Abstract
Convolutional neural networks (CNNs) for extracting structural information from structural magnetic resonance imaging (sMRI), combined with functional magnetic resonance imaging (fMRI) and neuropsychological features, has emerged as a pivotal tool for early diagnosis of Alzheimer’s disease (AD). However, the fixed-size convolutional kernels in CNNs have limitations in capturing global features, reducing the effectiveness of AD diagnosis. We introduced a group self-calibrated coordinate attention network (GSCANet) designed for the precise diagnosis of AD using multimodal data, including encompassing Haralick texture features, functional connectivity, and neuropsychological scores. GSCANet utilizes a parallel group self-calibrated module to enhance original spatial features, expanding the field of view and embedding spatial data into channel information through a coordinate attention module, which ensures long-term contextual interaction. In a four-classification comparison (AD vs. early MCI (EMCI) vs. late MCI (LMCI) vs. normal control (NC)), GSCANet demonstrated an accuracy of 78.70%. For the three-classification comparison (AD vs. MCI vs. NC), it achieved an accuracy of 83.33%. Moreover, our method exhibited impressive accuracies in the AD vs. NC (92.81%) and EMCI vs. LMCI (84.67%) classifications. GSCANet improves classification performance at different stages of AD by employing group self-calibrated to expand features receptive field and integrating coordinated attention to facilitate significant interactions among channels and spaces. Providing insights into AD mechanisms and showcasing scalability for various disease predictions.
Similar content being viewed by others
Introduction
With the increase in the aging population worldwide, Alzheimer’s disease (AD) has become a rapidly growing global public health concern1. The progression of AD is irreversible, but early diagnosis and intervention can effectively slow its onset and progression, thereby improving the quality of life for elderly patients2. The development of AD involves complex changes in brain structure and function, which are influenced by various factors, including the complex interactions among various mechanisms and the impact of the disease3. In terms of structure, individuals with AD commonly exhibit cortical thinning, reduced gray matter volume and hippocampal atrophy1. In terms of the brain’s functions, AD patients often show weakened connectivity in the default mode network4,5. However, current AD diagnostic studies are typically limited to a single modality, focusing either on brain structure or function; this approach only reflects part of the changes and fails to completely capture the unique pathological features of early AD patients6,7. Combining clinical data from brain imaging and neuropsychological assessments offers a comprehensive view of the brain, enhancing diagnostic accuracy and informing treatment strategies. This integrative approach can detect subtle brain changes that are not visible with a single imaging modality. By incorporating multimodal clinical information, our study aims to enhance the early detection and classification of AD, ultimately contributing to better patient outcomes.
Magnetic resonance imaging (MRI), functional magnetic resonance imaging (fMRI), and structural magnetic resonance imaging (sMRI) techniques, in conjunction with neuropsychological features, have emerged as critical tools for the early diagnosis of AD8,9. fMRI, which utilizes the blood-oxygen-level-dependent (BOLD) contrast10, serves as a precise tool for measuring neural activity and functional changes in the brain. Functional connectivity (FC) can be assessed by analyzing the correlation among BOLD signals from different brain regions, which sensitively detects functional changes in the brain associated with AD pathology11,12. sMRI enables a comprehensive examination of brain structures in individuals with AD13. Texture features, such as gray-level co-occurrence matrices (GLCMs), obtained from sMRI images, provide valuable information on gray-level directions and variations, which are significant for the diagnosis and prognosis of AD14,15,16. Additionally, neuropsychological assessment scales can evaluate individuals’ cognitive, emotional, and behavioral functions. The mini-mental state examination (MMSE) and the clinical dementia rating (CDR) are commonly-used convenient tools for the early clinical diagnosis of AD. These multimodal data can be used to obtain biological information in different dimensions of brain structure, brain function and cognitive level, resulting in new perspectives for additional disease research and intelligent diagnostic assistance as compared to single-modal brain structure or function information.
