Abstract
Dyskalemia is a common electrolyte abnormality. Since dyskalemia can cause fatal arrhythmias and cardiac arrest in severe cases, it is crucial to monitor serum potassium (K+) levels on time. We developed deep learning models to detect hyperkalemia (K+ ≥ 5.5 mEq/L) and hypokalemia (K+ < 3.5 mEq/L) from electrocardiograms (ECGs), which are noninvasive and can be quickly measured. The retrospective cohort study was conducted at two hospitals from 2006 to 2020. The training set, validation set, internal testing cohort, and external validation cohort comprised 310,449, 15,828, 23,849, and 130,415 ECG-K+ samples, respectively. Deep learning models demonstrated high diagnostic performance in detecting hyperkalemia (AUROC 0.929, 0.912, 0.887 with sensitivity 0.926, 0.924, 0.907 and specificity 0.706, 0.676, 0.635 for 12-lead, limb-lead, lead I ECGs) and hypokalemia (AUROC 0.925, 0.896, 0.885 with sensitivity 0.912, 0.896, 0.904 and specificity 0.790, 0.734, 0.694) in the internal testing cohort. The group predicted to be positive by the hyperkalemia model showed a lower 30-day survival rate compared to the negative group (p < 0.001), supporting the clinical efficacy of the model. We also compared the importance of ECG segments (P, QRS, and T) on dyskalemia prediction of the model for interpretability. By applying these models in clinical practice, it will be possible to diagnose dyskalemia simply and quickly, thereby contributing to the improvement of patient outcomes.
Similar content being viewed by others
Introduction
Potassium (K+) regulates multiple physiological processes and is necessary for the normal functioning of nerves, muscles, and the heart. However, diarrhea, vomiting, metabolic alkalosis, and various medications reduce serum K+ levels. Renal failure and medications (renin–angiotensin–aldosterone system [RAAS] inhibitors and nonsteroidal anti-inflammatory drugs) may increase K+ concentration because they inhibit K+ excretion1,2,3.
Hypokalemia (K+ < 3.5 mEq/L) is commonly observed in approximately 21% of hospitalized patients4. Hyperkalemia (K+ ≥ 5.5 mEq/L) is common in patients with chronic kidney disease (CKD) and heart failure; in particular, as the severity of CKD increases, the prevalence of hyperkalemia also increases1,5,6,7. Dyskalemia increases the risk of fatal arrhythmias, flaccid paralysis, hyporeflexia, and death; therefore, a prompt response is required8,9. In patients with cardiovascular disease, the risk of death increases even with moderate dyskalemia; therefore, caution is required3,10.
Therefore, monitoring dyskalemia is important. However, a blood test, which is the only way to monitor serum K+ levels, is invasive, takes time to confirm results, and causes inconvenience when repeatedly performed. The report that only 10% of patients are monitored through blood tests while administering dyskalemia-inducing drugs suggests that continuous monitoring of electrolytes through blood sampling is difficult11.
In contrast, an electrocardiogram (ECG) is noninvasive, fast, and repeatable and provides significant information about heart diseases12. Accordingly, ECG is used together for the differential diagnosis of hyperkalemia, and if changes suspicious for hyperkalemia are observed on ECG, immediate treatment without waiting for the test results is sometimes recommended.
However, even skilled physicians have had difficulty diagnosing dyskalemia with only minute changes in ECG13,14. Artificial intelligence (AI) based ECG analysis technology differentiates minute current differences in the heart that are difficult for humans to read, making it possible to predict future arrhythmias and detect heart failure. Therefore, active study is underway on its clinical applicability. In this study, using information from ECGs, serum K + concentration was predicted, and dyskalemia was diagnosed. The findings suggest that the AI-based technology has potential clinical utility.
Results
Study cohort description and demographic characteristics
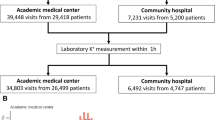
From January 1, 2006, to December 31, 2020, 440,835 ECG-serum K+ laboratory pairs from 196,976 patients were collected from Hallym University Sacred Heart Hospital (Hospital A) for deep learning model development, whereas Hallym University Kangnam Sacred Heart Hospital (Hospital B) contributed 304,332 ECG-serum K+ laboratory pairs from 140,517 patients, consisting external validation cohort. The summary of study cohorts can be found in Fig. 1. Following data split and exclusion criteria, the training set, validation set, internal testing cohort, and external validation cohort comprised 310,449, 15,828, 23,849, and 130,415 ECG-serum K+ laboratory samples, which were obtained from 152,016, 9,577, 14,382, and 82,710 individuals, respectively. In the training and validation sets, 3,710 and 252 patients with hyperkalemia with 5,931 and 359 ECGs, respectively, and 10,620 and 758 patients with hypokalemia with 19,789 and 1,195 ECGs, respectively, were included.
