Abstract
Recent evidence has demonstrated that abnormal expression and regulation of circular RNA (circRNAs) are implicated in the development and progression of various tumors. The aim of this study was to investigate the effects of circ_SMA4 in Gastrointestinal Stromal Tumors (GISTs) malignant progression. Human circRNAs microarray analysis was conducted to identify differentially expressed (DE) circRNAs in GISTs. The effect of circ_SMA4 on cell proliferation, invasion, migration, and apoptosis was assessed in both in vitro and in vivo settings. Dual-luciferase reporter assay, RT-qPCR, Western-blot, and rescue assay were employed to confirm the interaction between circ_SMA4/miR-494-3p/ KIT axis. The results revealed that circ_SMA4 was significantly upregulated in GISTs, and exhibited high diagnostic efficiency with an AUC of 0.9824 (P < 0.01). circ_SMA4 promoted cell proliferation, invasion, migration, while inhibiting apoptosis in GISTs cells, both in vitro and in vivo. Silencing circ_SMA4 partially inhibited GISTs malignant progression. Additionally, circ_SMA4 acted as a competing endogenous RNA (ceRNA) by targeting miR-494-3p, and KIT was identified as a functional gene for miR-494-3p in GISTs. Furthermore, the results confirmed that circ_SMA4/miR-494-3p/ KIT axis plays a role in activating the JAK/STAT signaling pathway in GISTs. Therefore, for the first time, we have identified and emphasized that circ_SMA4 is significantly upregulated and plays an oncogenic role in GISTs by sponging miR-494-3p to activate the KIT/JAK/STAT pathway. These findings underscore circ_SMA4 may serve as a novel diagnostic biomarker and therapeutic target for GISTs.
Similar content being viewed by others
Introduction
Gastrointestinal stromal tumors (GISTs) are infrequently encountered among gastrointestinal carcinomas, originating from mesenchymal tissue1. These tumors are identified by the presence of the CD117 receptor in cells and exhibit diverse biological phenotypes, ranging from benign to highly malignant2. Among the most prevalent non-epithelial neoplasms, GISTs are predominantly situated in the stomach (55.6%) and small intestine (31.8%)3. The preferred approach for treatment is radical surgery, and molecular targeted therapies, such as imatinib, have demonstrated efficacy in enhancing the survival rates of advanced patients harboring c-kit and/or PDGFRα mutations4,5. Despite this, the availability of effective tumor biomarkers for GISTs diagnosis and prognostication remains limited.
Circular RNA (circRNA) represents an emerging class of endogenous non-coding RNA characterized by a distinctive closed-loop structure, wherein the 3′ and 5′ ends are covalently linked6. This unique structure imparts resistance to exonucleases, rendering circRNAs more stable compared to traditional linear RNA, including lncRNA and miRNA7. Categorically, circRNAs can be classified into four types based on their source: exonic circRNAs (ecircRNA), intronic circRNA (ciRNA), exonic- intronic circRNA (EIciRNA), and intergenic circRNAs. Notably, 80% of circRNAs belong to the ecircRNA category8. The functional roles of circRNAs are diverse and impactful. They can function as microRNA (miRNA/miR) sponges, competitively binding to miRNA response elements to modulate downstream target gene expression9. This concept gained support with the identification of cerebellar degeneration-related protein (CDR1as), also known as CiRS-7, which unveiled circRNAs’ ability to function as competing endogenous RNAs (ceRNA) with over 60 miRNA binding sites10. Simultaneously, circRNAs exert influence at the post-translational level, impacting gene function. Intriguingly, a single circRNA can regulate multiple miRNAs, and reciprocally, the same miRNAs can regulate multiple mRNA genes, forming an extensive circRNA-miRNA-mRNA competitive network that significantly influences tumor development11. Additionally, certain circRNAs exhibit activities through interactions with proteins, and some ecircRNAs may participate in assembly and protein ribosome translation processes. Reports indicate that circRNAs are implicated in diverse biological processes, including signal transduction, transcription, cell cycle regulation, RNA-binding protein interactions, stress responses, protein metabolism, cellular immunity, and cell structure. Recent studies underscore the involvement of abnormal circRNA expression and regulation in the occurrence and progression of various tumors12. Consequently, circRNAs hold significant value as biomarkers for cancer diagnosis, prediction, and treatment response evaluation. Furthermore, they may serve as potential targets for innovative cancer treatments13.
Our preliminary study14 revealed that circ_SMA4 (hsa_circRNA_103870) was significantly up-regulated in GISTs (n = 20), and circ_SMA4 also showed high diagnostic values, and significantly associated with tumor size, mitotic figure, and malignant degrees in GISTs. We hypothesized that circ_SMA4 might be critical circRNA and may present as potential diagnostic biomarkers for GISTs. To validate our hypothesis, we established cell models with circ_SMA4 overexpression and knockdown to investigate the effect of circ_SMA4 on GISTs malignant progression. Its mechanism of action is also being explored in depth.
Materials and methods
Patients and tissue samples
Twenty pairs of GISTs and adjacent tissues were collected from The Second Xiangya Hospital of Central South University. All pathological specimens were confirmed by experienced pathologists and did not undergo pre-operative radiotherapy, chemotherapy, or imatinib targeted therapy. The clinicopathological features are detailed in Supplemental Table SI. Tissues were collected during surgical operations and promptly stored in liquid nitrogen. The present study was approved by the ethics committee of The Second Xiangya Hospital of Central South University, and informed consents were obtained from all participants. Total RNAs from each sample were extracted using the RNeasy Mini Kit (Qiagen, Hilden, Germany). Differences in circular RNAs (circRNA) between the two sample groups were analyzed using the Arraystar Human circRNA Array V2 (8 × 15 K; Arraystar).
Cell culture and treatments
The GISTs cell lines GIST-T1 and GIST-882 and Human Stomach smooth muscle cells (HGSMC) were obtained from the American Type Culture Collection (ATCC). These cell lines underwent regular authentication through Short Tandem Repeat (STR) analysis and were routinely screened for mycoplasma contamination to ensure their integrity. The cell lines were cultured in Dulbecco’s minimum essential medium supplemented with 10% fetal calf serum at 37 °C in a humidified atmosphere containing 5% CO2.
Reverse transcription-quantitative (RT-q) PCR
Total RNA extraction was conducted utilizing TRIzol® reagent. Subsequently, the synthesis of complementary DNA (cDNA) was performed using the PrimeScript™ RT-PCR Kit following the manufacturer’s protocol. The primer sequences for circular RNAs (circRNAs), microRNA (miRNA), and messenger RNA (mRNA) were custom-synthesized by Shanghai GeneChem Co., Ltd. GAPDH served as the internal reference for circRNAs and mRNA, while U6 was employed as the internal reference for miRNA. The thermocycling conditions involved an initial step at 95 °C for 15 min, followed by 40 cycles at 94 °C for 15 s, 55 °C for 30 s, and 70 °C for 30 s. Following the PCR procedure, the relative expression levels were determined using the 2−ΔΔCq method.