Recently, convolutional neural networks (CNNs) have progressively emerged as the leading and effective tools in medical image feature extraction and classification tasks17,18,19. However, current CNN architectures face considerable challenges that limit their application and performance in AD diagnosis. Specifically, the fixed-size convolutional kernels in CNNs constrain their ability to learn similar patterns of existing objects and lack sufficient receptive fields to capture global features20,21. This limitation reduces the effectiveness of the field of view for feature extraction, hindering the exchange of information on distant features outside the field of view, which may diminish the accuracy of feature recognition and mapping.
To address these challenges, this study introduces an innovative group self-calibrated coordinate attention network (GSCANet). GSCANet utilizes a group self-calibrated convolution module22 and a coordinate attention module23 to facilitate the identification of multiscale features and improve the exchange of information between feature maps. The group self-calibrated convolution module22 significantly improves the exchange of information between feature mappings, enabling more comprehensive feature extraction. This method employs differently sized convolutional kernels within the same network layer to identify multiscale features, thereby expanding the receptive field and enhancing the recognition accuracy of feature mapping20. Recent research has shown that expanding the receptive field has the potential to improve classification performance in AD diagnosis24,25. By mplementation of group self-calibrated convolution leads to more comprehensive and diverse set of AD brain image features, resulting in more accurate classification.
Furthermore, we employed the coordinate attention module assigns different weights to input features, effectively eliminating redundant information and accelerating CNN convergence, thereby improving model performance26. There are two primary types of attention mechanisms: channel-wise and spatial-wise. Channel-wise attention assigns different weights to feature map channels, enabling the network to prioritize specific attributes, such as brightness and texture. Spatial-wise attention allocates weights across spatial locations within the feature map, highlighting important areas, such as the hippocampus27, that contain critical information.However, the previous model ignored interactions between the channel and spatial attention28. To address this issue, we employed coordinate attention23 to compress spatial information into channel descriptors. Specifically, in this study, channel relations and long-term dependencies are encoded as precise positional information, sensitive to channel and spatial variations when capturing cross-channel information. Feature processing is coordinated and enhanced by selectively focusing on spatial and channel information to help suppress irrelevant and detailed information. We can effectively capture a more diverse and critical set of AD brain image features by integrating coordinate attention into our CNN model, achieving improved performance in classification tasks.
In conclusion, we proposed a CNN-based GSCANet model that incorporates group self-calibrated convolution and coordinate attention modules. The key contributions of our study are as follows:
-
(1)
We used multimodal features including GLCM, FC and neuropsychological features (CDR/MMSE) for classification as changes in the brain structure and function in AD are not independent. This approach provides a more comprehensive view of the complex relationships between brain structural morphology, functional dynamics, and cognitive impairments, improving classification accuracy compared to traditional single-modality methods that focus solely on structural MRI images. This is useful in capturing the complexity of pathological relationships and thus is better suited for classification.
-
(2)
We incorporated group self-calibrated convolution into our model to expand the receptive field through varied-sized convolutional kernels within the same layer. This technique enhances feature extraction by capturing a broader and more detailed representation of AD brain images.
-
(3)
We employed coordinate attention module by selectively focusing on critical spatial and channel information to effectively suppress less relevant details. This targeted focus enables the capture of a diverse and critical set of features, leading to improved performance in classifying AD brain images.
The overall workflow is shown in Fig. 1. We extracted features from multimodal data which were input into the GSCANet model. Subsequently, the model was applied to the multi-classification study of AD, MCI and NC using ten-fold cross-validation to evaluate its performance.
Materials and methods
Subjects
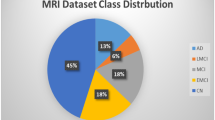
Two sources of data were utilized in this study: the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (http://adni.loni.usc.edu/) and the database from the First Hospital of Jilin University. The ADNI database provided baseline MRI scans of 637 participants (143 AD, 264 MCI, and 143 NC). The MCI group comprised 92 EMCI and 172 LMCI participants based on the severity of the disease symptoms.
