Abstract
Cardiac cellular fate transition holds remarkable promise for the treatment of ischemic heart disease. We report that overexpressing two transcription factors, Sall4 and Gata4, which play distinct and overlapping roles in both pluripotent stem cell reprogramming and embryonic heart development, induces a fraction of stem-like cells in rodent cardiac fibroblasts that exhibit unlimited ex vivo expandability with clonogenicity. Transcriptomic and phenotypic analyses reveal that around 32 ± 6.4% of the expanding cells express Nkx2.5, while 13 ± 3.6% express Oct4. Activated signaling pathways like PI3K/Akt, Hippo, Wnt, and multiple epigenetic modification enzymes are also detected. Under suitable conditions, these cells demonstrate a high susceptibility to differentiating into cardiomyocyte, endothelial cell, and extracardiac neuron-like cells. The presence of partially pluripotent-like cells is characterized by alkaline phosphatase staining, germ layer marker expression, and tumor formation in injected mice (n = 5). Additionally, significant stem-like fate transitions and cardiogenic abilities are induced in human cardiac fibroblasts, but not in rat or human skin fibroblasts. Molecularly, we identify that SALL4 and GATA4 physically interact and synergistically stimulate the promoters of pluripotency genes but repress fibrogenic gene, which correlates with a primitive transition process. Together, this study uncovers a new cardiac regenerative mechanism that could potentially advance therapeutic endeavors and tissue engineering.
Similar content being viewed by others
Introduction
Cardiovascular disease remains the leading cause of death globally1. Because the adult human heart cannot regenerate itself after cardiomyocyte death, resulting scars irreversibly damage contractile function and resist conventional therapy. While direct conversion of scar fibroblasts into induced cardiomyocytes (iCMs) provides a promising strategy for myocardium regeneration, the low efficiency of generating functional iCMs remains a significant limitation2,3,4,5. Alternatively, the reprogramming of induced cardiac progenitor cells (iCPCs) offers the option of producing not only proliferating cardiomyocytes, but also other cardiac lineages required for tissue repair6,7. Yet, the iCPC induction process often requires complex factors and labor-intensive procedures, and the generated iCPCs may exhibit heterogeneity and tumorigenicity upon stimuli. Furthermore, no successful in situ iCPC induction attempts have been reported to date.
As an additional strategy, inducing cell transition to a more primitive or dedifferentiated state has been explored in promoting progenitor regeneration, repair, and rejuvenation in systems such as the heart, neurons, skeletal muscle, and the liver8,9,10,11,12,13,14,15. By targeting specific tissues, this approach to “partial reprogramming” facilitates a smooth and natural cellular reversion and re-specification within the local tissue microenvironment, thereby boosting their regenerative activity. To date, most studies have utilized temporally controlled expressions of pluripotency genes, i.e., Oct3/4, Klf4, Sox2 and cMyc (OKSM) to maximize the effectiveness and mitigate the risk of inducing full pluripotency. Yet, alternative factors have also been reported. Chandrakanthan et al. applied the combination of growth factor PDGF-AB and 5-Azacytidine in mature bone and fat cells, inducing multipotent stem cells that contributed to tissue regeneration in vivo16. In this regard, targeted factors and refined strategies, especially in the realm of myocardial regeneration, still require exploration.
The spalt-like gene family member Sall4 is a unique stem cell factor that acts to maintain embryonic stem cell (ESC) self-renewal and pluripotency17,18,19,20. We previously have reported its potent role in regulating its own expression as well as the expression of Oct421. In fact, ectopic expression of Sall4, Nanog, Esrrb, and Lin2822, or Sall4, Sall1, Utf1, Nanog and Myc23 in embryonic fibroblasts reprogrammed them into induced pluripotent stem cells (iPSCs) in the absence of Oct4. While the GATA family members can also induce pluripotency in somatic cells by substituting for Oct4, RNA and ATAC sequencing assays identified Sall4, but not Oct4, as a direct downstream target of Gata4 and Gata6. Thus, Sall4 serves as a bridge linking the lineage specifying GATA family to the pluripotency circuit24. Additionally, both Sall4 and Gata4 play significant roles in the process of resident CPCs, iCM and/or iCPC reprogramming, as well as heart development25,26,27,28,29. In the developing murine heart, Sall4 expression is detected in the left ventricle and interventricular septum, regulated by Tbx5, and the two interact both positively and negatively to regulate heart morphogenesis and patterning30,31,32. Furthermore, mutations of Sall4 have been associated with heart septal defects similar to those caused by Gata4 and Tbx5 mutations32,33,34. Taken together, these discoveries pinpoint a potential role for Sall4 and the Gata family members in cardiac cellular fate transition.
To explore this potential, we previously have conducted screening experiments substituting Sall4 for the iCM reprogramming factors Gata4, Mef2c, and Tbx5. Intriguingly, in primary cardiac fibroblasts isolated from both neonatal and adult mice and rats, the overexpression of Sall4 with Gata4 and Mef2C induced rapidly-dividing stem-like clusters expressing Oct4. Moreover, our pilot study using a rat coronary ligation model for myocardial infarction (MI) suggests that both Sall4 and Gata4 improve cardiac function recovery and reverse remodeling (unpublished data and35,36). Thus, the present work seeks to deeply characterize the cardiac fibroblast transition process for potential regenerative applications. Our study demonstrates that elevated expressions of Sall4 and Gata4 are capable of inducing cellular conversion towards partially multipotent stem-like cells that acquire cardiogenic and also extracardiac potency.
Materials and methods
Tissue collection and cell culture
All animal experiments were conducted under a protocol approved by the Baylor College of Medicine (BCM) Institutional Animal Care and Use Committee (IACUC) and all methods were carried out in accordance with the NIH guidelines (Guide for the care and use of laboratory animals). Rat cardiac fibroblasts (RCFs) were harvested from 6- to 8‐week‐old male Sprague‐Dawley rats (Envigo, NJ) and cultured using a standard procedure as the group previously described36,37. During procedures, animal pups were euthanized with CO2 inhalation followed by decapitation. Adult euthanasia was achieved by isoflurane anesthesia followed by exsanguination, in accordance with American Veterinary Medical Association guidelines. All studies were conducted and are reported in compliance with relevant elements of ARRIVE guidelines. Additionally, adult cardiac fibroblasts (HCFs) were purchased from PromoCell (cat#c12375), and adult human (HDF, cat#2320) and rat (RDF, cat#R2320) dermal fibroblasts were obtained from ScienCell.
Viral vectors and stem-like cluster induction
Lentiviral vectors each encoding Gata4 (G), Mef2c (M), Tbx5 (T), or GFP, were prepared by BCM’s Gene Vector36,37. Sall4 (S), Sall4a, and Sall4bi lentiviral vectors were described previously38,39. Adenoviral vectors encoding SALL4 or GATA4 were purchased from VectorBuilder. For stem-like cell reprogramming, viral vectors were added to 5 × 104 cells/well in a 24-well plate with a multiplicity of infection (MOI) of 20, unless otherwise specified, with or without polybrene (5 µg/µl). One day after transduction, the medium was replaced with a modified iCM medium containing DMEM/Medium-199 (4:1), 10% fetal bovine serum (FBS), 1 × β-mercaptoethanol (Gibco), L-ascorbic acid (10 µg/ml), and 1% penicillin/streptomycin. In RCFs, aggregated stem-like clusters typically appeared at days 10–14 and then steadily expanded. When the cultures reached confluence, they were trypsinized and passed at 1:4 ~ 5 every 4–5 days to establish individual lines.
qRT-PCR
Total RNA was prepared using a RNeasy Mini kit (Qiagen, cat#74106), and cDNA synthesized using iScript RT Supermix (Biorad, cat #1708841). Relative quantification of RNA was performed using SYBR green Supermix (Biorad, cat#1725124). PCR products were detected in the QuantStudio 5 Real-Time PCR System (Thermo Fisher Scientific), and mRNA levels were normalized by comparative ΔΔCT method. Primers used in this study are listed in Supplemental Table S1.