Cell Counting Kit-8 (CCK-8)
Cells were plated at a density of 2000 cells per well in a 96-well plate and exposed to Pembrolizumab (0.0–1.0 µM) for a duration of 24 h. Assessment of cell viability was conducted by measuring absorbance at 492 nm following incubation with CCK-8 solution. The formula employed for calculating Cell Viability was:
This experiment was repeated three times for robustness and reliability.
Matrigel invasion assay
For the experiment, a Transwell system featuring an 8-µm pore size from Corning was employed. In this setup, the upper chamber of the Transwell was seeded with 2 × 10^4 cells cultured in 500 µL of serum-free medium, while the bottom well was supplemented with 1 mL of complete culture medium containing 10% serum. After undergoing a 48-hour incubation period in a dedicated incubator, the cells were fixed using 4% paraformaldehyde and subsequently stained with crystal violet solution. To eliminate any remaining cells that had not migrated, a cotton swab was utilized. The remaining cells were then observed and imaged under a microscope to ensure thorough analysis and documentation. This process was conducted with attention to detail and precision.
Wound healing assay
To evaluate cell migration, a wound healing assay was conducted. Following trypsinization and cell counting, 5 × 10^5 cells were seeded into each well of a pre-marked 6-well plate with evenly spaced horizontal lines. Once cells reached confluence, perpendicular scratches were carefully made across the marked lines. Subsequently, PBS was employed to remove any cellular debris, and the cells were cultured in McCoy’s 5 A medium with low serum at 37 ℃ and 5% CO2.The widths of the scratches were observed and documented at 0-, 24-, and 48- hours post-scratching using an inverted biological microscope (CX53, Olympus, Japan). Three fields of view were captured at each time point, ensuring a comprehensive assessment of the wound healing process. This meticulous procedure provides valuable insights into cell migration dynamics over the specified time intervals.
Flow cytometry analysis for Annexin V/PI assay
For apoptosis assays, the Annexin V-FITC Apoptosis Detection Kit (BD, San Jose, CA, USA) was employed. Following the manufacturer’s protocol, THP-1 cells were harvested, resuspended in 100 µL of binding buffer, and incubated with 5 µL of annexin V-FITC and 10 µL of propidium iodide (PI) for 15 min in the dark at room temperature. Subsequently, 400 µL of binding buffer was added, and the stained cells were assessed by flow cytometry after 1 h. This meticulous procedure ensures accurate detection and quantification of apoptotic cells, contributing to a comprehensive analysis of cellular responses.
Dual-luciferase reporter assay
HEK293T cells at 30% confluence were carefully seeded into 24-well plates. In each well, a combination of the reporter plasmid (100 ng, containing 34 signaling pathways), pRL-CMV (5 ng), and the specified gene-expressing plasmids (Flag-USP5, 500 ng) or an empty vector was transiently transfected into the HEK293T cells. This transfection process aimed to introduce the relevant genetic elements into the cells. Following a 48-hour incubation period, luciferase reporter assays were conducted using a dual luciferase assay kit (E1960, Promega, Beijing). This assay provides a sensitive and quantitative measure of gene expression and signaling pathway activities. The experimental design and execution were meticulous to ensure accurate and reliable results.
Western-blot assay
Cell lysis was carried out using SDS lysis buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, and 10% glycerol) at 95 °C for 10 min. The total protein was subsequently separated through 10% SDS-PAGE and transferred onto PVDF membranes (IPVH00010; Millipore, Billerica, MA, USA). Following transfer, the membranes were blocked with 5% skim milk for 1 h to prevent nonspecific binding. Primary antibodies were then applied to the membranes and allowed to incubate overnight at 4 °C, facilitating specific binding to target proteins. Subsequently, the membranes were thoroughly washed in TBST before being incubated with HRP-labelled secondary antibodies at room temperature for 1 h, enabling the detection of primary antibody-bound proteins. Chemiluminescence of the protein bands was evaluated using the Tanon 5500 chemiluminescence image analysis system (Tanon, Shanghai, China). GAPDH was utilized as the internal control, ensuring normalization of protein expression levels. This method ensures a robust and precise analysis of protein content and expression in the experimental samples.
Mice subcutaneous xenograft in vivo
To assess the anti-tumor efficacy of circRNA on GISTs cells, we employed a subcutaneous xenograft model in nude mice. GIST-T1 and GIST-882 cells, labeled with fluorescence, were subcutaneously injected, with each experimental group comprising a minimum of 5 mice. The determination of animal numbers per group was meticulously performed through power analysis to ensure statistical robustness. Randomization of animals into experimental groups was carried out for each study, minimizing potential bias. It’s important to note that no blinding of experiment groups was applied in this study. Tumor volume was diligently monitored on a weekly basis using the formula V = 1/2×Width^2×Length. The growth curve, derived from length and time, provided a dynamic representation of tumor development. Following a 5-week period, fluorescence imaging was conducted to visualize the subcutaneous xenografts in mice. Subsequently, the mice were euthanized by cervical dislocation in accordance with ARRIVE guidelines to ensure humane and ethical treatment. And the subcutaneous xenografts were excised and measured for size and weight. Histological changes, Ki-67 staining, and TUNEL assay were performed on the subcutaneous tumor tissues to comprehensively examine the impact of circRNA on GISTs cells. This multifaceted analysis aimed to provide a thorough understanding of the therapeutic effects at both macroscopic and microscopic levels.
Statistical analysis
The data are expressed as the mean ± standard deviation unless otherwise specified. Statistical analyses were performed using SPSS 17.0 software (SPSS Inc., Chicago, IL, USA), for more information, please visit [SPSS official website] (https://www.ibm.com/products/spss-statistics). And GraphPad Prism 7 (GraphPad Software, Inc., La Jolla, CA, USA). Unpaired two-tailed Student’s t-test was applied to assess differences in cell viability, cell migration, invasion ability, cell apoptosis level, xenograft tumor volume, and xenograft tumor weight among various groups. The paired two-tailed Student’s t-test was employed to compare the expression of circ_SMA4 between GISTs tissues and their paired adjacent tissues. Spearman’s rho was utilized to evaluate the correlation between different variables. The chi-square test was conducted to analyze the association between circ_SMA4 expression and the clinicopathological features of the patients. Kaplan-Meier curves and log-rank tests were utilized to compare the survival of patients with high and low circ_SMA4 expression. A significance level of P < 0.05 was considered statistically significant.