We divided the internal testing and external validation cohorts according to the presence or absence of dyskalemia: the patients with hypokalemia, the normal K+ patients, and the patients with hyperkalemia. We compared the demographic characteristics (age, sex), ECG numeric features (PR interval, QRS duration, QT interval, QTc interval, P-wave axis, R-wave axis, T-wave axis), and eGFR levels of the patients among the groups. Table 1 summarizes the patient demographics of each group from the internal testing and external validation cohort. In both cohorts, significant differences were found among the groups for most variables.
Deep learning model performance
We developed DeepECG-Hyperkalemia models that diagnose whether the serum potassium level is 5.5 mEq/L or higher and DeepECG-Hypokalemia models that diagnose whether the serum potassium level is lower than 3.5 mEq/L. Tables 2 and 3 summarize the performance of DeepECG-Hyperkalemia and DeepECG-Hypokalemia, respectively, in the internal testing and external validation cohorts (Supplement Table 1 and 2).
The area under the receiver operating curves (AUROC) of the DeepECG-Hyperkalemia model were 0.929 (95% CI: 0.916–0.941), 0.912 (95% CI, 0.898–0.925), and 0.887 (95% CI, 0.871–0.902) for the 12-lead, limb-lead, and lead I models, respectively, in the internal testing cohort. In the external validation cohort, the AUROCs were 0.923 (95% CI, 0.914–0.931), 0.910 (95% CI, 0.902–0.918), and 0.881 (95% CI, 0.872–0.891), respectively. These values were similar to those obtained in the internal testing cohort, suggesting that the model is capable of generalizing data that have not been previously observed. The performance decreased for the models of reduced number of input leads, as confirmed by the ROC curve comparison plot shown in Fig. 2A,B.
A similar pattern was observed in the DeepECG-Hypokalemia model. The AUROCs in the internal testing cohort were 0.925 (95% CI, 0.919–0.930), 0.896 (95% CI, 0.889–0.903), and 0.885 (95% CI, 0.878–0.892) for the 12-lead, limb-lead, and lead I models, respectively. In the external validation cohort, the AUROCs were 0.913 (95% CI, 0.910–0.916), 0.883 (95% CI, 0.879–0.887), and 0.870 (95% CI, 0.867–0.874), respectively. The ROC curve comparison plots in Fig. 2C,D also showed the highest performance in the 12-lead model and the lowest performance in the lead I model.
Subgroup analysis
We further examined the dyskalemia diagnostic performance across various subgroups in the external validation cohort. Notably, estimated glomerular filtration rate (eGFR) status, age, sex, heart rate, QTc value, and the existence of atrial fibrillation or flutter showed interactions with the performance of the DeepECG-Hyperkalemia 12-lead model (Supplement Table 3). However, the DeepECG-Hypokalemia model did not interact with sex, age, or eGFR status (Supplement Table 4). In both models, abnormal ECG findings, such as tachycardia, atrial fibrillation, and flutter, had poorer performance than those without these findings.
Thirty-day all-cause mortality analysis
To assess the clinical efficacy of the DeepECG-Hyperkalemia model, we conducted a 30-day survival analysis using the Kaplan–Meier estimation method on the external validation cohort. Specifically, we analyzed the survival function of the cohort over 30 days following their last ECG and serum K+ test. The external validation cohort was divided into groups based on their ground-truth label of hyperkalemia and the prediction of the DeepECG-Hyperkalemia model.
The survival rate was significantly lower in the hyperkalemia group than in the non-hyperkalemia group (p < 0.001, Fig. 3A). Furthermore, the survival rate was significantly lower (p < 0.001) when the DeepECG-Hyperkalemia model made a positive prediction (Fig. 3B). To specify the clinical efficacy of the model for mortality prediction, it is important to note the comparison between false-positive and true-negative groups of patients without hyperkalemia (Fig. 3C). The survival rate of the group predicted to be positive was lower than that of the group predicted to be negative (p < 0.001).