Transcriptomic analysis
Bulk RNA sequencing (RNA-seq) was performed by Novogene Corporation. Messenger RNA was purified from total RNA using poly-T oligo-attached magnetic beads. The first strand cDNA was synthesized using random hexamer primers, followed by the second strand cDNA synthesis using dTTP for non-directional library. Quantified libraries are pooled and sequenced on Illumina platforms. For bioinformatics analysis, raw data (raw reads) in fastq format was processed through in-house perl scripts. Index of the reference genome was built using Hisat2 v2.0.5 and paired-end clean reads were aligned to the reference genome. Differentially expressed genes were determined using the DESeq2 R package (1.20.0)40. The resulting P-values were adjusted using Benjamini and Hochberg’s approach. Statistical enrichment of differential expression genes in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Gene Ontology (GO) were implemented using the clusterProfiler R package41.
Immunofluorescence (IF) staining
Cell cultures were fixed in multiwell plates with 4% paraformaldehyde (PFA) or ice-cold methanol, followed by permeabilization with 0.1% Triton and blocked in 1% bovine serum albumin (BSA). IF staining was performed using antibodies as described in Supplemental Table S2. Following fluorophore-conjugated secondary antibody staining and DAPI counterstaining, images were captured and analyzed with EVOS M5000 Imaging System or BioTek Cytation 5 Imaging Multi-Mode Readers.
High-throughput imaging and quantitative analysis
Multiwell plates with IF staining were automatically imaged on a Yokogawa CV8000 high throughput spinning disk confocal microscope using a 10 × objective, at BCM’s Integrated Microscopy Core. For each well, 4–6 fields of view were captured depending on the cell density. Images were saved as 16bit greyscale TIFF files and analyzed using CellPathfinder. Nuclei were segmented based on the DAPI channel using the automated algorithm; pseudocell boundaries were generated from the nuclear mask by expanding it 20 μm maximum. Objects touching the borders of the image were eliminated from further analysis. From the cell mask, total intensity for the antibodies was calculated for each cell. Based on visual inspection a set of intensity thresholds were identified for each experiment and each antibody that allowed calculating % positive cells in the population.
Cardiac lineage differentiation
To induce cardiomyocytes, GS cells were trypsinized and aggregated in an Ultra-Low Binding culture plate, using iCM medium supplemented with 10 µM ICG-001 (AdooQ BioScience), 100ng/ml BMP4 (Peprotech), and EGM™-2 Bullet solution (Lonza, cat#CC-3162) at a 3:1 volume ratio. After 4 days, cells were pelleted, dissociated, and replated in a regular dish using either the same solution, or Cardiomyocyte Differentiation Medium SCM102 (Sigma) for up to 7 days. In separate experiments, the STEMdiff™ Cardiomyocyte Maintenance (CMM) Kit (STEMCELL Technologies, cat#05020), supplemented with 1% FBS, was applied. In some other experiments, the PSC Cardiomyocyte Differentiation Kit (Gibco, cat#A2921201) B solution was used in accordance with the vendor’s instructions. To promote EC- or SMC-like cell differentiation, cells exposed to the SCM102 medium for 3 days were treated with either EGM™-2 with 0.5% FBS, or Rat Pulmonary Artery SMC Media with 2% FBS (Celprogen, cat#M12111-04) for additional 5 days. For human cardiomyocyte differentiation, GS-transduced HCFs at days 14 to 21 were replaced with CMM medium supplemented with 1% FBS for an additional 10–14 days.
Co-culture experiment and contractility analysis
Wild-type cardiomyocytes were isolated from 2- to 5-day-old mouse pups using the Neonatal Heart Dissociation Kit (Miltenyi Biotec, cat#30-098-373) and cultured as described42. When cells reached ~ 60% confluency, GFP-labelled co-cultures were plated onto neonatal cardiomyocytes on day 3 at a ratio of approximately 1:3. The next day, media was changed to Cardiac Myocyte Medium (ScienCell, cat#6101) with 0.5% FBS. To measure cell shortening, contracting GS-CMs on a glass-bottom dish was assessed using a video-based edge detection system (IonOptix, Ireland), and data analyzed using IonWizard software as described42.
Characterization of cell pluripotency
Alkaline phosphatase (ALP) staining was performed using Vector® Red AP Substrate (Vector Labs, cat#SK-5100). Germ layer marker mRNA expression was detected by RT-PCR and agarose gel electrophoresis. For teratoma formation assays, cells were harvested and re-suspended in phosphate-buffered saline (PBS). A 106 cell suspension in 70 µl was mixed with 30ul Matrigel (Corning, cat#354248) and injected subcutaneously into NSG mice (Jackson Laboratory, stock#005557). Tumor structures were dissected and stained with H&E and designed antibodies, and evaluated by a pathologist at BCM’s Pathology & Histology Core. In addition, neural-like cells were induced using a NeuroCult™ Proliferation Kit following recommended procedures (STEMCELL Technologies, cat#05702).
Luciferase assays
Promoter constructs for NANOG (#25900), SOX2 (#101761), SNAIL (#31694) and cMYC (#114724) were purchased from Addgene. The OCT4 promoter construct and the 293T cell transfection procedures were previously described21. Transfection Carrier DNA (Promega, cat#E4881) was used as a mock treatment and for balancing total DNA amount. Promoter activity was measured using Renilla-Glo® and Luciferase Assay Systems (Promega, cat#E2710 & cat#E1500).
Flow cytometry
Antibodies and reagents are listed in Supplemental Table S2. Cells were fixed and permeabilized using CytoFix/CytoPerm™ solution (BD, cat#554722) as instructed. Blocking was performed with 5% goat serum in Perm/Washing buffer. Cells were stained with primary and secondary antibodies following the manufacturer’s recommendations and analyzed using a BD FACSCanto II sorter with FlowJo software.
Co-immunoprecipitation (co-IP) and Western blotting
293T cells were transfected with plasmids encoding Gata4 or HA-tagged Sall4a, Sall4b, and a truncated Sall4a (termed Δ2Sall4)43, using Lipofectamine™ 2000 (Invitrogen, cat#11668027). Cells were harvested after 72 h, and proteins were prepared using CelLyticTM MT cell lysis reagent (Sigma, cat#C3228). Co-IPs were performed using the Dynabeads® protein G immunoprecipitation kit (Invitrogen, cat#10007D). Western blots were performed with antibodies against HA and Gata4 as listed in Supplemental Table S2.
Chromatin immunoprecipitation (ChIP) assay
HEK293 cells were transfected with HA-Sall4b and Gata4 plasmids as described above. ChIP was performed 48 h post-transfection using antibodies targeting Sall4, the HA tag, and Gata4 with the ChIP-IT Express Enzymatic Kit (Active Motif, cat#53009). Two chromatin shearing conditions were tested, and IgG was used as a negative control to confirm specificity. Input DNA and immunoprecipitated DNA were analyzed by qPCR using SYBR Green Supermix as described above. Percent input values were calculated to normalize the results. The primers and antibodies used in these assays are listed in Supplemental Tables S1 and S2, respectively.
Statistical analysis
Data were expressed as mean ± standard deviation of the mean (S.D.), unless otherwise stated. The unpaired t-test was used to determine the significance of differences between two groups. One-way ANOVA was used in Prism GraphPad to determine the significance of differences when more than two groups were compared. A value of p < 0.05 or lower was considered statistically significant. Details regarding number of biological replicates, and sample sizes are reported in the main text and figure legend.