Results
Diagnosis and prognosis values of circ_SMA4 in GISTs
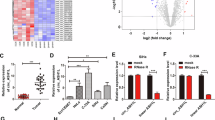
Differential expression profiles of circRNAs in GISTs were successfully established. The cluster heat map in Supplemental Fig. 1A illustrates the top eight upregulated circRNAs (Supplemental Table SII, fold change > 4, P < 0.05) and the top eight downregulated circRNAs (Supplemental Table SIII, fold change > 3, P < 0.05). Validation through RT-qPCR in 20 GISTs and adjacent tissue pairs confirmed significant upregulation of circ_SMA4 (circRNA_103870) in GISTs tissues (FC = 4.06) compared to adjacent tissues (P < 0.05; Supplemental Fig. 1B). To explore the correlation of circ_SMA4 expression with clinical-pathologic features, the 20 GISTs tissue were divided into higher and lower circRNA expression groups based on the median circ_SMA4 expression. The statistical analysis revealed no significant correlation between circ_SMA4 and the following clinicopathological features: age, gender, tumor size (cm), mitotic figure (HPF), or malignant degrees and type, as well as gene mutation status (P > 0.05). Chi-square analysis revealed a positive association between circ_SMA4 expression and mitotic figures, malignant degrees (P < 0.05; Supplemental Fig. 1C), while no significant relation was observed with tumor size (P > 0.05; Supplemental Fig. 1C). Furthermore, the Receiver Operating Characteristic (ROC) curve analysis highlighted the high diagnostic efficiency of circ_SMA4, with an Area Under the Curve (AUC) of 0.9824 (P < 0.01; Figure Supplemental Fig. 1D).
circ_SMA4 plays an oncogenic role in GISTs
Upon querying the circBase database, has_circRNA_103870 (circBase ID: hsa_circ_0072805) was identified to be located on chromosome 5:69206201–69,206,389. It represents one of the most prevalent circRNAs derived from exons and is associated with the gene symbol SMA4; therefore, has_circRNA_103870 is also referred to as circ_SMA4. In our study, GISTs cell lines GIST-T1 and GIST-882 were chosen as parental cells, and Human Stomach smooth muscle cells (HGSMC) served as a control. The results of the RT-qPCR assay revealed a significant upregulation of circ_SMA4 in GIST-T1 and GIST-882 cells compared to HGSMC cells (P < 0.05; Supplemental Fig. 2A). Also, a slightly lower expression of circ_SMA4 was observed in GIST-T1 cells compared to GIST-882 cells, although this difference was not statistically significant (P > 0.05; Supplemental Fig. 2B).
Additionally, we constructed overexpression and knockdown lentiviral vectors, as well as negative control (NC) vectors for circ_SMA4. Subsequently, these vectors were stably transfected into GIST-T1 and GIST-882 cells respectively, successfully establishing circ_SMA4 overexpression and knockdown cellular models (P < 0.05; Fig. 1A). Furthermore, CCK-8 assay, Annexin V-FITC/PI assay, Transwell chamber assay, Cell scratch assay assays were performed to explore the impact of circ_SMA4 on cell proliferation and apoptosis in GISTs. The CCK-8 assay revealed that circ_SMA4 overexpression significantly promoted GISTs cell proliferation, while circ_SMA4 knockdown dramatically inhibited GISTs cell proliferation (P < 0.05; Fig. 1B). The Annexin V-FITC/PI assay demonstrated that circ_SMA4 overexpression significantly suppressed GISTs cell apoptosis, whereas circ_SMA4 knockdown significantly promoted GISTs cell apoptosis (P < 0.05; Fig. 1C). Subsequent assays, including Transwell chamber and cell scratch assays, suggested that circ_SMA4 overexpression markedly promoted GISTs cell invasion and migration, while circ_SMA4 knockdown dramatically inhibited GISTs cell invasion and migration (P < 0.05; Fig. 2A-B). Collectively, these findings indicate that circ_SMA4 promotes GISTs proliferation, invasion, and migration while inhibiting apoptosis, thereby playing an oncogenic role in GISTs.
circ_SMA4 Promotes GISTs Cell Proliferation and Inhibits Cell Apoptosis In Vitro. (A) Successful establishment of circ_SMA4 overexpression and knockdown cellular models. The transfection efficiency of each group was confirmed through RT-qPCR. (B) Cell counting kit-8 (CCK8) assay and (C) Annexin V-FITC/PI assay was performed to analyze the effect of circ_SMA4 on GISTs cell proliferation and apoptosis. n = 3. *P < 0.05, **P < 0.01. RT-qPCR, reverse transcription- quantitative PCR. OE: OverExpression. shRNA: short hairpin RNA. NC: negative control. OD, Optical Density. circ_SMA4: circ_103870.
circ_SMA4 Promotes GISTs cell Invasion and Migration In Vitro. (D) Transwell chamber assay (D) Cell scratch assay was performed to analyze cell invasion and migration ability. n = 3. *P < 0.05, **P < 0.01. RT-qPCR, reverse transcription- quantitative PCR. OE: OverExpression. shRNA: short hairpin RNA. NC: negative control. OD, Optical Density. circ_SMA4: circ_103870.
circ_SMA4 serves as an Efficient miRNA Sponge for miR-494-3p
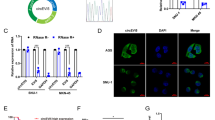
CircRNAs are known to function as miRNA sponges, regulating the expression of specific target genes. In our study, we identified hsa-miR-494-3p, hsa-miR-1271-5p, hsa-miR-96-5p, hsa-miR-410-5p, and hsa-miR-661 as the top five targeted miRNAs for circ_SMA4 (Fig. 3A). A schematic representation of the potential binding sites between op five targeted miRNAs and the circ_SMA4-3’-UTR is presented in Fig. 3B. Among them, notably, hsa-miR-494-3p exhibited the highest binding score (Context++ score) with circ_SMA4, prompting the selection of miR-494-3p for further exploration in our study.
circ_SMA4 serves as an Efficient miRNA Sponge for miR-494-3p in GISTs. (A) The top five targeted miRNAs by circ_SMA4. (B) A schematic representation of the potential binding sites between op five targeted miRNAs and the circ_SMA4-3’-UTR. (C-D) RT-qPCR analysis demonstrating the downregulation of miR-494-3p in GIST-T1 and GIST-882 cells lines and tissues, compared to HGSMC cells and adjacent tissues. (E) Pearson’s correlation analysis showing a negative correlation between circ_SMA4 and miR-494-3p expression in GISTs tissues. n = 20. (F) Dual-luciferase reporter assay confirmed that miR-494-3p could competitively targeted circ_SMA4 (HEK-293T). (F) RT-qPCR assay demonstrated that miR-494-3p expression was significantly and negatively regulated by circ_SMA4 expression in GISTs cells. n = 3. *P < 0.05, **P < 0.01, na: no statistical significance. RT-qPCR, reverse transcription- quantitative PCR. OE: OverExpression. shRNA: short hairpin RNA. NC: negative control. OD, Optical Density. circ_SMA4: circ_103870.
