. T. & Earnshaw, W. C. Seh1 targets GATOR2 and Nup153 to mitotic chromosomes. J Cell Sci. 131(9), jcs213140 (2018).
Hindupur, S. K. et al. The protein histidine phosphatase LHPP is a tumour suppressor. Nature. 555(7698), 678–682 (2018).
Liu, J. et al. Metabolic enzyme LDHA activates Rac1 GTPase as a noncanonical mechanism to promote cancer. Nat. Metab. 4(12), 1830–1846 (2022).
Huang, J. et al. PPFIA4 promotes colon cancer cell proliferation and migration by enhancing tumor glycolysis. Front. Oncol. 11, 653200 (2021).
Ichikawa, M., Scott, D. A., Losfeld, M. E. & Freeze, H. H. The metabolic origins of mannose in glycoproteins. J. Biol. Chem. 289(10), 6751–6761 (2014).
Xie, Z. et al. ZBTB20 is essential for cochlear maturation and hearing in mice. Proc. Natl. Acad. Sci. U. S. A. 120(24), e2220867120 (2023).
Wang, Z. H., Zhang, Y. Z., Wang, Y. S. & Ma, X. X. Identification of novel cell glycolysis related gene signature predicting survival in patients with endometrial cancer. Cancer Cell Int. 19, 296 (2019).
Fukushi, A., Kim, H. D., Chang, Y. C. & Kim, C. H. Revisited metabolic control and reprogramming cancers by means of the Warburg effect in tumor cells. Int. J. Mol. Sci. 23(17), 10037 (2022).
Cottereau, A. S. et al. Risk stratification in diffuse large B-cell lymphoma using lesion dissemination and metabolic tumor burden calculated from baseline PET/CT†. Ann. Oncol. 32(3), 404–411 (2021).
Aktan, M. et al. Prognostic value of pre-treatment 18F-FDG PET uptake for nasopharyngeal carcinoma. Radiol. Med. https://doi.org/10.1007/s11547-017-0837-4 (2017).
Kitajima, K. et al. Prognostic value of FDG PET imaging in patients with laryngeal cancer. PLoS One. 9(5), e96999 (2014).
Na, F. et al. Primary tumor standardized uptake value measured on F18-Fluorodeoxyglucose positron emission tomography is of prediction value for survival and local control in non-small-cell lung cancer receiving radiotherapy: Meta-analysis. J. Thorac. Oncol. 9(6), 834–842 (2014).
Kim, S. J., Choi, J. Y., Ahn, Y. C., Ahn, M. J. & Moon, S. H. The prognostic value of radiomic features from pre- and post-treatment 18F-FDG PET imaging in patients with nasopharyngeal carcinoma. Sci. Rep. 13(1), 8462 (2023).
Zheng, Y. et al. The preliminary study of 18F-FLT micro-PET/CT in predicting radiosensitivity of human nasopharyngeal carcinoma xenografts. Ann. Nucl. Med. 29(1), 29–36 (2015).
Heiden, B. T. et al. 18F-FDG PET intensity correlates with a hypoxic gene signature and other oncogenic abnormalities in operable non-small cell lung cancer. PLoS One. 13(7), e0199970 (2018).
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324(5930), 1029–1033 (2009).
Graham, K. & Unger, E. Overcoming tumor hypoxia as a barrier to radiotherapy, chemotherapy and immunotherapy in cancer treatment. Int. J. Nanomed. 13, 6049–6058 (2018).
Zhao, L. et al. Usefulness of [18F]fluorodeoxyglucose PET/CT for evaluating the PD-L1 status in nasopharyngeal carcinoma. Eur. J. Nucl. Med. Mol. Imaging 47(5), 1065–1074 (2020).
Kist-de-Ruijter, L. et al. Whole-body CD8+ T cell visualization before and during cancer immunotherapy: A phase 1/2 trial. Nat. Med. 28(12), 2601–2610 (2022).
Kheshtchin, N. & Hadjati, J. Targeting hypoxia and hypoxia-inducible factor-1 in the tumor microenvironment for optimal cancer immunotherapy. J. Cell Physiol. 237(2), 1285–1298 (2022).
Bader, J. E., Voss, K. & Rathmell, J. C. Targeting metabolism to improve the tumor microenvironment for cancer immunotherapy. Mol. Cell. 78(6), 1019–1033 (2020).
Acknowledgements
We thank Medjaden Inc. for scientific editing and proofreading of this manuscript.
Funding
This work was supported by research projects for the the Fujian Province Joint Funds for the Innovation of Science and Technology (2023Y9427, 2023Y9409). Fujian Provincial Cancer Hospital Talent Development Program (2022YNG02).
Author information
Authors and Affiliations
Contributions
Study concept and design: CB C, ZD F. Acquisition, analysis, or interpretation of data: All authors. Drafting of the manuscript: JM D, LQ. Critical revision of the manuscript for important intellectual content: All authors. Statistical analysis:YH L,XB Z,CX H,JB H. Study supervision: CB C, ZD F.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study was approved by the Ethical Committee of Fujian Cancer Hospital (YKT2020-011-01) and conducted in accordance with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. Patient identifiers such as names were not collected; instead, patients were given a numerical identifier. Informed consent was obtained from all participants or their parents/legal guardians (for those younger than 18 years). For confidentiality, the patients’ charts were used only within the confines of the records department, and only the investigators and study assistants had access to the files.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ding, J., Liqian, Lin, Y. et al. Baseline SUVmax is correlated with tumor hypoxia and patient outcomes in nasopharyngeal carcinoma. Sci Rep 14, 20157 (2024). https://doi.org/10.1038/s41598-024-71191-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-71191-y
Keywords
This article is cited by
-
18F-NOTA-FAPI-04 PET/CT metabolic parameters combined with hematological biomarkers predict pathological complete response after neoadjuvant chemoradiotherapy in esophageal squamous cell carcinoma
European Journal of Nuclear Medicine and Molecular Imaging (2025)