(A) Kaplan–Meier curve for mortality at 30-day follow up according to the hyperkalemia (Total ECGs = 82,984). (B) Kaplan–Meier curve for mortality at 30-day follow up according to the DeepECG-Hyperkalemia diagnosis result. (C) Kaplan–Meier curve for mortality at 30-day follow up according to the DeepECG-Hyperkalemia diagnosis among patients with actual K+ more than 5.5 mEq/L—The patients classified as DeepECG-Hyperkalemia positive showed worse 30-day survival (Total ECGs = 82,320).
Model interpretability through electrocardiogram feature masking
Although the deep learning model guarantees excellent performance, it is basically a black-box model, so it is not known what characteristics the diagnostic results depend on. To interpret the dyskalemia detection process of the models, we masked the main features (P wave, QRS complex, and T wave) of the ECG waveforms and observed the effects of the absence of each feature on the dyskalemia detection models. Supplement Table 5 summarizes the AUROCs of the 12-lead models evaluated on the feature-masked ECGs from the external validation cohort.
Notably, the presence or absence of a P wave had little effect on the model performance. Both the hyperkalemia and hypokalemia models showed little decline in AUROC (from 0.923 to 0.918 and from 0.913 to 0.912, respectively), but only slightly increased the positively predicted ratio of the hyperkalemia model. In contrast, when we masked the QRS complex and T-wave features, the AUROC decreased for both the hyperkalemia (from 0.923 to 0.770 when masking QRS and from 0.923 to 0.759 when masking T wave) and hypokalemia (from 0.913 to 0.763 when masking QRS and from 0.913 to 0.792 when masking T wave) models.
Interestingly, there were differences in the effects of the feature masking on the models. When we masked the QRS complex in the hyperkalemia model, the positively predicted ratio significantly increased, which led to a significant decrease in the specificity and an increase in the sensitivity. However, when the T wave was masked, both the sensitivity and the specificity declined. Hypokalemia worked in the opposite direction. In the case of the QRS complex masking, both the sensitivity and the specificity were reduced; however, in the case of the T-wave masking, the specificity was lowered, whereas the sensitivity was improved.
To investigate the different effects of ECG masking on the models, we examined the distribution of changes in the model output probabilities resulting from masking the ECG features, as shown in Supplement Fig. 1. In the case of the hyperkalemia model, QRS complex masking tended to move the output value of the model in the positive direction, whereas T-wave masking had a slight tendency to move in the positive direction. However, the roles of QRS complex masking and T wave masking were switched in the hypokalemia model. Meanwhile, the change in the probability value by P-wave masking formed a high density of approximately zero in both models, suggesting an insignificant effect.
Discussion
In this study, using the DeepECG-Hyperkalemia and DeepECG-Hypokalemia models, 12-lead, limb-lead, and lead I models showed high performances for dyskalemia in both the internal testing and external validation cohorts. Although the performance decreased as the number of input leads decreased, some performances were maintained, even in lead I alone. Particularly, changes in the QRS complex and T wave were important and meaningful in predicting dyskalemia through a model masking the P wave, QRS complex, and T wave.
Previous studies in which experienced physicians diagnosed dyskalemia using only ECG data demonstrated poor diagnostic accuracy, and there has been difficulty in diagnosing dyskalemia by observing minute changes in the ECG caused by dyskalemia13,15,16. Studies proving its usefulness by diagnosing hyperkalemia or calculating K+ values without a blood test through single-lead ECG13,17,18 and recently reported deep learning algorithm studies for predicting dyskalemia using ECG19 have shown better performance. However, these studies had limitations in terms of selection bias and generalization, in that they were conducted at only one center. There was no multicenter validation, and only patients who visited the emergency department were included. However, in this study, the 12-lead, limb-lead, and lead I models showed good performance even in the external validation cohort.
It is also meaningful that a subgroup analysis was performed to predict hyperkalemia. In the presence of abnormal HR, QT interval, atrial fibrillation, or flutter, performance decreased, whereas the PR interval and QRS duration did not have a significant effect. The performance of DeepECG-Hyperkalemia was high in advanced CKD, and the AUROC values observed in GFR < 60 or 45 mL/min/1.73 m2 showed results comparable to those of a previous study20. This suggests that this model can be particularly useful in patients with CKD at high risk of hyperkalemia. In contrast, the performance of DeepECG-Hypokalemia was not significantly affected by renal function.