Subsequent assays, RT-qPCR assay demonstrated that miR-494-3p was significantly downregulated in GIST-T1 and GIST-882 cells (P < 0.05; Fig. 3C) and GISTs tissues (n = 20; P < 0.05; Fig. 3D), compared to HGSMC cells and adjacent tissues. Pearson’s correlation analysis highlighted a negative correlation between circ_SMA4 and miR-494-3p expression in GISTs tissues (n = 20; r=-0.55, P = 0.01; Fig. 3E). Additionally, a dual-luciferase reporter assay was performed to verify the respective binding sites (MRE)-based circ_SMA4-miR-494-3p interaction. The results demonstrated a significant reduction in luciferase activity in HEK-293T cells co-transfected with pmirGLO-circ_SMA4-wt and miR-494-3p mimic compared to cells co-transfected with pmirGLO-circ_SMA4-wt and miR-494-3p NC (P < 0.05; Fig. 3F). However, no significant change in luciferase activity was observed in cells co-transfected with pmirGLO-circ_SMA4-mut and miR-494-3p mimic or pmirGLO-circ_SMA4-mut and miR-494-3p NC (P > 0.05; Fig. 3F). Moreover, miR-494-3p expression was found to be significantly and negatively regulated by circ_SMA4 expression in GIST-T1 and GIST-882 cells (P < 0.05; Fig. 3G).
Furthermore, in order to investigate the impact of miR-494-3p on GISTs, we successfully constructed miR-494-3p mimic and inhibitor and subsequently stably transfected them into GIST-T1 and GIST-882 cells using LipofectamineTM3000 (P < 0.05; Fig. 4A). The ensuing CCK-8 assay revealed that miR-494-3p overexpression significantly inhibited the proliferation of GISTs cells, while miR-494-3p knockdown markedly promoted GISTs cell proliferation (P < 0.05; Fig. 4B). The Annexin V-FITC/PI assay demonstrated that miR-494-3p overexpression significantly promoted GISTs cell apoptosis, whereas miR-494-3p knockdown significantly suppressed GISTs cell apoptosis (P < 0.05; Fig. 4C). The cumulative evidence indicates that miR-494-3p serves as one of the functional target miRNAs of circ_SMA4, manifesting a miRNA sponge effect. Moreover, miR-494-3p assumes the role of a tumor suppressor in GISTs.
miR-494-3p plays a tumor suppressor role in GISTs. (A) Successful establishment of miR-494-3p overexpression and knockdown cellular models: miR-494-3p mimic and inhibitor and subsequently stably transfected them into GIST-T1 and GIST-882 cells, respectively. The transfection efficiency was assessed by RT-qPCR. (B) Cell counting kit-8 (CCK8) assay and (C) Annexin V-FITC/PI assay was performed to analyze the effect of miR-494-3p on GISTs cell proliferation and apoptosis. n = 3. *P < 0.05, **P < 0.01. RT-qPCR, reverse transcription- quantitative PCR. OE: OverExpression. shRNA: short hairpin RNA. NC: negative control. OD, Optical Density. circ_SMA4: circ_103870.
KIT identified as a functional target gene for miR-494-3p
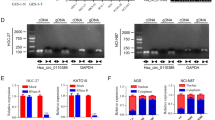
The potential targets of miR-494-3p were predicted through three miRNA target gene databases (miRBase, TargetScan, and miRanda), KIT, a well-known proto-oncogene, is simultaneously predicted by three miRNA databases (as shown in Fig. 5A). Based on microRNA Response Element (MRE) analysis, miR-494-3p was found to have potential binding sites within the 3’-UTR region of KIT (as shown in Fig. 5B).
KIT identified as a functional target gene for miR-494-3p in GISTs. (A) KIT was simultaneously predicted by three miRNA databases (miRBase, TargetScan, and miRanda). (A) Schematic representation of potential binding sites between miR-494-3p and KIT. (C-D) RT-qPCR analysis demonstrating the upregulation of KIT mRNA in GIST-T1 and GIST-882 cells lines and tissues, compared to HGSMC cells and adjacent tissues. (E) Pearson’s correlation analysis showing a negative correlation between KIT mRNA and miR-494-3p expression in GISTs tissues. n = 20. (F) Dual-luciferase reporter assay confirmed that miR-494-3p could competitively targeted wt 3’UTR sequence of KIT (HEK-293T). RT-qPCR (G) and Western-blot (H) assay demonstrated that KIT mRNA and protein expression was significantly and negatively regulated by miR-494-3p expression in GISTs cells. n = 3. *P < 0.05, **P < 0.01, na: no statistical significance. RT-qPCR, reverse transcription- quantitative PCR. OE: OverExpression. shRNA: short hairpin RNA. NC: negative control. OD, Optical Density. circ_SMA4: circ_103870.
In subsequent assays, RT-qPCR analysis revealed a significant upregulation of KIT mRNA in GIST-T1 and GIST-882 cells (P < 0.05; Fig. 5C) and GISTs tissues (n = 20; P < 0.05; Fig. 5D), compared to HGSMC cells and adjacent tissues. Pearson’s correlation analysis highlighted a negative correlation between miR-494-3p and KIT mRNA expression in GISTs tissues (n = 20; r=-0.51, P = 0.02; Fig. 5E). Additionally, a dual-luciferase reporter assay was conducted to confirm the KIT -miR-494-3p interaction based on the respective binding sites (MRE). The results showed a significant reduction in luciferase activity in HEK-293T cells co-transfected with pmirGLO-KIT-wt and miR-494-3p mimic compared to cells co-transfected with pmirGLO-KIT-wt and miR-494-3p NC (P < 0.05; Fig. 5F). However, no significant change in luciferase activity was observed in cells co-transfected with pmirGLO-KIT -mut and miR-494-3p mimic or pmirGLO-circ_SMA4-mut and miR-494-3p NC (P > 0.05; Fig. 5F). Furthermore, both KIT mRNA and protein expression were found to be significantly and negatively regulated by miR-494-3p in GIST-882 and GIST-T1 cells (P < 0.05; Fig. 5G-H).
Moreover, to explore the impact of KIT on GISTs, we successfully constructed lentiviral vector-mediated cellular models with KIT overexpression (KIT OE) and silencing (sh-KIT) (P < 0.05; Fig. 6A). The CCK-8 assay revealed that KIT overexpression significantly enhanced GISTs cell proliferation, while KIT knockdown markedly inhibited GISTs cell proliferation (P < 0.05; Fig. 6B). The Annexin V-FITC/PI assay demonstrated that KIT overexpression significantly suppressed GISTs cell apoptosis, whereas KIT knockdown significantly promoted GISTs cell apoptosis (P < 0.05; Fig. 6C). The cumulative evidence supports KIT as a functional target gene for miR-494-3p and further substantiates the oncogenic role of KIT in GISTs.