Results
Emergence of aggregated stem-like clusters in cardiac fibroblasts overexpressed with Sall4 and Gata4
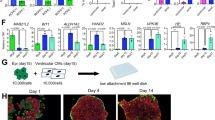
To characterize Sall4’s role in cardiac fibroblast transition, we first focused on adult rat cells (RCFs) given our intended plan of in-depth rat MI model studies. RCFs were prepared using the lab’s standard protocol and validated through their ubiquitous expression of two fibroblast-specific markers COL1A1 and PDGFRα (Supplementary Figure S1). In these cells, various combinations of Gata4 (G), Mef2c (M), Tbx5 (T), and Sall4 (the b isoform; S) were screened. Two weeks after lentivirus transduction and growth in a modified iCM medium, aggregated clusters with irregular shapes and sizes were observed in RCFs treated with GMTS, GMS, and GS, but not in those treated with GFP control, GMT, or other vector combinations (Fig. 1A; Supplementary Figure S2a; n ≥ 3 separate transductions for each group). We then focused on the GS group, which was further validated using the Sall4a and Sall4bi vectors (Supplementary Figure S2b, n > 2). Typically, about 1 to 3 clusters were visible per well in a 24-well dish, which developed into well-formed colonies containing different numbers of cells in secondary cultures (Fig. 1B). Subsequently, these cells expanded unlimitedly ex vivo, enabling us to establish numerous lines from each group at various passages. By measuring population doubling time (PDT) at different passages, the GS group cells (hereafter termed GS cells) constantly exhibited a nearly 6-fold higher proliferation rate than GFP control-treated cells (p3), highlighting their significant self-renewal (Fig. 1B-C).
Stem-like clusters induced by Gata4 and Sall4-based transcription factors. (A) Bright field and fluorescence images showing aggregated clusters following overexpression of indicated factors in RCFs for up to two weeks (from n ≥ 3 separate transduction experiments for each group). The GFP and GMT group cells served as controls. A phase/fluorescence overlay image is shown for the GS group cells. (B) Top: Images of GS cell colonies containing different numbers of cells in secondary cultures. Bottom: Expanding GS cell colonies at indicated passages. (C) Cell population doubling time data showing significantly accelerated GS cell growth compared to GFP control RCFs. Ns: not significant, ***p < 0.001; n = 3.
The expanding cells express cardiac progenitor (CPC) and pluripotency markers
We next conducted qRT-PCR assays. Compared to GFP control RCFs, the GS cells displayed significantly increased mRNAs of pluripotency genes Oct4, Lin28, and Sox2, an endo/mesodermal gene Sox17, and CPC marker genes Nkx2.5, Flk1, and Isl1. In contrast, fibroblast related genes Tcf21, Fsp1, Postn, and Col1a2 were downregulated (Fig. 2A). Consistently, significant expressions of pluripotency and CPC marker proteins were detected by single and dual antibody immunofluorescence (IF) staining. While some clustered cells co-expressed NKX2.5 and OCT4, the levels of NKX2.5 were noticeably higher (Fig. 2B and Supplementary Figure S3). This is consistent with flow cytometry data showing that 32.0 ± 6.4% of the cells were Nkx2.5+, 29.2 ± 15.0% were Flk1+, and 13.1 ± 3.6% were Oct4+ (Fig. 2C). Notably, the percentages of Nanog+ and SSEA1+ cells were below 5%, suggesting that a significant portion of GS cells may develop cardiac progenitor-like features, while a subset attains certain levels of pluripotency.
Expression of pluripotency and cardiac progenitor markers in GS cells. (A) Fold mRNA changes for indicated genes in GS cells (p9) versus GFP-treated RCFs. Data was normalized using β-actin as the reference gene; ***p < 0.001, **p < 0.01; n = 3 separate cultures. (B) Single and dual antibody IF staining depicting indicated marker proteins (in red) in GS cells (data collected from passages 6–12). Cell nuclei were counterstained with DAPI (in blue). (C) Flow cytometric dot plots showing indicated marker expressions in GS cells vs. GFP control RCFs, with bar graph percentage values presented separately. More than 8000 events were analyzed. *p < 0.05, **p < 0.01, ***p < 0.01; n = 3.
Differentiation of GS cells into cardiovascular cell types
To determine the CPC-like features, we initially aggregated the GS cells in an ICG-001 (a Wnt/β-catenin inhibitor)-based solution for 4 days, followed by plating them in either the same solution or in a rodent CPC-targeted cardiomyocyte differentiation medium (SCM102, see Methods) for 5 days. These treatments induced rapid changes in cell morphology with up to 48.0% cTNT expressing cells, as determined by high-throughput IF imaging and quantification. Many cTNT+ cells also expressed other two cardiomyocyte markers, cTNI and CXCR4 (Fig. 3A i– iii). Additionally, some cell fractions expressed cardiovascular lineage markers CD31 and Calponin, at varying levels, indicating a multi-lineage differentiation process (Fig. 3A-iv, and Supplementary Figure S4). In prolonged culture, however, these cells displayed stressed structures. We then investigated the STEMdiff™ cardiomyocyte maintenance medium (CMM), which similarly induced 38.2 ± 8.3% cTNT+ cells within 5 days, as analyzed by flow cytometry. Moreover, sarcomere organization was observed in both cTNT+ and α-actinin+ cells, indicative of a maturation process (Fig. 3B-C). In comparison, approximately 4.9 ± 1.1% cTNT+ cells were detected by flow cytometry in GMT-transdifferentiated RCFs over 2 weeks (Supplementary Figures S5a-b).
Differentiation of GS cells into cardiovascular cell types. (A) (i) Bright field image of aggregated GS cells in the ICG-001-based solution at day 3 (n > 12). (ii-iii) IF staining of GS-CMs with dual antibodies (n > 4 separate experiments), with bar graph showing the percentages of cTNT+ cells by high-throughput imaging analysis (iv; n = 3 staining/group). (B) Sarcomeric structures observed in cTNT+ and α-actinin+ cells (see 40× images). (C) Flow cytometric dot plots illustrating the distribution of cTNT+ cells in GS cells vs. GS-CMs, with bar graphs showing the percentage values separately (n ≥ 3 treatments). (D) Top: co-cultured GS-CM clusters, but not GMT or GFP treated RCFs, demonstrated contractility (images here, see Video Files). Bottom: representative cell length cycling for each group is displayed (n > 3). (E) Differentiated EC-like cells stained with indicated single or dual antibodies (i ~ ii). Multiple CD31+ cells formed a lumen structure (iii ~ iv; n ≥ 3). Percentages of cells expressing each cardiac trilineage marker protein are shown (n = 3 staining/group). (F) Differentiated GS-SMCs expressed indicated marker proteins with quantification analyses of trilineage markers (n = 3 staining/group).
While none of the above treatments induced spontaneous cell contraction, we next re-infected the growing GS cells with GFP lentivirus, followed by aggregating the cells and then directly seeding them onto sparsely cultured murine neonatal cardiomyocytes (MNCMs, GFP-), under a low-serum condition. In parallel, the GMT or GFP group RCFs were replated under the same condition. Within 7 days, the latter two group cells displayed stressed survival and disrupted MNCM growth and contraction by bright field and fluorescence microscopy. In contrast, the GS cell-differentiated cardiomyocyte-like cells (GS-CMs, GFP+) exhibited clustered growth, with around 25% of total GFP + cells contracting synchronous with MNCMs, and some displayed rhythmic shortening (Fig. 3D and Supplementary Videos). Furthermore, IF staining for such prepared cTNT+ GS cells showed a similar distribution pattern to the beating GFP + GS-CMs (Supplementary Figure S6 and Videos 3 - 5). Together, these findings suggest that appropriate environmental conditions, as might be found in the myocardium, could readily induce the GS-converted fibroblasts into functional cardiomyocytes.