The First Hospital of Jilin University contributed data from 195 participants (60 AD, 62 MCI, and 73 NC). This study was approved by the ethics committee of the First Hospital of Jilin University. All procedures were conducted in accordance with the Helsinki declaration. The dataset included participants with AD, MCI, and NC subjects. All participants signed a written informed consent before the experiment and participated in cognitive psychological evaluations, including the MMSE and the CDR. Combined with the scale information, the diagnosis and enrolment of the participants were completed by experienced clinical neurologists.
MRI acquisition
All ADNI participants were scanned using a Philips 3T MRI scanner with the following parameters: TR = 3000 ms, TE = 30 ms, flip angle = 90º, number of layers = 48, layer thickness = 3.3 mm, FOV = 256 × 256 mm, and 140 time points. For T1 weighted imaging (T1w), the parameters acquired were TR = 8.9 ms, TE = 3.9 ms, flip angle = 8°, slice thickness = 1 mm, and FOV = 256 × 256 mm.
Participants recruited from the First Hospital of Jilin University were scanned using a 3T field strength Siemens MRI equipped with a standard head coil to acquire resting-state MRI images of the brain. Prior to acquisition of the experimental data, the participants were instructed to keep their eyes closed and remain awake during the acquisition process. MRI images of all the participants with both T1w as well as rs-fMRI were obtained. The fMRI parameters acquired were TR = 2500 ms, TE = 27 ms, flip angle = 90º, and FOV = 230 × 230 mm. T1w parameters acquired were TR = 8.5 ms, TE = 3.3 ms, flip angle = 12º, and FOV = 256 × 256 mm.
Data preprocessing
Visual quality control is conducted on T1w MRI images. Images of low quality, marked by incomplete brain coverage, low signal-to-noise ratio, or apparent visible artifacts, are excluded from the analysis. Preprocessing of the T1w MRI images is performed using the Statistical Parametric Mapping (SPM) (version 12)29 software. The preprocessing involves procedures such as skull stripping, denoising, aligning the T1w MRI with the Montreal Neurological Institute (MNI) space, and resampling to a resolution of 1 × 1 × 1 mm, all of which facilitate subsequent analysis.
The preprocessing protocol for fMRI data is implemented using SPM 12.029. The protocol includes removal of the initial ten time points, slice timing correction, motion correction, registration and spatially normalized to the MNI template. Linear drift is corrected, followed by the application of a bandpass filter in the range 0.01–0.1 Hz. Covariates, including six motion parameters as well as the mean signal from the white matter and cerebrospinal fluid, are removed to mitigate their potential influence. Finally, a visual quality control check is performed to discard any incorrect alignments from the dataset.
Haralick texture features extraction
GLCM describes images by quantifying the relationship between pairs of gray values in grayscale images. Haralick30 defined 14 statistical parameters to characterize texture features. In this study, we focused on visualizing a specific parameter (Fig. 2). Ten feature values, including those from the original image, were employed to describe the texture information of the brain image. These feature values were selected based on their effectiveness in differentiating between normal and abnormal brain tissues and were used in subsequent analyses.
Consider an image I with pixel coordinates (x, y) and a pixel gray value li, and its neighboring pixel with coordinates (x + Δx, y + Δy) and a pixel gray value lj. GLCM counts the number of occurrences of pixel pairs with a grayscale value (\(\:{l}_{i}\), \(\:{l}_{j}\)) along a direction θ, separated by d steps, and denoted as M(i, j/d, θ). Typically, the values of θ are 0°, 45°, 90°, or 135°. GLCM reflects the grayscale variation of the image. For an image I with L gray levels, each element in the matrix of dimension L × L can be represented by C(\(\:{\text{l}}_{\text{i}}\), \(\:{\text{l}}_{\text{j}}\)). The corresponding formula is shown in (1).
After calculating the GLCM, it was normalized to obtain P(i, j), given by (2)
.
Using this approach, we extracted features from the image by computing frequencies of specific gray voxel value pairs. Nine features were extracted from T1w MRI using the GLCM feature extraction method.