Patients with CKD easily experience impaired renal excretion of K+ due to decreased renal function and easily develop dyskalemia owing to the use of RAAS blockers or diuretics. Refractory hyperkalemia may be an indication for dialysis treatment10. Moreover, if patients with CKD have cardiovascular complications, they are significantly vulnerable to dyskalemia and may experience fatal consequences3,10. Several medications used for the treatment of cardiovascular diseases can also promote dyskalemia21. Thus, serum K+ levels are critical for the use, adjustment, and discontinuation of medications; and monitoring serum K+ levels and maintaining an appropriate concentration are emphasized22. However, periodic blood tests are invasive and time-consuming, and their repeatability is limited in patients with chronic diseases and poor vascular conditions. As demonstrated in this study, if drug-induced hyperkalemia can be detected quickly and easily through ECG, it is expected to be remarkably useful in clinical practice and will greatly help in improving patient prognosis.
The 30-day survival rate was significantly lower in patients with false-positive prediction of hyperkalemia, which is consistent with the results of recent studies reporting more comorbidities and worse outcomes in patients with false-positive results on AI-assisted ECG23. This finding suggests that the DeepECG-Hyperkalemia model can predict death by detecting minute ECG changes following a gradual increase in serum K+ concentrations. Recently, cardiac arrest prediction models using deep learning algorithms have been applied in clinical practice24,25,26. In addition, if the deep learning model of this study can be used together, it is expected to increase the accuracy of cardiac arrest prediction, thereby contributing to the improvement of patient prognosis. However, additional studies are required to verify its clinical efficacy.
This study has some limitations. First, this was a retrospective study, but the robustness of the results was increased by targeting many patients over a long period of time. Second, no investigation was conducted on the history of medications that affect serum K+ levels or whether treatment for dyskalemia was present. Therefore, it is necessary to evaluate the accuracy and clinical applicability of these models in prospective clinical studies. Third, as the cause of death was not analyzed, further studies are required to evaluate the accuracy, mechanism, and clinical usefulness of this model for predicting the risk of death.
In conclusion, we demonstrated the high diagnostic performance of deep learning models for the noninvasive screening of dyskalemia based on ECGs. By applying these models in clinical practice, it will be possible to diagnose dyskalemia simply and quickly, thereby contributing to the improvement of patient outcomes. A multicenter prospective study is required, and the possibility of remote monitoring needs to be verified.
Methods
Ethics statement
This retrospective database study was approved by our institutional review board (IRB). The requirement for informed consent from the study subjects was waived by the IRB of Hallym University Sacred Heart Hospital (No. HALLYM 2021-07-011-001) and Hallym University Kangnam Sacred Heart Hospital (No. HKS 2022-05-015) owing to its retrospective study design. This study was conducted by the principles of the Declaration of Helsinki.
Study population
The study was conducted based on electronic medical record data from 2006 to 2020, which contained a history of ECG measurements and blood laboratory tests from Hospital A and Hospital B. The study cohorts consisted of the patients who conducted serum K + concentration tests before and after 24 h of the ECG measurement. We excluded the patients whose age is under 18. Patients with pacemakers or implantable cardiac defibrillators or receiving cardiac resynchronized therapy were also excluded.
Data acquisition and preparation
We extracted the ECG data of the study cohorts in raw XML format through the GE MUSE database system of the institutions. Each ECG was 10 s long, sampled at 500 Hz or 250 Hz with 16-bit resolution. The ECG waveforms were extracted from the XML data and resampled at 250 Hz. We applied a bandpass digital filter (0.67 Hz–40 Hz) to the extracted ECG waveforms to eliminate the baseline wandering and signal noise that could potentially affect model training. We also collected the machine measurements of the ECGs (e.g., heart rates, PR intervals, QRS durations, QTc, and diagnoses). We used this information to find ECG abnormalities (e.g., bradycardia, tachycardia, wide QRS complex, prolonged QT, and atrial fibrillation) for the subgroup analyses. The other information (e.g., gender, age, lab test results) was acquired from the EMR database.