To further clarify cardiac multilineage potency, we next treated the SCM102 medium-exposed GS cells with either an endothelial cell (EC) medium EGM-2, or a rat smooth muscle cell (SMC) maintenance medium (see Methods). This treatment markedly activated lineage-specific markers within 7 days, including CD31 and VE-cadherin for ECs, and Myh11, Calponin, and αSMA for SMCs. High-throughput IF imaging analysis revealed up to 84.2 ± 3.6% CD31+ and 64.3 ± 17.5% Calponin+ cells in the differentiated EC and SMC-like populations, respectively. Moreover, the differentiated CD31+ cells exhibited typical endothelial morphological changes and lumen-like EC structures (Fig. 3E, F). Collectively, these findings suggest that GS cells acquired multipotency, capable of readily differentiating into at least various cardiac lineage phenotypes.
Transcriptomic analysis for GS cells and following cardiac differentiation
To comprehensively elucidate the shifts in gene expression profiles, bulk RNA sequencing was conducted for each differentiated GS cells. Consistent with the qRT-PCR data, undifferentiated GS cells displayed significant gene expressions associated with pluripotency (e.g., Pou5f1, Lin28a, Klf4, Myc, Sall4, Sall1, Utf1) and cardiac specification (e.g., Nkx2.5, Isl1, Isl2, Hcn4, Sox17, Fgf10, Pax3, Ets2), as compared to parental RCFs, whereas fibroblast genes (e.g., S100a4/Flsp1, Postn, Col1a2, Ddr2, Tcf21, Wt1) were downregulated (Fig. 4A; see Supplementary Excel File for full contents). Signaling pathways involved in these transitions, such as PI3K/Akt, Hippo, cytokine-cytokine receptor interaction, Wnt, and TGF-beta, were also activated (Fig. 4B). Additionally, epigenetic enzymes implicated in Sall4 and Gata4’s regulatory functions, including the DNA methyltransferases (DNMTs) Dnmt3b and Dnmt3l, the HDAC/NuRD complex component HDAC1, and histone deacetylase SIRT143,44, were identified, indicative of related epigenetic regulations (Fig. 4A). In comparison, the CMM medium-treated GS cells (designated as GSdiff1) exhibited activated genes related to cardiac differentiation (Myh6, Myh7b, Actc1, Actn2, Tnni1, Tnni3, Mybpc3, Pln), while CMM and EGM-2 co-treated cells (GSdiff2; 1:1 volume ratio) for 3 days expressed both cardiomyocyte (Tnnt2, Ryr2, Gja1, Myoz2) and vascular endothelial cell associated genes (vWF, Prom1/Cd133, Etv4, Gja4, Flt1/Vegfr-1, Gata2, Tal1, Esm1) (Fig. 4A). In summary, a significant proportion of differentially expressed genes were identified among each GS cell treatment group and the control RCFs, as illustrated in the Venn diagram (Fig. 4C). Hence, the GS cells are highly prone to cardiomyocyte and endothelial cell differentiation upon specific stimuli, and suitable growth conditions could boost their phenotype progression and cellular interaction46,47,48.
Transcriptomic analysis of GS cells and following differentiation. (A) Heatmaps of RNA-seq data illustrating a list of differentially expressed genes in GS cells (p12), GSdiff1, GSdiff2, and RCF group cells (n = 3 preparations for each group). (B) KEGG pathway analysis revealed a total of 19 significantly enriched pathways in GS cells compared to control RCFs based on adjusted p values (padj). The numbers of differentially expressed genes are shown. (C) Venn diagram showing the total numbers of overlapping and differentially expressed genes among each GS cell treatment group and control RCFs.
Partial pluripotent properties in the expanding cells and extracardiac lineage differentiation
The transcriptomic and phenotypic data prompted us to characterize the presence of pluripotent cells. In GS cells from various transduction experiments (n > 5) and across different passages, robust alkaline phosphatase (ALP) expression was consistently detected, contrasting with the basal level in GFP control RCFs (Fig. 5A). We then grew the GS cells in an embryoid body media for 5 days, and detected mRNA expression of germ layer markers for endoderm (Afp, Sox17), mesoderm (Flk1, Gata4), and ectoderm (Nestin, Sox1, NFH) (Fig. 5B and Supplementary Figure S7). Next, we collected the GS cultures (p6 to 8) and subcutaneously injected them into the hind leg of immunodeficient NSG mice (G/S: n = 3; and G/S-a isoform: n = 2). After 12–14 weeks, tumor formation was observed in all five recipients, but not in the GFP control cell-injected animals (n = 2). Pathological examinations revealed that most tumor tissues represent poorly differentiated chondrosarcoma (cartilage) or adenocarcinoma (glandular), distinguishing them from teratoma (Fig. 5C)49. Therefore, the GS cells under current conditions still lack full pluripotency, although refined manipulations, such as colony pickup or treatment with rat PCS-specific media, could be explored to boost bona fide iPSC induction. Nevertheless, we detected nerves in the tissues via βIII-tubulin (TUJ-1) antibody staining, and TUJ-1+ cells were also induced using a neuron progenitor medium in culture (Fig. 5C-D), supporting the notion that the GS cells possess a broad range of differentiation abilities beyond cardiovascular lineages.
Assessment of pluripotency and differentiation of extracardiac lineages. (A) Images of scattered and intensive alkaline phosphatase expression seen in clustered GS cells but not in the control RCFs (from n > 4 separate viral transduction experiments). (B) The GS cells were aggregated in an EB media and then detected for indicated gene expressions by RT-PCR and agarose gel electrophoresis. Images are cropped from two parts of raw gel data, outlined in red in Supplementary Figure S7; n ≥ 3 separate experiments. (C) The GS cells developed tumors in injected NSG mice. Representative H&E staining images of sectioned tumors showing differentiated glandular and cartilage tissues. Neuron-like cells were detected by IHC staining using a βIII-tubulin (TUJ-1) antibody. (D) Representative Tuj1 IF staining comparing parental fibroblasts with GS cell-derived neuron-like cells on day 6, n > 3 separate experiments.
Stem-like feature induction and cardiac differentiation in human cardiac fibroblasts (HCFs)
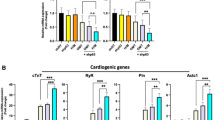
To determine whether the Sall4/Gata4 approach applies to clinically important human models, we next investigated a commercial HCF line (PromoCell). Cells were transduced with G/S vectors and maintained in the same iCM media for up to 3 weeks. At this stage, no apparent stem-like clusters were noticed. However, robust ALP staining was detected in nearly 50% of the transduced cells, contrasting with the background levels in GFP-treated control HCFs (Fig. 6A, n > 4 separate transduction experiments). Upregulated mRNA expressions of OCT4, NANOG, LIN28, SOX2, and downregulated POSTN were also identified (Fig. 6B, n = 3). In subsequent cultures, some aggregated cells continued to grow into enlarged clusters, expressing pluripotency proteins TRA-1-60 and OCT4, as well as CPC markers NKX2.5 and FLK1, as detected by single and double antibody IF staining. Flow cytometry assays likewise revealed 12.7 ± 2.5% of TRA-1-60+ and 20.9 ± 5.2% of NKX2.5+ cells, respectively, supporting concurrent primitive state transition and cardiac specification processes (Fig. 6C-D; Supplementary Figure S8). However, unlike findings from RCFs, the induced clusters were difficult to sustain expansion beyond 5 ~ 6 passages, and their surrounding cells displayed stressed growth, despite the application of a human PSC maintenance medium Essential 8 (Gibco). These observations suggest that while Sall4/Gata4 similarly induce a primitive cell fate transition in HCFs, distinct regulatory mechanisms and manipulation requirement exist that warrant further elucidation. Nevertheless, when these GS-induced HCFs were switched to CMM media for an additional 10 days, significant expressions of cTNT, cTNI, and α−actin marker proteins were detected with visible sarcomeric structures by IF staining (Fig. 6E). Hence, these results highlight again Sall4/Gata4’s effect in inducing a primitive stage transition in HCFs, endowed with strong cardiogenic potential.