KIT act as oncogenic role in GISTs. (A) Successful establishment of KIT overexpression and knockdown cellular models. The transfection efficiency was assessed by RT-qPCR. (B) Cell counting kit-8 (CCK8) assay and (C) Annexin V-FITC/PI assay was performed to analyze the effect of KIT on GISTs cell proliferation and apoptosis. n = 3. *P < 0.05, **P < 0.01. RT-qPCR, reverse transcription- quantitative PCR. OE: OverExpression. shRNA: short hairpin RNA. NC: negative control. OD, Optical Density. circ_SMA4: circ_103870.
circ_SMA4 promotes GISTs progression by sponging miR-494-3p and activating KIT/JAK/STAT pathway
Rescue experiments were conducted to authenticate the role of the circ_SMA4/miR-494-3p/KIT axis in GISTs. Findings from CCK-8 assays revealed that circ_SMA4 overexpression enhanced GISTs cell proliferation, which was counteracted by treatment with miR-30a-3p mimic. Conversely, circ_SMA4 silencing restrained GISTs cell proliferation, but this inhibition was reversed following treatment with KIT overexpression (P < 0.05; Fig. 7A). KIT mRNA and protein expression, assessed by RT-qPCR and Western blot assays, respectively, significantly increased in the circ_SMA4 overexpression group, but these effects were counteracted by miR-30a-3p mimic treatment (P < 0.05; Fig. 7B-C). Conversely, KIT mRNA and protein expression markedly decreased in the sh-circ_SMA4 group, and these effects were mitigated by KIT overexpression (P < 0.05; Fig. 7B-C).
circ_SMA4 promotes GISTs progression by sponging miR-494-3p and activating KIT/JAK/STAT pathway. GIST-T1 cells were co-transfected with the circ_SMA4 OE vector and miR-494-3p mimics. GIST-882 cells were also co-transfected with sh-circ_SMA4 and KIT OE. (A) Cell counting kit-8 (CCK8) and (B) RT-qPCR and (C) Western-blot assay showed the effects of circ_SMA4/miR-494-3p/KIT axis on cell proliferation and KIT mRNA and protein expression in GISTs. (D-E) JAK/STAT signaling pathway can activated by the circ_SMA4/miR-494-3p/KIT axis in GISTs. n = 3. *P < 0.05, **P < 0.01, na: no statistical significance. RT-qPCR, reverse transcription- quantitative PCR. OE: OverExpression. shRNA: short hairpin RNA. NC: negative control. OD, Optical Density. circ_SMA4: circ_103870.
Moreover, our investigation focused on the downstream JAK/STAT signaling pathway regulated by the KIT gene. Western blot assay results indicated a significant increase in p-JAK and p-STAT protein expression in the circ_SMA4 overexpression group, which was reversed by miR-30a-3p mimic treatment (P < 0.05; Fig. 7D-E). Conversely, p-JAK and p-STAT protein expression notably decreased in the sh-circ_SMA4 group, and these effects were mitigated by KIT overexpression (P < 0.05; Fig. 7D-E). Notably, these alterations did not impact the expression of JAK and STAT proteins. The cumulative evidence suggests that circ_SMA4 promotes GISTs progression by sponging miR-494-3p and activating the KIT/JAK/STAT pathway.
circ_SMA4 promotes GISTs progression in vivo
The in vivo mice xenograft tumor model, as depicted in Fig. 8A, was established to delve deeper into the oncogenic role of circ_SMA4 in GISTs. Following this, 1 × 106 GIST-T1 and GIST-882 cells, fluorescently labeled, were subcutaneously injected into the armpits of nude mice. Upon comparative analysis with the control group, noteworthy increases were observed in the tumor growth rate, tumor weight, and tumor volume in the circ_SMA4 overexpressing group, whereas these parameters experienced a significant reduction in the sh-circ_SMA4 group (P < 0.05; Fig. 8B-C). The proliferative index of GIST-T1 and GIST-882 cells in vivo was assessed using Ki-67 staining in xenograft tumor samples. The results highlighted a substantial increase in the percentage of Ki-67-positive cells in the circ_SMA4 overexpressing group and a corresponding decrease in the sh-circ_SMA4 group (P < 0.05; Fig. 8D).
Oncogenic role of circ_SMA4 in GISTs progression in vivo. (A) Establishment of xenograft tumor model in nude mice. After 5 weeks of feeding, the mice were euthanized, and nude mice were taken out to measure their size and weighed. (B) Xenograft tumor growth curves and (C) Xenograft tumor weight revealed that circ_SMA4 promotes GISTs cell proliferation in vivo. (D) The percentage of Ki67-positive cells in xenograft tumors was measured. (E) TUNEL assay was performed to determine the cell apoptosis rate in Xenograft tumors. The expression of circ_SMA4 (F), miR-494-3p (G) and KIT mRNA (H) in Xenograft tumors were detected by RT-qPCR assay.n = 3. *P < 0.05, **P < 0.01, na: no statistical significance. RT-qPCR, reverse transcription- quantitative PCR. OE: OverExpression. shRNA: short hairpin RNA. NC: negative control. OD, Optical Density. circ_SMA4: circ_103870.
Moreover, TUNEL apoptosis assays conducted with xenograft tumor samples provided evidence that circ_SMA4 overexpression inhibited apoptosis in GIST-T1 cells in vivo, while circ_SMA4 silencing promoted apoptosis in GIST-882 cells in vivo (P < 0.05; Fig. 8E). Additionally, the RT-qPCR assay unveiled a significant upregulation of circ_SMA4 and KIT mRNA expression of xenograft tumor samples in the circ_SMA4 overexpressing group, accompanied by a marked downregulation in the sh-circ_SMA4 group (P < 0.05; Fig. 8F/H). Conversely, miR-494-3p expression of xenograft tumor samples exhibited a significant downregulation in the circ_SMA4 overexpressing group and a substantial upregulation in the sh-circ_SMA4 group (P < 0.05; Fig. 8G). These comprehensive findings, consistent with the in vitro results, strongly suggest that circ_SMA4 plays a pivotal oncogenic role in GISTs in vivo.
Discussion
Gastrointestinal stromal tumors (GISTs) are a diverse group of tumors found throughout the gastrointestinal tract, varying in malignancy15. Initially categorized as smooth muscle tumors, they were later reclassified as gastric stromal tumors due to atypical smooth muscle differentiation observed in some cases16. GISTs are now recognized as originating from interstitial cells of Cajal, leading to their classification as mesenchymal tumors emerging in the submucosa17. This reclassification marked a new era, with reports before 1993 predominantly referring to GISTs when discussing gastric and intestinal smooth muscle tumors18. The primary approach for GISTs typically involves radical surgical intervention, which remains the cornerstone of treatment. Complete resection of the tumor, whenever feasible, is the preferred strategy as it offers the best chance for long-term disease control and potentially even cure19. However, for patients with advanced or metastatic disease, especially those harboring c-kit and/or PDGFRα mutations, additional therapeutic modalities are often necessary.