Hospital A’s data were used for the deep learning model development and were divided into a training set for model learning, a validation set for model selection, and a test set for internal testing. Each set of patients was exclusively grouped. Hospital B’s data were used for the external validation. As serum K+ levels change according to various external factors, only blood chemistry test data within an hour before and after the ECG measurement were included in the validation, internal testing, and external validation cohorts. For a better generalization performance, the ECG-serum K+ test time interval was extended to 12 h for the training set to increase the training data.
Deep learning model development
We developed a DeepECG-Hyperkalemia model that diagnosed whether the serum K+ level was ≥ 5.5 mEq/L and a DeepECG-Hypokalemia model that diagnosed whether the serum K+ level was < 3.5 mEq/L using 12-lead ECG. Considering the adoption of mobile ECG devices, we also developed the limb-lead and single-lead (lead I) models. We employed SE-ResNet architecture14,15, a variation of the convolutional neural network13. It is an end-to-end model capable of directly processing raw signals obtained from ECG measuring devices, eliminating the need for an explicit feature extraction procedure. The model consisted of 34 layers. When the raw ECG signal was input from the first convolution layer, the embedding feature was obtained through four SE-ResNet blocks, followed by one fully connected output layer and a sigmoid activation function.
We developed six distinct models, categorized based on the number of input signal channels (12-lead, limb-lead, and lead I) and the types of disease being addresed (hyperkalemia and hypokalemia). Each of these six models was trained independently to perform the corresponding binary classification task. The models were trained via backpropagation using the stochastic gradient descent method27 to minimize the cross-entropy between the final output value and ground-truth label. This work was implemented using Python programming, in which the model implementation and learning codes were based on the PyTorch library28.
Model evaluation
We chose the models which showed the best performance in the validation set. Considering the class imbalance of the data, the AUROC was used as a model selection criterion. Operating points were selected based on the validation set. We considered two operating points: the point where the Youden J index29 was maximized (Youden J) and the point where the specificity was maximized while the sensitivity satisfied 0.9 (high sensitivity). Based on the selected models and operating points, we observed the AUROC, sensitivity, specificity, positive predictive value, and negative predictive value in the internal and external validation cohorts. Subgroup analysis was performed on the external validation cohort.
Deep learning model interpretability
Existing studies in deep learning have used a method of visualizing a saliency map using the activation values of the model30 or learning an explanatory model for a specific data point and observing the importance of each characteristic31,32. These local interpretability methods are used to interpret the predictions of a model for each specific data point rather than interpreting the model as a whole.
We proposed a method of directly masking the main features (P wave, QRS complex, and T wave) of an ECG waveform and investigated the importance of these features. We utilized the deep learning-based segmentation method to delineate the ECG features33. The model was trained using an open ECG dataset labeled by clinicians to accurately delineate these ECG features34. Based on the ECG delineation results, the raw values of the features to be masked were replaced with the median of the raw values of the baseline segments. Supplement Fig. 2 shows examples of the masking of each feature. By observing how the distribution of the model output values changed when the P wave, QRS complex, and T wave were masked, we aimed to interpret the impact of each feature on the prediction of the dyskalemia models in a broader scope.
Statistical analyses
For the significance test of the characteristics comparison, we performed the chi-squared test for the proportion of sex and the one-way analysis of variance for the rest of the variables. Considering that multiple hypotheses were tested simultaneously, we adjusted the significance level via the Bonferroni correction method to control the overall Type I error. The bootstrapping method35 was used to estimate variances in model performances. The estimated variances were used to calculate the confidence intervals of classification performance metrics. The bootstrapping method was also employed to perform significance tests of the AUROC comparisons between the subgroups. P-values were calculated using z-statistics for comparison between two groups and chi-squared statistics for comparisons among three groups36. The significance of the differences in survival probability was assessed using the log-rank test.
Data availability
All data are available after request to JNA, MP, YL, and SGK. The code is available online at https://github.com/bakqui/ecg-dyskalemia. Please note that this is a re-implemented version of the original code, licensed for non-commercial use. We have verified that the reported results are reproducible with this version.
Change history
08 April 2025
An Editorial Expression of Concern to this paper has been published: https://doi.org/10.1038/s41598-025-96414-8
References
Einhorn, L. M. et al. The frequency of hyperkalemia and its significance in chronic kidney disease. Arch. Intern. Med. 169, 1156–1162. https://doi.org/10.1001/archinternmed.2009.132 (2009).