Stem-like feature transitions in HCFs and cardiac differentiation. (A) Representative ALP staining in GS-overexpressed HCFs (GS-HCFs, 14-day and 21-day experiments), versus GFP control cells (n > 4). (B) qRT-PCR analysis of mRNA expressions in GS-HCFs (21-day) vs. non-infected mock HCFs. Data was normalized using GAPDH as the reference gene. ***p < 0.001, **p < 0.01; n = 3. UD: CT value undetected. 40/40: MOI numbers used for G and S vectors. (C) Single and dual antibody IF staining showing expression of indicated pluripotency and CPC marker proteins in enlarged GS-HCF at p2 (n > 3 separate experiments). (D) Representative flow cytometric dot plots and bar graph showing the proportions of positively stained cells in indicated groups (n = 3 separate experiments). (E) Single and dual antibody IF staining showing indicated marker expressions. Cells with sarcomeric structures in cTNT+ cells are presented (40image), with bar graph quantification estimated in indicated HCF groups. n ≥ 3 separate experiments. **p < 0.01.
Inefficient cellular fate conversion in skin fibroblasts
To assess whether Sall4/Gata4 may exert a broad influence across different fibroblast types, we next overexpressed the G/S vectors in dermal fibroblasts obtained from both adult rats (RDFs) and humans (HDFs). In RDFs, no clustered growth or colony expansion was observed after 3 weeks of culture and subsequent passaging. ALP screening revealed negligible staining, indicating a lack of significant activation (Fig. 7A; n > 3 separate experiments). Further, qRT-PCR analysis showed insignificant upregulation of stem/progenitor and cardiac specification genes Oct4, Nanog, Sox2, Sox17, Nkx2.5, and Isl1 (Fig. 7B; p < 0.01, n = 3). In HDFs, similar negligible ALP staining results were observed. Moreover, comparative qRT-PCR screening analysis using GS-overexpressed HDFs vs. HCFs showed negligible gene expression changes for stemness and cardiac specification genes OCT4, LIN28, NANOG, SOX17, and NKX2.5 (Fig. 7C-D; n > 2 separate experiments). Collectively, these data suggest that the impact of Sall4/Gata4 is likely confined to certain fibroblast origins, such as those in the heart.
Inefficient transition effects in skin fibroblasts. (A) A phase/fluorescence overlay image depicting G/S vector (GFP+) expression in infected RDFs (left), with negligible ALP staining after 3 weeks of culture (right); n > 3 experiments. (B) Fold mRNA changes for indicated genes in GFP vs. GS-overexpressed RDFs. ***p < 0.001, **p < 0.01; n = 3. (C) Representative GFP+ vector expression and ALP staining images in GS-overexpressed HDFs (n > 3). (D) Comparative qRT-PCR analysis of indicated mRNA expressions in HDFs versus HCFs overexpressed with GFP (mock) or G/S factors. ADV: adenovirus; 30/40: MOI numbers for G and S vectors, respectively (n > 2).
SALL4 and GATA4 interact to synergistically regulate cell transition-related genes
We proceeded to assess how the cellular transition process was initiated. Luciferase assays were first conducted using promoter constructs for pluripotency genes OCT4, NANOG, MYC, and a fibrogenic gene SNAIL. These constructs were separately transfected into 293T cells, along with different combinations of Sall4 and Gata4 vectors for 72 h. While overexpression of either SALL4 or GATA4 activated or repressed these promoters to varying degrees, co-expressing the two exerted a more significant synergistic effect on all of them (Fig. 8A). These results are consistent with the transcription data (Figs. 4A and 6B) and support previous findings of Sall4 and Gata4’s effects on these genes36,44. Inspired by these complementary and synergistic findings, we next designed and conducted a co-immunoprecipitation (co-IP) assay. Using an HA antibody, we successfully immunoprecipitated the SALL4 protein complex, which was then detected by an anti-GATA4 antibody (Fig. 8B). Conversely, a GATA4 antibody-immunoprecipitated complex was strongly detected by an anti-HA antibody, exhibiting the presence of the SALL4b protein (Supplementary Figure S9, Gel #2). Additionally, within the nuclei of transduced HCFs and RCFs, IF staining revealed overlapping distributions of SALL4 and GATA4 proteins (Fig. 8C). Next, we performed ChIP-qPCR using primers designed from SALL4-enriched sequences identified in our ChIP-ChIP studies21. Data demonstrated correlated binding patterns and levels of GATA4 and SALL4 on the promoter regions of POU5F1 (OCT4), MYC, and NKX2.5 genes (Fig. 8D). These findings, combined with the transcriptomic data, suggest that elevated SALL4/GATA4 form a regulatory complex to epigenetically and transcriptionally regulate essential stemness and fibrogenic genes, thereby activating subsequent cellular transition processes.
SALL4 and GATA4 interact to regulate cell transition related genes. (A) Relative fold induction of luciferase expression in transfected 293T cells, reflecting the activity of indicated promoters following SALL4 and/or GATA4 overexpression. Luciferase expression levels were normalized to mock treated conditions; ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05, n = 3 separate transfection experiments. (B) 293T cells were transfected with indicated plasmids. Expression of HA-tagged proteins following pull-down was validated by Western blotting (i). Co-IP was performed using an anti-HA (ii) or anti-Gata4 antibody (iii) and detected by Western using an anti-Gata4 antibody. Images are cropped from two sets of raw blot data, as outlined in red in Supplementary Figure S9. (C) Representative IF images indicating overlapping distributions of SALL4 (red) and GATA4 (yellow) proteins within the nuclei of transduced RCFs and HCFs. Images were captured using EVOS M5000 microscope at 40× magnification. (D) The upper panels show SALL4-enriched bindings identified from ChIP-ChIP assays on promoter regions of the indicated genes, along with primer designs. Log2 values indicate enrichment of DNA by the target protein21. The lower panels present qPCR-amplified DNA levels as relative percentages of input, correlating with the indicated antibodies. Data represent consistent patterns from two ChIP experiments conducted under different conditions.
Discussion
In this study, we present a novel role for Sall4/Gata4 interaction in transitioning cardiac fibroblasts into a multipotent primitive stage endowed with cardiogenic potential. The induced cells contain at least partially pluripotent cells, cardiac precursors, and intermediate fractions, which are highly susceptible to cardiovascular differentiation. These findings are in line with Sall4 and Gata4’s functions in diverse contexts, including ESC maintenance, iPSC reprogramming, the human cardiac progenitor process (Sall4+/Isl1+)25,26, second heart field progenitor and cardiomyocyte proliferation (Sall1/Sall4/Myocd/SRF)27, and transdifferentiation of iCMs in MI-injured fibroblasts28,29. In further characterizing the clonality and multipotency of Sall4/Gata4-induced cells, including their cardiogenic multi-, bi-, or unipotency, we attempted a limiting dilution assay in 96-well plates. However, we repeatedly observed retarded single-cell growth, which hindered subsequent analyses (data not shown). We speculate that proliferating fibroblasts in the GS cell cultures may have acted as feeders, critically contributing to partial cellular reprogramming, maintenance, and differentiation processes. Future studies, including optimized limiting dilution conditions and temporal single-cell RNA-sequencing assays, are needed to decode the cellular phenotypes and identify different cell subpopulations or differentiation states.