In recent years, molecular targeted therapies have revolutionized the management of GISTs, particularly for patients who are not amenable to surgery or who have recurrent or metastatic disease20. Imatinib, a tyrosine kinase inhibitor, has emerged as a pivotal therapeutic agent in this context. By selectively inhibiting the activity of mutant c-kit and PDGFRα receptors, imatinib effectively suppresses tumor growth and prolongs survival in these patients. Its success in clinical trials has led to its widespread adoption as a first-line treatment for advanced GISTs21. Despite the efficacy of targeted therapies like imatinib, challenges persist in accurately diagnosing and predicting the behavior of GISTs. Currently, the repertoire of effective tumor biomarkers for GISTs diagnosis and prognostication remains limited. This underscores the need for ongoing research efforts aimed at identifying novel biomarkers that can aid in early detection, risk stratification, and treatment optimization for patients with GISTs22. Additionally, the development of personalized medicine approaches, tailored to the specific molecular characteristics of individual tumors, holds promise for further improving outcomes in this challenging disease landscape23.
Circular RNAs (circRNAs) are significant contributors to cancer biology, playing diverse roles in tumorigenesis, tumor progression, and metastasis. These non-coding RNAs feature a covalently closed loop structure, making them resistant to exonucleases and providing stability24. Dysregulated expression patterns of circRNAs in various cancer types compared to normal tissues underscore their relevance in cancer. Studies suggest circRNAs act as competing endogenous RNAs (ceRNAs), sequestering microRNAs (miRNAs) to relieve repression of miRNA target genes, thereby promoting oncogenic signaling pathways25. Additionally, circRNAs interact with RNA-binding proteins (RBPs), influencing RNA processing, stability, and translation, contributing to cancer pathogenesis26. Recent researches27,28,29 highlights the involvement of circRNAs in GISTs, influencing tumorigenesis, progression, and metastasis. Dysregulated expression of circRNAs in GISTs compared to normal gastrointestinal tissues suggests their potential as diagnostic or prognostic biomarkers. CircRNAs modulate key cellular processes in GISTs development, including proliferation, apoptosis, migration, invasion, and epithelial-to-mesenchymal transition (EMT), functioning as ceRNAs by sequestering miRNAs and influencing target gene expression in oncogenic pathways. Aberrant circRNA expression correlates with clinicopathological features, tumor stage, metastasis, and patient prognosis in GISTs, suggesting their utility as potential biomarkers for diagnosis, prognosis, and therapeutic response prediction. In summary, circRNAs play significant roles in GISTs pathogenesis and may serve as novel targets for diagnosis, prognosis, and therapeutic intervention. Further research is needed to elucidate the specific roles and underlying mechanisms of circRNAs in GISTs tumorigenesis and progression.
Our preliminary study14 revealed that circ_SMA4 (hsa_circRNA_103870) was significantly up-regulated in GISTs (n = 20), and circ_SMA4 also showed high diagnostic values, and significantly associated with tumor size, mitotic figure, and malignant degrees in GISTs. In this study, we established circ_SMA4 overexpression and knockdown cell models to investigate its effects on the biological behavior of GISTs cells. Our findings revealed that circ_SMA4 promotes cell proliferation, invasion, and migration while inhibiting apoptosis in GISTs cells, both in vitro and in vivo. Silencing circ_SMA4 partially inhibited the malignant progression of GISTs. As far as we know, this is the first such report for the oncogene role of circ_SMA4 in GISTs. Additionally, through querying the circBase database, we identified has_circRNA_103870 (circBase ID: hsa_circ_0072805) located on chromosome 5:69206201–69,206,389, representing one of the most prevalent circRNAs derived from exons and associated with the gene symbol SMA4. To date, there are no reports on circ_SMA4 (has_circRNA_103870) in the current literature.
Moreover, our study elucidated the molecular mechanism underlying the oncogenic function of circ_SMA4 in GISTs. Current research suggests that circRNAs primarily function as competitive endogenous RNAs (ceRNAs) by binding to microRNAs (miRNAs), thereby silencing or degrading the target miRNAs and relieving their regulation on downstream target genes, thus exerting regulatory functions. We identified circ_SMA4 as a ceRNA that binds to miR-494-3p, thereby sequestering it and relieving its suppression of KIT expression. More recently, miR-494-3p has emerged as a significant member of the miR-494 family. It originates from the 3’-arm of the miR-494 precursor and is located on chromosome 12q32. Current research indicates that miR-494-3p primarily participates as a tumor suppressor gene in the occurrence, development, and drug resistance processes of cancer. For instance, Faversani et al.30 found that miR-494-3p is a novel driver of lung carcinogenesis. Kim et al.31 reported that RAS-stimulated release of exosomes miR-494-3p promotes the osteolytic bone metastasis of breast cancer cells. Kaźmierczak et al.32 reported that elevated expression of miR-494-3p is associated with resistance to Osimertinib in EGFR T790M-positive NSCLC. However, there is currently no research on miR-494-3p in GISTs. Our study confirmed the tumor-suppressive role of miR-494-3p in GISTs through circ_SMA4, further substantiating the critical role of the miR-494-3p family in cancer regulation.
Furthermore, KIT, a well-known oncogene in GISTs, was confirmed as a functional target of miR-494-3p in GISTs cells. Our research findings indicate that circ_SMA4-induced silencing of miR-494-3p results in the activation of KIT. KIT, also known as c-KIT, located on chromosome 4q12, is an oncogene that encodes a type III transmembrane receptor tyrosine kinase with tyrosine kinase activity32. The receptor tyrosine kinase c-KIT dimerizes and autophosphorylates upon binding with its ligand stem cell factor (SCF), also known as mast cell growth factor (MGF). The genetic underpinning of GISTs growth involves activation of c-KIT, resulting in the constitutional activation of receptor tyrosine kinases, serving as the driving force behind tumor development33. Activation of c-KIT enhances intracellular signaling through JAK/STAT pathways ultimately leading to cell proliferation and survival. Our research further reveals that upon activation of KIT, the JAK/STAT signaling pathway is further activated in GISTs34. The JAK/STAT signaling pathway plays a crucial role in the occurrence and development of GISTs35. Activation of this pathway, often initiated by aberrant KIT signaling due to mutations, leads to the phosphorylation and activation of Janus kinases (JAK) and subsequent phosphorylation of Signal Transducer and Activator of Transcription (STAT) proteins36. These activated STAT proteins then translocate to the nucleus, where they regulate the transcription of target genes involved in cell proliferation, survival, and differentiation37. Dysregulation of the JAK/STAT pathway contributes to the uncontrolled growth and survival of GISTs cells, making it a significant pathway for targeted therapeutic interventions in GISTs38,39.