Goyal, A. et al. Serum potassium levels and mortality in acute myocardial infarction. JAMA 307, 157–164. https://doi.org/10.1001/jama.2011.1967 (2012).
Gennari, F. J. Hypokalemia. N. Engl. J. Med. 339, 451–458. https://doi.org/10.1056/NEJM199808133390707 (1998).
Conway, R., Creagh, D., Byrne, D. G., O’Riordan, D. & Silke, B. Serum potassium levels as an outcome determinant in acute medical admissions. Clin. Med. (Lond.) 15, 239–243. https://doi.org/10.7861/clinmedicine.15-3-239 (2015).
Betts, K. A. et al. The prevalence of hyperkalemia in the United States. Curr. Med. Res. Opin. 34, 971–978. https://doi.org/10.1080/03007995.2018.1433141 (2018).
Nakhoul, G. N. et al. Serum potassium, end-stage renal disease and mortality in chronic kidney disease. Am. J. Nephrol. 41, 456–463. https://doi.org/10.1159/000437151 (2015).
Aldahl, M. et al. Associations of serum potassium levels with mortality in chronic heart failure patients. Eur. Heart J. 38, 2890–2896. https://doi.org/10.1093/eurheartj/ehx460 (2017).
Kovesdy, C. P. et al. Serum potassium and adverse outcomes across the range of kidney function: A CKD Prognosis Consortium meta-analysis. Eur. Heart J. 39, 1535–1542. https://doi.org/10.1093/eurheartj/ehy100 (2018).
Jensen, H. K., Brabrand, M., Vinholt, P. J., Hallas, J. & Lassen, A. T. Hypokalemia in acute medical patients: risk factors and prognosis. Am. J. Med. 128, 60–67. https://doi.org/10.1016/j.amjmed.2014.07.022 (2015).
An, J. N. et al. Severe hyperkalemia requiring hospitalization: Predictors of mortality. Crit. Care 16, R225. https://doi.org/10.1186/cc11872 (2012).
Schmidt, M. et al. Adherence to guidelines for creatinine and potassium monitoring and discontinuation following renin-angiotensin system blockade: A UK general practice-based cohort study. BMJ Open 7, e012818. https://doi.org/10.1136/bmjopen-2016-012818 (2017).
Diercks, D. B., Shumaik, G. M., Harrigan, R. A., Brady, W. J. & Chan, T. C. Electrocardiographic manifestations: Electrolyte abnormalities. J. Emerg. Med. 27, 153–160. https://doi.org/10.1016/j.jemermed.2004.04.006 (2004).
Lin, C. S. et al. A deep-learning algorithm (ECG12Net) for detecting hypokalemia and hyperkalemia by electrocardiography: Algorithm development. JMIR Med. Inform. 8, e15931. https://doi.org/10.2196/15931 (2020).
Van Mieghem, C., Sabbe, M. & Knockaert, D. The clinical value of the ECG in noncardiac conditions. Chest 125, 1561–1576. https://doi.org/10.1378/chest.125.4.1561 (2004).
Rafique, Z. et al. Can physicians detect hyperkalemia based on the electrocardiogram?. Am. J. Emerg. Med. 38, 105–108. https://doi.org/10.1016/j.ajem.2019.04.036 (2020).
Wrenn, K. D., Slovis, B. S. & Slovis, C. M. The ability of physicians to predict electrolyte deficiency from the ECG. Ann. Emerg. Med. 19, 580–583. https://doi.org/10.1016/s0196-0644(05)82194-8 (1990).
Attia, Z. I. et al. Novel bloodless potassium determination using a signal-processed single-lead ECG. J. Am. Heart Assoc. https://doi.org/10.1161/JAHA.115.002746 (2016).
Urtnasan, E. et al. Noninvasive screening tool for hyperkalemia using a single-lead electrocardiogram and deep learning: development and usability study. JMIR Med. Inform. 10, e34724. https://doi.org/10.2196/34724 (2022).
Lou, Y. S. et al. Development and validation of a dynamic deep learning algorithm using electrocardiogram to predict dyskalaemias in patients with multiple visits. Eur. Heart J. Digit. Health 4, 22–32. https://doi.org/10.1093/ehjdh/ztac072 (2023).