In RCFs, the incidence of primarily induced stem-like clusters was relatively low, with 1 to 3 clusters observed across 8 out of 11 separate transduction experiments. However, given the high phenotypic heterogeneity among resident cardiac fibroblasts50,51,52, Sall4/Gata4 may preferentially reprogram certain subfractions with high plasticity. Various progenitor-like fractions have been characterized based on markers such as Sca1, Isl1, Pdgfrα, Flk1, and Thy153,54,55,56,57. Accordingly, the composition and activities of such cell subtypes in primary cultures may markedly impact the reprogramming time course and outcomes. As an example, exogenous Oct4 with either Sox2 or Klf4 partially reprogramed Thy1+/Sca1+ fibroblasts to a primitive stage concurrently expressing mixed lineage markers58. Further research, e.g., using individually sorted cell subtypes or with refined induction conditions, may help define these putative co-relationships, facilitating improved and targeted applications.
Following transgene overexpression, we detect protein interactions between SALL4 and GATA4. Both have been known to dynamically interact with diverse partners such as OCT4, NANOG, NKX2.5, TBX5, MYOCD, SOX2, SALL1, etc., depending on cellular contexts and functions19,30,59,60,61. Based on gene expression data (Figs. 4A and 7D, and 8A), we propose that elevated levels of SALL4/GATA4 in cardiac fibroblasts primarily activate key pluripotency genes but repress fibroblast networks, leading to an iPSC-like feature transition. During this process, however, the interplay of cellular epigenetic memory62 and signaling crosstalk with neighboring cells, such as relevant growth factor and cytokine interactions, facilitate the expression of cardiogenic genes (e.g., Sox17, Nkx2.5, Isl1), driving subsequent cardiac specification and lineage regeneration (Fig. 9). Supporting this, we observed robust Sox17 expression at both mRNA and protein levels (Figs. 2A, 4A and 7D, and Supplementary Figure S10). Oct4 and Sox17 have been proposed to steer pluripotent cell progression toward a cardiac fate63. Further, Sall4, along with Sox17, acts as a partner of Oct4 to mediate Oct4’s cardiogenic function64. In this context, comprehensive analyses such as Sall4/Gata4-mediated genome-wide DNA binding, epigenetic landscape changes, including DNA methylation patterns and histone modifications, as well as single-cell level transcriptomics, would help uncover detailed regulatory pathways and discover new enhancers or inhibitors in this process.
While our data support Sall4/Gata4-induced primitive fate transitions in both RCFs and HCFs, there are notable differences between the two systems. In HCFs, nearly half of the primarily induced cells strongly expressed ALP, yet they exhibited limited expandability and clonogenicity. Additionally, when transferred to STEMdiff™ CMM media, they rapidly differentiated into a cardiomyocyte phenotype. These discrepancies likely arise from the variations in genetic settings, epigenetic landscapes, cellular context, signaling pathways, and self-renewal mechanisms. This is consistent with the variability frequently observed in rodent and human iCM and iPSC reprogramming studies65,66. On the other hand, both human and rat skin fibroblasts showed ineffective conversions, which may be due to specific epigenetic landscapes unique to cardiac fibroblasts. Moreover, cardiac fibroblasts naturally express many cardiogenic genes, such as Gata4, Tbx20, Mef2c, and Nkx2-5, which are not present in skin fibroblasts67,68. Nevertheless, these findings may suggest several therapeutic advantages of Sall4/Gata4, such as enhanced cardiogenic ability, cardiac tissue specificity, decreased tumorigenic risk, and reduced fibrosis. It will be essential to carefully evaluate these aspects in various treatment models. Due to limitations related to the availability of patient cardiac fibroblasts and the limited expandability of GS-induced HCFs, this study did not determine contractility in differentiated cardiomyocyte-like cells. Additionally, other cardiac or extracardiac potencies were not assessed. For a better understanding of mechanisms and to enhance in vitro applications, including cardiac tissue engineering, drug screening, and organoid development, it is important to establish optimized differentiation protocols and functional clarifications, such as ventricular or atrial features, calcium dynamics, and electrophysiological properties, in both human and rat systems.
Unlike the OSKM factors, both Sall4 and Gata4 play significant roles in cardiac progenitors and embryonic heart morphogenesis. Gene therapy utilizing these two factors may facilitate streamlined in situ cardiac regeneration. More research is needed to ascertain the lineage regeneration selectivity, efficacy, and safety of Sall4/Gata4 in rodent and large animal models of myocardial infarction in vivo. In this regard, the observed extracardiac neuron-like differentiation may provide additional benefits, as cardiac innervation has been shown to improve cardiac repair through neurohormonal regulation, cellular signaling, and angiogenesis etc69,70,71. These aspects will be the focus of our next studies.
In summary, our study identifies a novel, non-OSKM approach to convert cardiac fibroblasts toward a multipotent primitive state with cardiogenic potential, potentially offering a new strategy in the field of myocardium regeneration. Additionally, it broadens our understanding of Sall4’s diverse roles across different stem/progenitor cell systems.
Data availability
Multiple induced expandable cell lines are generated in this study, available from the corresponding author upon reasonable request. RNA sequencing datasets produced from these cells under different conditions have been deposited at GEO with the accession number GSE268997. The token number is: ypifmauojbunpgp. Any requests should be directed to the corresponding author.
Change history
09 November 2024
The original online version of this Article was revised: In the original version of this Article, the Supplementary Videos, which were included with the initial submission, were omitted from the Supplementary Information files. The correct Supplementary Information files now accompany the original Article.
References
Roth, G. A. et al. Global burden of cardiovascular diseases and risk factors, 1990–2019: Update from the GBD 2019 study. J. Am. Coll. Cardiol. 76, 2982–3021. https://doi.org/10.1016/j.jacc.2020.11.010 (2020).
Cahill, T. J., Choudhury, R. P. & Riley, P. R. Heart regeneration and repair after myocardial infarction: translational opportunities for novel therapeutics. Nat. Rev. Drug Discov. 16, 699–717. https://doi.org/10.1038/nrd.2017.106 (2017).
Garry, G. A., Bassel-Duby, R. & Olson, E. N. Direct reprogramming as a route to cardiac repair. Semin. Cell. Dev. Biol. 122, 3–13. https://doi.org/10.1016/j.semcdb.2021.05.019 (2022).
Patel, V., Mathison, M., Singh, V. P., Yang, J. & Rosengart, T. K. Direct cardiac cellular reprogramming for cardiac regeneration. Curr. Treat. Options Cardiovasc. Med. 18, 58. https://doi.org/10.1007/s11936-016-0480-8 (2016).
Perveen, S., Vanni, R., Lo Iacono, M., Rastaldo, R. & Giachino, C. Direct reprogramming of resident non-myocyte cells and its potential for in vivo cardiac regeneration. Cells. 12. https://doi.org/10.3390/cells12081166 (2023).
He, X. et al. Advances in cellular reprogramming-based approaches for heart regenerative repair. Cells. 11. https://doi.org/10.3390/cells11233914 (2022).
Xu, J., Lian, W., Li, L. & Huang, Z. Generation of induced cardiac progenitor cells via somatic reprogramming. Oncotarget. 8, 29442–29457. https://doi.org/10.18632/oncotarget.15272 (2017).
Chen, Y. et al. Reversible reprogramming of cardiomyocytes to a fetal state drives heart regeneration in mice. Science. 373, 1537–1540. https://doi.org/10.1126/science.abg5159 (2021).
Hishida, T. et al. In vivo partial cellular reprogramming enhances liver plasticity and regeneration. Cell. Rep. 39, 110730. https://doi.org/10.1016/j.celrep.2022.110730 (2022).