Based on our current research evidence, it is evident that during the occurrence and progression of GISTs, circ_SMA4 is activated. Upregulation of circ_SMA4 leads to the silencing or degradation of its target miR-494-3p, thereby relieving the inhibition on its target gene KIT, resulting in the activation of KIT. The activated KIT gene further activates downstream signaling pathways, particularly the JAK/STAT pathway, ultimately driving the malignant progression of GISTs. However, whether the activation of KIT by circ_SMA4 is correlated with KIT gene mutations (from quantitative to qualitative changes), or if the activation of KIT by circ_SMA4 may represent an adaptive response before KIT gene mutations in GISTs, remains uncertain and warrants further investigation. Nonetheless, the functional role of the circ_SMA4/miR-494-3p/KIT/JAK/STAT pathway in the occurrence and development of GISTs is clearly established. These findings have several important implications. Firstly, our study highlights circ_SMA4 as a potential diagnostic biomarker for GISTs, given its consistent upregulation in GISTs tissues. Detection of circ_SMA4 levels may aid in the early diagnosis and monitoring of GISTs patients. Secondly, targeting circ_SMA4 or its downstream signaling pathway, such as the miR-494-3p/ KIT/JAK/STAT axis, may offer a promising therapeutic strategy for GISTs treatment. Inhibition of circ_SMA4 expression or interference with its interaction with miR-494-3p could potentially suppress GISTs cell proliferation and metastasis, providing a new avenue for therapeutic intervention.
However, there are several limitations to our study. First, only 20 patients were enrolled in our study, which makes the sample size relatively small, and the results show an association rather than a definite causal relationship. Additionally, the analysis of the relationship between clinical factors and circRNAs needs to be supported by larger samples. Secondly, the critical circRNA circ_SMA4 chosen in this study has numerous downstream target miRNAs. We only observed the effect of circ_SMA4 on miR-494-3p, which is a significant limitation. Thirdly, in terms of the regulation of downstream gene signaling pathways of KIT, our research mainly focused on the JAK/STAT signaling pathway. Other pathways, such as the PI3K/AKT/mTOR and RAS/RAF/MEK/ERK pathways, were not further explored. Furthermore, although current research suggests that most circular RNAs are predominantly enriched in the cytoplasm, we did not explicitly determine the localization of circ_SMA4 in this study. In future research, we plan to further clarify the subcellular localization of circ_SMA4 using ISH and nucleocytoplasmic separation techniques. Therefore, in our future work, further studies with larger groups of patients, the improvement of the circ_SMA4-miRNA-mRNA regulatory network, and further validation in vivo and in vitro also need to be explored.
Conclusion
In summary, our study marks the first comprehensive exploration into the role of circ_SMA4 in GISTs. We have underscored its significant upregulation and unveiled its oncogenic function in GISTs, where it acts by sequestering miR-494-3p, thereby activating the KIT/JAK/STAT pathway. These findings illuminate circ_SMA4’s potential as a novel diagnostic biomarker and therapeutic target for GISTs. However, further investigation into the precise mechanisms by which circ_SMA4 mediates oncogenesis in GISTs is imperative to fully realize its clinical implications and therapeutic potential. Furthermore, as our understanding of the intricate regulatory roles of circRNAs continues to evolve, there lies the promise of unveiling innovative strategies to enhance the efficacy of targeted therapies for GISTs patients. To fully harness the clinical significance of circRNAs in this context, additional research efforts and clinical validation are essential.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.The datasets generated for this study can be found in NCBI GEO accession GSE147303.
References
Keung, E. Z., Raut, C. P. & Rutkowski, P. The Landmark Series: systemic therapy for Resectable gastrointestinal stromal tumors. Ann. Surg. Oncol.27 (10), 3659–3671 (2020).
Farag, S., Smith, M. J., Fotiadis, N., Constantinidou, A. & Jones, R. L. Revolutions in treatment options in gastrointestinal stromal tumours (GISTs): the latest updates. Curr. Treat. Options Oncol.21 (7), 55 (2020).
Rasheed, M. W. et al. Clinicopathological and immunohistochemical characterization of gastrointestinal stromal tumour at four tertiary health centers in Nigeria using CD117, DOG1, and human epidermal growth factor receptor-2 biomarkers. Ann. Afr. Med.22 (4), 501–507 (2023).
He, Y., Da, M., Fan, C. & Tao, P. Unexpected reaction of wild-type gastrointestinal stromal tumor to imatinib: case report and literature review. Front. Oncol.13, 1334784 (2024).
Cicala, C. M., Olivares-Rivas, I., Aguirre-Carrillo, J. A. & Serrano, C. KIT/PDGFRA inhibitors for the treatment of gastrointestinal stromal tumors: getting to the gist of the problem. Expert Opin. Investig Drugs.14, 1–12 (2024).
Liu, A. et al. Isoliquiritigenin inhibits circ0030018 to suppress glioma tumorigenesis via the miR-1236/HER2 signaling pathway. MedComm 2023;4:e282. (2020).
Ngo, L. H. et al. Nuclear export of circular RNA. Nature 14. (2024).
Heydarnia, E. et al. Circular RNAs and cervical cancer: friends or foes? A landscape on circRNA-mediated regulation of key signaling pathways involved in the onset and progression of HPV-related cervical neoplasms. Cell. Commun. Signal.22 (1), 107 (2024).
Saleem, A. et al. Biological role and regulation of circular RNA as an emerging biomarker and potential therapeutic target for cancer. Mol. Biol. Rep.51 (1), 296 (2024).
Pietrzak, J., Świechowski, R., Wosiak, A., Wcisło, S. & Balcerczak, E. ADAMTS Gene-Derived circRNA molecules in Non-small-cell Lung Cancer: expression profiling, clinical correlations and survival analysis. Int. J. Mol. Sci.25 (3), 1897 (2024).
Saleh, R. O. et al. A therapeutical insight into the correlation between circRNAs and signaling pathways involved in cancer pathogenesis. Med. Oncol.41 (3), 69 (2024).
Yi, J. et al. CircMYBL2 facilitates hepatocellular carcinoma progression by regulating E2F1 expression. Oncol. Res.32, 1129–1139 (2024).
To, K. K. W., Huang, Z., Zhang, H., Ashby, C. R. Jr & Fu, L. Utilizing non-coding RNA-mediated regulation of ATP binding cassette (ABC) transporters to overcome multidrug resistance to cancer chemotherapy. Drug Resist. Updat. 73, 101058 (2024).
Zou, F. W. et al. Identification of CircRNA-miRNA-mRNA Regulatory Network in gastrointestinal stromal tumor. Front. Genet. 11, 403 (2020).
Caturegli, I. & Raut, C. P. Gastrointestinal stromal tumors and the General Surgeon. Surg. Clin. North. Am. 102 (4), 625–636 (2022).
Blay, J. Y., Kang, Y. K., Nishida, T. & von Mehren, M. Gastrointestinal stromal tumours. Nat. Rev. Dis. Primers. 7 (1), 22 (2021).
Beham, A. W., Schaefer, I. M., Schüler, P., Cameron, S. & Ghadimi, B. M. Gastrointestinal stromal tumors. Int. J. Colorectal Dis. 27 (6), 689–700 (2012).
Zhang, C. et al. Radiomics analysis of contrast-enhanced computerized tomography for differentiation of gastric schwannomas from gastric gastrointestinal stromal tumors. J. Cancer Res. Clin. Oncol. 150 (2), 87 (2024).