The mean value, Mean, is a statistical parameter, used to describe the degree of regularity of texture, defined as.
where \(\:P(i,j)\) is the normalized GLCM, the standard deviation, Std, which is a measure of the deviation of the true image from the Mean is defined as
Contrast (Con) is another statistical parameter that quantifies the local variation present in the image and is given by.
Dissimilarity (Dis) is a measure of the total amount of local contrast variation present in the image, which is defined as.
Homogeneity (Hom) is used to quantify the local smoothness in an image and is defined as.
The angular second moment (ASM) is a metric used to describe the uniformity of the image grayscale distribution and texture thickness, which can be calculated using.
Energy (SE) is used as a measure of whether a textured pattern is relatively homogeneous and regularly varying; it is defined as.
The maximum value (Max) is used to describe the maximum value of the image grayscale pair calculated using GLCM, which is given by
.
Entropy (ENT), used to quantify the randomness and complexity of the image; it is defined as
.
Functional connectivity feature extraction
Previous studies have demonstrated FC changes in patients with AD. These connectivity alterations can be explored by measuring both global and local network topology. We calculated the Pearson correlation coefficients between the time series of resting-state fMRI (rs-fMRI) of each brain region and those of other regions using the Gretna toolkit and the AAL 90 template. This resulted in a 90 × 90 resting-state FC (rs-FC) matrix (Fig. 3). In the FC network, each brain region is represented as a node, with the strength of connections described as edges. After constructing the FC network, graph theory is employed to calculate both the local and global properties of the brain network. The properties include the average clustering coefficient, average shortest path length, clustering coefficient, small worldness, local efficiency, assortativity, and synchronization31.These parameters will be the functional features in the GSCAnet model. The definition and calculation formula of functional connection features are as follows:
The clustering coefficient (aCp) is used to characterize the degree of clustering among all nodes in the network, indicating local connectivity of the image. The average clustering coefficient is calculated by averaging over the clustering coefficients of all nodes in the network (Fig. 3)
where n is the number of connection edges that exist between node i and its neighboring nodes.
The shortest path length (aLp) is the average of the shortest path lengths of all pairs of nodes in the network. The aLp is a measure of the optimal path for information transfer from one node to another node in the network; it reflects the global information transfer capability of the network. The calculation formula is given by
where Li, j is the length of all paths between nodes i and j, and min{Li, j} is the shortest path length between nodes i and j.
The local efficiency (\(\:{E}_{i-loc}\)) of a node represents the tolerance of the network for this node (i.e., the effect of removing this node on the transmission efficiency of the sub-network). It is given by
where \(\:{\text{L}}_{\text{j},\text{k}}\) is the shortest path length between each pair of nodes directly adjacent to node i. \(\:{\text{N}}_{{\text{G}}_{\text{i}}}\) is the total number of node pairs present in the sub-network directly adjacent to node i.
Network local efficiency (aEloc) is the average of the local efficiency of all nodes, which is given by.
Assortativity is used to analyze the likelihood that similar nodes tend to connect to each other.
Synchronization refers to the degree to which changes in the connectivity of nodes in a network are similar when the network undergoes disturbances or perturbations.
Neuropsychological scores feature extraction
Cognitive data were extracted from the “ADNIMERGE” file, which incorporates merged data sets from ADNI 1/GO/2. The file contains clinical data and numeric summaries to assess cognitive function over time. The neuropsychological variables used in the analysis of cognitive changes were the clinical medical dementia rating sum of boxes (CDR) and mini-mental state examination (MMSE). All participants from the First Hospital of Jilin University underwent psychological cognitive assessments, including the MMSE and CDR. We performed a one-way analysis of variance (ANOVA) for each of the two scales across the groups in both the databases. The results are summarized in Table 1. Significant differences were observed between the groups on both scales.
GSCANet model
CNNs feature local area connectivity and weight-sharing properties. The receptive field represents the size of the region where the pixel points on the output feature map of each layer of the network correspond to the input image. Selecting a receptive field that is too small leads to information loss, whereas a very large receptive field results in computational overload and information redundancy. To address this, we propose a new method that uses group convolution with group self-calibrated and coordinated attention, enabling efficient expansion of the receptive field without additional computational costs.