Galloway, C. D. et al. Development and validation of a deep-learning model to screen for hyperkalemia from the electrocardiogram. JAMA Cardiol. 4, 428–436. https://doi.org/10.1001/jamacardio.2019.0640 (2019).
Ferreira, J. P. et al. Abnormalities of potassium in heart failure: JACC state-of-the-art review. J. Am. Coll. Cardiol. 75, 2836–2850. https://doi.org/10.1016/j.jacc.2020.04.021 (2020).
Raebel, M. A. et al. Diabetes and drug-associated hyperkalemia: Effect of potassium monitoring. J. Gen. Intern. Med. 25, 326–333. https://doi.org/10.1007/s11606-009-1228-x (2010).
Lin, C. et al. Point-of-care artificial intelligence-enabled ECG for dyskalemia: A retrospective cohort analysis for accuracy and outcome prediction. NPJ Digit. Med. 5, 8. https://doi.org/10.1038/s41746-021-00550-0 (2022).
Lee, Y. et al. Deep learning in the medical domain: Predicting cardiac arrest using deep learning. Acute Crit. Care 33, 117–120. https://doi.org/10.4266/acc.2018.00290 (2018).
Kwon, J. M., Lee, Y., Lee, Y., Lee, S. & Park, J. An algorithm based on deep learning for predicting in-hospital cardiac arrest. J. Am. Heart Assoc. https://doi.org/10.1161/JAHA.118.008678 (2018).
Cho, K. J. et al. Detecting patient deterioration using artificial intelligence in a rapid response system. Crit. Care Med. 48, e285–e289. https://doi.org/10.1097/CCM.0000000000004236 (2020).
Dean, J. et al. in Advances in neural information processing systems.
Paszke, A. et al. in Advances in neural information processing systems.
Ruopp, M. D., Perkins, N. J., Whitcomb, B. W. & Schisterman, E. F. Youden Index and optimal cut-point estimated from observations affected by a lower limit of detection. Biometr. J. J. Math. Methods Biosci. 50, 419–430 (2008).
Selvaraju, R. R. et al. in Proceedings of the IEEE international conference on computer vision. 618–626.
Ribeiro, M. T., Singh, S. & Guestrin, C. in Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining. 1135–1144.
Hughes, J. W. et al. Performance of a convolutional neural network and explainability technique for 12-lead electrocardiogram interpretation. JAMA Cardiol. 6, 1285–1295 (2021).
Ji, Y. et al. Abstract 13745: Exploring the segment-wise explainability of electrocardiogram on deep learning-based models for acute myocardial infarction. Circulation 148, A13745–A13745. https://doi.org/10.1161/circ.148.suppl_1.13745 (2023).
Kalyakulina, A. I. et al. LUDB: A new open-access validation tool for electrocardiogram delineation algorithms. IEEE Access 8, 186181–186190 (2020).
Tibshirani, R. J. & Efron, B. An introduction to the bootstrap. Monogr. Stat. Appl. Probab. 57, 1–436 (1994).
DeLong, E. R., DeLong, D. M. & Clarke-Pearson, D. L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 837–845 (1988).
Acknowledgements
This study was supported by a grant from Korean Nephrology Research Foundation (BAXTER, 2020).
Author information
Authors and Affiliations
Contributions
J.N.A. primarily conceptualized and designed the study, contributed to acquisition, analysis, and interpretation of data, and was involved in drafting the manuscript. S.J., M.C., Y.L., and S.G.K. participated in the conception and design of the study. J.N.A., D.H.K., D.G.S., J.-K.K, H.-S.L., Y.R.S., and S.G.K and collected and managed the experimental data. J.N.A., M.P., S.J., M.C., D.H.K., Y.N., Y.L., and S.G.K. were involved in data analysis, and interpretation. J.N.A. and M.P. drafted the manuscript, while S.J., M.C., D.H.K., Y.N., Y.L., and S.G.K. critically revised it for important intellectual content. J.N.A., M.P., S.J., M.C., Y.L., and S.G.K. conducted statistical analyses. All authors reviewed and approved the final version of the manuscript for publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
An, J.N., Park, M., Joo, S. et al. Development of deep learning algorithm for detecting dyskalemia based on electrocardiogram. Sci Rep 14, 22868 (2024). https://doi.org/10.1038/s41598-024-71562-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-71562-5