Munteanu, A. et al. Clinical and morphological characteristics of gastrointestinal stromal tumor. Chirurgia (Bucur). 118 (6), 618–623 (2023).
Jiang, Z. et al. Cell-permeable PI3 kinase competitive peptide inhibits KIT mutant mediated tumorigenesis of gastrointestinal stromal tumor (GIST). Mol. Biol. Rep. 51 (1), 98 (2024).
Serrano, C. et al. Novel trial designs for patients with gastrointestinal stromal tumor. ESMO Open. 9 (1), 102218 (2024).
Calderillo-Ruíz, G. et al. Carbajal-López B. Genomic profiling in GIST: implications in clinical outcome and future challenges. Neoplasia. 48, 100959 (2024).
Nishida, T. et al. Molecular and clinicopathological features of KIT/PDGFRA wild-type gastrointestinal stromal tumors. Cancer Sci. 5. (2024).
Nasser, J. S., Altahoo, N., Almosawi, S., Alhermi, A. & Butler, A. E. The role of MicroRNA, Long non-coding RNA and circular RNA in the pathogenesis of polycystic ovary syndrome: a Literature Review. Int. J. Mol. Sci. 25 (2), 903 (2024).
Su, Z. et al. Regulation of angiogenesis by non-coding RNAs in Cancer. Biomolecules. 14 (1), 60 (2024).
Zhang, Z., Gao, Z., Fang, H., Zhao, Y. & Xing, R. Therapeutic importance and diagnostic function of circRNAs in urological cancers: from metastasis to drug resistance. Cancer Metastasis Rev. 22, (2024).
Sui, S. et al. Circ-CCS enhances autophagy during imatinib resistance of gastrointestinal stromal tumor by regulating miR-197-3p/ATG10 signaling. J. Cancer Res. Ther. 18 (5), 1338–1345 (2022).
Chunyan-Zou, Y. H. Xiukun-Chai, Chenxi-He, Dongqiang-Zhao. Potential value of circular RNA circTBC1D4 in gastrointestinal stromal tumors. J. Immunol. Res. 2022, 9019097 (2022).
Jia, N. et al. CeRNA expression profiling identifies KIT-Related circRNA-miRNA-mRNA networks in gastrointestinal stromal tumour. Front. Genet. 10, 825 (2019).
Faversani, A. et al. Mir-494-3p is a novel tumor driver of lung carcinogenesis. Oncotarget. 8 (5), 7231–7247 (2017).
Kim, O. et al. RASstimulated release of exosomal mir-494-3p promotes the osteolytic bone metastasis of breast cancer cells. Int. J. Mol. Med. 52 (3), 84 (2023).
Kaźmierczak, D. et al. Elevated expression of mir-494-3p is associated with resistance to osimertinib in EGFR T790M-positive non-small cell lung cancer. Transl Lung Cancer Res. 11 (5), 722–734 (2022).
von Mehren, M. & Joensuu, H. Gastrointestinal stromal tumors. J. Clin. Oncol. 36 (2), 136–143 (2018).
Mechahougui, H., Michael, M. & Friedlaender, A. Precision Oncology in gastrointestinal stromal tumors. Curr. Oncol. 30 (5), 4648–4662 (2023).
Hu, X., Li, J., Fu, M., Zhao, X. & Wang, W. The JAK/STAT signaling pathway: from bench to clinic. Signal. Transduct. Target. Ther. 6 (1), 402 (2021).
Paner, G. P. et al. Analysis of signal transducer and activator of transcription 3 (STAT3) in gastrointestinal stromal tumors. Anticancer Res. 23, 2253–2260 (2003).
Bauer, S., Duensing, A., Demetri, G. D. & Fletcher, J. A. KIT oncogenic signaling mechanisms in imatinib-resistant gastrointestinal stromal tumor: PI3-kinase/AKT is a crucial survival pathway. Oncogene. 26 (54), 7560–7568 (2007).
Cao, J. et al. Genome-scale CRISPR-Cas9 knockout screening in gastrointestinal stromal tumor with Imatinib resistance. Mol. Cancer. 17 (1), 121 (2018).
Wang, Z. et al. The circROBO1/KLF5/FUS feedback loop regulates the liver metastasis of breast cancer by inhibiting the selective autophagy of afadin. Mol. Cancer. 21, 29 (2022).
Acknowledgements
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.There was no clinical trial in the manuscript.Author Contribution declaration: Fang-wen Zou: Bioscientific experiments; Data base screening; Paper drafting. Yi-fang Tang and Xiaojing Li: Bioscientific experiments. Cong Liu and Chenhao Wu: Bioscientific experiments. Chao-Yuan Liu: Statistical analysis. Lei-yi Zhang: Study designation; Statistical analysis, Paper reviewing.
Funding
This project was granted from the National Natural Science Foundation of China (grant number: 82202923), Changsha Science Foundation for Distinguished Young (grant number: kq2306012), Natural Science Foundation of Changsha (grant number: kq2208334)), Soft Science Project of Changsha (grant number: kh2302033)), Health Research Project of Hunan Provincial Health Commission (grant number: W20243014), the Scientific Research Launch Project for new employees of the Second Xiangya Hospital of Central South University, and National Natural Science Foundation of Hunan (grant number: 2022JJ30866), and Key guidance project of Hunan Provincial Health Commission (grant number: 202204013949), and National Natural Science Foundation of Hunan (grant number: 2024JJ9224), and Key guidance project of Hunan Provincial Health Commission (grant number: 202204013949), and Educational Teaching Reform Research Project at Central South University in 2023 (grant number: 2023jy098), and Beijing science and technology innovation medical development foundation (grant number: KC2023JX-0186PQ005).
Author information
Authors and Affiliations
Contributions
Fang-wen Zou: Bioscientific experiments (RT-qPCR, CCK-8, Matrigel invasion assay, Wound healing assay, Annexin V/PI assay, Dual-luciferase reporter assay); Data base screening; Paper drafting. Yi-fang Tang and Xiaojing Li: Bioscientific experiments (Mice Subcutaneous Xenograft in vivo). Cong Liu and Chenhao Wu: Bioscientific experiments (Dual-luciferase reporter assay, Western-blot assay). Lei-yi Zhang: Study designation; Statistical analysis, Paper reviewing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study was approved by the institutional review board of Medical Ethics Committee of The Second Xiangya Hospital of Central South University in accordance with the Declaration of Helsinki and the Basel Declaration, and written informed consent was obtained from all participants.
Consent for publication
Authors declare each has approved this article to be published.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zou, Fw., Tang, Yf., Li, X. et al. circ_SMA4 promotes gastrointestinal stromal tumors malignant progression by sponging miR-494-3p/KIT axis and activating JAK/STAT pathway. Sci Rep 14, 22004 (2024). https://doi.org/10.1038/s41598-024-73393-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-73393-w