Wang, C. et al. In vivo partial reprogramming of myofibers promotes muscle regeneration by remodeling the stem cell niche. Nat. Commun. 12, 3094. https://doi.org/10.1038/s41467-021-23353-z (2021).
Hong, Y. J. et al. In vivo generation of neural stem cells through teratoma formation. Stem Cells Dev. 25, 1311–1317. https://doi.org/10.1089/scd.2016.0124 (2016).
Chuang, W. et al. Partial reprogramming of pluripotent stem cell-derived cardiomyocytes into neurons. Sci. Rep. 7, 44840. https://doi.org/10.1038/srep44840 (2017).
Puri, D. & Wagner, W. Epigenetic rejuvenation by partial reprogramming. Bioessays. 45, e2200208. https://doi.org/10.1002/bies.202200208 (2023).
Lehmann, M. et al. Partial reprogramming as an emerging strategy for safe induced cell generation and rejuvenation. Curr. Gene Ther. 19, 248–254. https://doi.org/10.2174/1566523219666190902154511 (2019).
Doeser, M. C., Scholer, H. R. & Wu, G. Reduction of fibrosis and scar formation by partial reprogramming in vivo. Stem Cells. 36, 1216–1225. https://doi.org/10.1002/stem.2842 (2018).
Chandrakanthan, V. et al. PDGF-AB and 5-azacytidine induce conversion of somatic cells into tissue-regenerative multipotent stem cells. Proc. Natl. Acad. Sci. U S A. 113, E2306–2315. https://doi.org/10.1073/pnas.1518244113 (2016).
Yang, J. et al. Genome-wide analysis reveals Sall4 to be a major regulator of pluripotency in murine-embryonic stem cells. Proc. Natl. Acad. Sci. U S A. 105, 19756–19761. https://doi.org/10.1073/pnas.0809321105 (2008).
Zhang, J. et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat. Cell. Biol. 8, 1114–1123. https://doi.org/10.1038/ncb1481 (2006).
Tanimura, N., Saito, M., Ebisuya, M., Nishida, E. & Ishikawa, F. Stemness-related factor Sall4 interacts with transcription factors Oct-3/4 and Sox2 and occupies Oct-Sox elements in mouse embryonic stem cells. J. Biol. Chem. 288, 5027–5038. https://doi.org/10.1074/jbc.M112.411173 (2013).
Zhou, Q., Chipperfield, H., Melton, D. A. & Wong, W. H. A gene regulatory network in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U S A. 104, 16438–16443. https://doi.org/10.1073/pnas.0701014104 (2007).
Yang, J., Gao, C., Chai, L. & Ma, Y. A novel SALL4/OCT4 transcriptional feedback network for pluripotency of embryonic stem cells. PLoS One. 5, e10766. https://doi.org/10.1371/journal.pone.0010766 (2010).
Buganim, Y. et al. The developmental potential of iPSCs is greatly influenced by reprogramming factor selection. Cell. Stem Cell. 15, 295–309. https://doi.org/10.1016/j.stem.2014.07.003 (2014).
Mansour, A. A. et al. The H3K27 demethylase utx regulates somatic and germ cell epigenetic reprogramming. Nature. 488, 409–413. https://doi.org/10.1038/nature11272 (2012).
Shu, J. et al. GATA family members as inducers for cellular reprogramming to pluripotency. Cell. Res. 25, 169–180. https://doi.org/10.1038/cr.2015.6 (2015).
Monteon, A., Hughes, L., Camberos, V. & Kearns-Jonker, M. Identification of SALL4 expressing Islet-1 + cardiovascular progenitor cell clones. Int. J. Mol. Sci. 24. https://doi.org/10.3390/ijms24021780 (2023).
Morita, Y. et al. Sall1 transiently marks undifferentiated heart precursors and regulates their fate. J. Mol. Cell. Cardiol. 92, 158–162. https://doi.org/10.1016/j.yjmcc.2016.02.008 (2016).
Katano, W. et al. Sall1 and Sall4 cooperatively interact with Myocd and SRF to promote cardiomyocyte proliferation by regulating CDK and cyclin genes. Development. 150. https://doi.org/10.1242/dev.201913 (2023).
Zhao, H. et al. Sall4 and Myocd empower direct cardiac reprogramming from adult cardiac fibroblasts after Injury. Front. Cell. Dev. Biol. 9, 608367. https://doi.org/10.3389/fcell.2021.608367 (2021).
Wu, J. et al. Improved factor combination for in vivo reprogramming of cardiac myofibroblast to cardiomyocyte-like cell with dual recombinase tracing. Circulation. 148, 1728–1731. https://doi.org/10.1161/CIRCULATIONAHA.122.062810 (2023).
Koshiba-Takeuchi, K. et al. Cooperative and antagonistic interactions between Sall4 and Tbx5 pattern the mouse limb and heart. Nat. Genet. 38, 175–183. https://doi.org/10.1038/ng1707 (2006).
Harvey, S. A. & Logan, M. P. sall4 acts downstream of tbx5 and is required for pectoral fin outgrowth. Development. 133, 1165–1173. https://doi.org/10.1242/dev.02259 (2006).
Cecchetto, A. et al. From molecular mechanisms of cardiac development to genetic substrate of congenital heart diseases. Future Cardiol. 6, 373–393. https://doi.org/10.2217/fca.10.10 (2010).
Bruneau, B. G. Signaling and transcriptional networks in heart development and regeneration. Cold Spring Harb. Perspect. Biol. 5, a008292. https://doi.org/10.1101/cshperspect.a008292 (2013).
McCulley, D. J. & Black, B. L. Transcription factor pathways and congenital heart disease. Curr. Top. Dev. Biol. 100, 253–277. https://doi.org/10.1016/B978-0-12-387786-4.00008-7 (2012).
Yang, L. et al. Sall4 blocks cardiac trans-differentiation but stimulates cardiac stem-like cell (iPSC) generation and improve post MI function in vivo. Circ. Res. 123, A34. https://doi.org/10.1161/res.123.suppl_1.349 (2018).
Mathison, M. et al. Cardiac reprogramming factor Gata4 reduces postinfarct cardiac fibrosis through direct repression of the profibrotic mediator snail. J. Thorac. Cardiovasc. Surg. 154, 1601–1610 e1603. https://doi.org/10.1016/j.jtcvs.2017.06.035 (2017).
Singh, V. P. et al. Enhanced generation of Induced cardiomyocytes using a small-molecule cocktail to overcome barriers to cardiac cellular reprogramming. J. Am. Heart Assoc. 9, e015686. https://doi.org/10.1161/JAHA.119.015686 (2020).
Liu, L. et al. Knockdown of SALL4 protein enhances all-trans retinoic acid-induced cellular differentiation in acute myeloid leukemia cells. J. Biol. Chem. 290, 10599–10609. https://doi.org/10.1074/jbc.M114.634790 (2015).
Yang, J. et al. Enhanced self-renewal of hematopoietic stem/progenitor cells mediated by the stem cell gene Sall4. J. Hematol. Oncol. 4. https://doi.org/10.1186/1756-8722-4-38 (2011).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11 (R106). https://doi.org/10.1186/gb-2010-11-10-r106 (2010).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Singh, V. P. et al. Hippo pathway effector Tead1 induces cardiac fibroblast to cardiomyocyte reprogramming. J. Am. Heart Assoc. 10, e022659. https://doi.org/10.1161/JAHA.121.022659 (2021).
Yang, J., Corsello, T. R. & Ma, Y. Stem cell gene SALL4 suppresses transcription through recruitment of DNA methyltransferases. J. Biol. Chem. 287, 1996–2005. https://doi.org/10.1074/jbc.M111.308734 (2012).
Yang, J. SALL4 as a transcriptional and epigenetic regulator in normal and leukemic hematopoiesis. Biomark. Res. 6, 1. https://doi.org/10.1186/s40364-017-0115-6 (2018).
Wang, L. et al. GATA-binding protein 4 (GATA-4) and T-cell acute leukemia 1 (TAL1) regulate myogenic differentiation and erythropoietin response via cross-talk with Sirtuin1 (Sirt1). J. Biol. Chem. 287, 30157–30169. https://doi.org/10.1074/jbc.M112.376640 (2012).
Anto Michel, N., Ljubojevic-Holzer, S., Bugger, H. & Zirlik, A. Cellular heterogeneity of the heart. Front. Cardiovasc. Med. 9, 868466. https://doi.org/10.3389/fcvm.2022.868466 (2022).
Yucel, N. et al. Cardiac endothelial cells maintain open chromatin and expression of cardiomyocyte myofibrillar genes. Elife. 9. https://doi.org/10.7554/eLife.55730 (2020).
Colliva, A., Braga, L., Giacca, M. & Zacchigna, S. Endothelial cell-cardiomyocyte crosstalk in heart development and disease. J. Physiol. 598, 2923–2939. https://doi.org/10.1113/JP276758 (2020).
Chan, E. M. et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat. Biotechnol. 27, 1033–1037. https://doi.org/10.1038/nbt.1580 (2009).
Furtado, M. B. et al. Cardiogenic genes expressed in cardiac fibroblasts contribute to heart development and repair. Circ. Res. 114, 1422–1434. https://doi.org/10.1161/CIRCRESAHA.114.302530 (2014).
Tallquist, M. D. Cardiac fibroblast diversity. Annu. Rev. Physiol. 82, 63–78. https://doi.org/10.1146/annurev-physiol-021119-034527 (2020).
Lendahl, U., Muhl, L. & Betsholtz, C. Identification, discrimination and heterogeneity of fibroblasts. Nat. Commun. 13, 3409. https://doi.org/10.1038/s41467-022-30633-9 (2022).
Barreto, S., Hamel, L., Schiatti, T., Yang, Y. & George, V. Cardiac progenitor cells from stem cells: Learning from genetics and biomaterials. Cells. 8. https://doi.org/10.3390/cells8121536 (2019).
Yu, J. et al. Topological arrangement of cardiac fibroblasts regulates Cellular plasticity. Circ. Res. 123, 73–85. https://doi.org/10.1161/CIRCRESAHA.118.312589 (2018).
Moretti, A. et al. Multipotent embryonic isl1 + progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 127, 1151–1165. https://doi.org/10.1016/j.cell.2006.10.029 (2006).
Mehanna, R. A. et al. Cardiac stem cells: Current knowledge and future prospects. World J. Stem Cells. 14, 1–40. https://doi.org/10.4252/wjsc.v14.i1.1 (2022).
Chong, J. J. et al. Adult cardiac-resident MSC-like stem cells with a proepicardial origin. Cell. Stem Cell. 9, 527–540. https://doi.org/10.1016/j.stem.2011.10.002 (2011).
Nemajerova, A., Kim, S. Y., Petrenko, O. & Moll, U. M. Two-factor reprogramming of somatic cells to pluripotent stem cells reveals partial functional redundancy of Sox2 and Klf4. Cell. Death Differ. 19, 1268–1276. https://doi.org/10.1038/cdd.2012.45 (2012).
Durocher, D., Charron, F., Warren, R., Schwartz, R. J. & Nemer, M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 16, 5687–5696. https://doi.org/10.1093/emboj/16.18.5687 (1997).
Maitra, M. et al. Interaction of Gata4 and Gata6 with Tbx5 is critical for normal cardiac development. Dev. Biol. 326, 368–377. https://doi.org/10.1016/j.ydbio.2008.11.004 (2009).
Sakaki-Yumoto, M. et al. The murine homolog of SALL4, a causative gene in Okihiro syndrome, is essential for embryonic stem cell proliferation, and cooperates with Sall1 in anorectal, heart, brain and kidney development. Development. 133, 3005–3013. https://doi.org/10.1242/dev.02457 (2006).
Kim, K. et al. Epigenetic memory in induced pluripotent stem cells. Nature. 467, 285–290. https://doi.org/10.1038/nature09342 (2010).
Stefanovic, S. et al. Interplay of Oct4 with Sox2 and Sox17: A molecular switch from stem cell pluripotency to specifying a cardiac fate. J. Cell. Biol. 186, 665–673. https://doi.org/10.1083/jcb.200901040 (2009).
Abboud, N. et al. A cohesin-OCT4 complex mediates Sox enhancers to prime an early embryonic lineage. Nat. Commun. 6, 6749. https://doi.org/10.1038/ncomms7749 (2015).
Schnerch, A., Cerdan, C., & Bhatia M. Distinguishing between mouse and human pluripotent stem cell regulation: The best laid plans of mice and men. Stem Cells. 28, 419–430. https://doi.org/10.1002/stem.298 (2010).
Garry, G. A., Bassel-Duby, R. & Olson N. E. Direct reprogramming as a route to cardiac repair. Semin. Cell Dev. Biol. 122, 3–13. https://doi.org/10.1016/j.semcdb.2021.05.019 (2022).
Tao, Y. et al. Robust small molecule-aided cardiac reprogramming systems selective to cardiac fibroblasts. iScience, 26, 108466. https://doi.org/10.1016/j.isci.2023.108466 (2023).
Furtado, M. B., et al. Cardiogenic genes expressed in cardiac fibroblasts contribute to heart development and repair. Circ. Res., 114, 1422–1434. https://doi.org/10.1161/CIRCRESAHA.114.302530 (2014).
Gong, G. Q. et al. A small-molecule PI3Kalpha activator for cardioprotection and neuroregeneration. Nature. 618, 159–168. https://doi.org/10.1038/s41586-023-05972-2 (2023).
Brandt, E. B., Bashar, S. J. & Mahmoud, A. I. Stimulating ideas for heart regeneration: The future of nerve-directed heart therapy. Bioelectron. Med. 5, 8. https://doi.org/10.1186/s42234-019-0024-0 (2019).
Mahmoud, A. I. et al. Nerves regulate cardiomyocyte proliferation and heart regeneration. Dev. Cell. 34, 387–399. https://doi.org/10.1016/j.devcel.2015.06.017 (2015).
Acknowledgements
This work was supported by Michael E. DeBakey Department of Surgery Faculty Award (Yang) and in part by NIH grant R01HL121294‐01A1 (Rosengart). Imaging and quantification were supported by the Integrated Microscopy Core at Baylor College of Medicine and the Center for Advanced Microscopy and Image Informatics (CAMII) with funding from NIH (DK56338, CA125123, ES030285, S10OD030414), and CPRIT (RP150578, RP170719). We thank the Cytometry and Cell Sorting Core with funding from the National Institutes of Health (P30 AI036211, P30 CA125123, and S10 RR024574), the Pathology and Histology Core (HTAP) with grant from NCI (NCICA125123) and the Dan L. Duncan Cancer Center.
Author information
Authors and Affiliations
Contributions
J.Y. conceived and designed the study. H.G., S.P., J.Y. performed experiments, H.G., S.P., B.D., J.Y. performed data analysis. B.D. and A.P. assisted with experimental procedures. J.Y. wrote the manuscript, with input from all authors. M.M., T.M., T.K.R. assisted with study design and manuscript editing. J.Y. and T.K.R. provided funding. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gao, H., Pathan, S., Dixon, B.R.E.A. et al. Sall4 and Gata4 induce cardiac fibroblast transition towards a partially multipotent state with cardiogenic potential. Sci Rep 14, 24182 (2024). https://doi.org/10.1038/s41598-024-73975-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-73975-8