The model framework is shown in Fig. 4A. It follows a series structure, where the outputs from each layer are the inputs to the next layer. The overall deep-learning model comprises one convolutional layer, one batch normalization layer, one maximum pooling layer, two activation layers, 16 residual blocks, one fully connected layer, one flatten layer, and a softmax layer. In this study, the GLCM extracted in the preliminary stage is fed into the model. Initially, the convolutional layer extracts different features from the input. The normalization layer addresses the issues of gradient explosion and vanishing problems. To minimize redundant features, the improved linear unit is used as the nonlinear activation function, while the maximum pooling layer is utilized for multiscale feature learning32. Additionally, the model employs residual blocks in the classical deep residual network (ResNet), which enhances its robustness by deepening the layers and eliminating the less effective ones. The Resblock module is illustrated in Fig. 4B. Each residual block has three convolutional layers, including group self-calibrated coordinated attention convolutions (Fig. 4C) and coordinated attention (Fig. 4D). The flatten layer incorporates multidimensional data into a single dimension, combining it with numerical information (FC features, MMSE scores, and CDR scores) to combine image and numerical data. Finally, classification of subjects is achieved using these multimodal features through the fully connected layer followed by the softmax layer.
Schematic diagram of the GSCANet model architecture. (A) Overall modeling framework based on diagnostic and predictive AD. (B) Residual blocks. (C) Group self-calibrated coordinate attention module. (D) Coordinate attention module. Abbreviations: Cov, convolutional layer; BatchNorm, batch normalization layer; ReLU, linear unit; Resblock, Residual block; FC, fully connected layer.
Group self-calibrated operation
Figure 4C illustrates the group self-calibrated convolutional framework. This framework splits the input data into multiple groups for parallel processing, with each group possessing the same structure, with the convolution filter of each group divided into multiple non-uniform parts. Each component is trained with different feature extraction methods to calibrate features across multiple scales. Instead of uniformly processing all features in the original space, self-calibrated convolution22 first splits the input data into two parts. One part performs direct feature mapping, whereas the other performs feature extraction from the global brain after down-sampling to enhance the signal-to-noise ratio, increase the receptive field, and reduce dimensionality. The group self-calibrated operation uses the lower dimensional patterns of the latter filtered transform to calibrate the convolutional transform of the former filter. In this study, the group self-calibrated convolution enables effective information communication with the filter following multiple heterogeneous convolution operations. This approach enables precise localization of target features and enhances feature discrimination, particularly when the convolutional receptive field expands. Finally, the features processed by the group self-calibration are input to the attention mechanism.
The group self-calibrated convolution comprises four heterogeneous filters—K1, K2, K3, K4—that are used to process the input data X. To achieve this, X is split into two parts, X1 and X2. In the first part, the original spatial information is preserved, and the features are directly mapped using K1, which helps prevent feature loss. Specifically, this is computed as Y1 = F1(X1), where F1(X1) = X1·K1. In the second part, the remaining three filters are applied to down sample X2 and generate Y2. This operation implements self-calibration from the entire dataset and performs calibrated operations within Y2. Finally, Y1 and Y2 are concatenated and subjected to attention coordination to enable multiscale information interactions in remote contexts. This leads to the extraction of relevant features from the data.
Self-calibrated convolution enhances the connection between individual feature maps by averaging pooling operations, where the latter capture the contextual information of features to reduce overfitting. Therefore, the average pooling operation in 3D space was performed for a given input X2, according to M2 = AvgPool3d(X2). The feature mapping of the averaged pooled features M2 was based on the K2 filter, \(\:{X}_{2}^{{\prime\:}}={F}_{2}\left({M}_{2}\right)\). The original X2 was activated using the sigmoidal activation function along with \(\:{X}_{2}^{{\prime\:}}\) after the following feature mapping, \(\:{X}_{2}^{{\prime\:}{\prime\:}}=S({X}_{2}+{X}_{2}^{{\prime\:}})\). \(\:{X}_{2}^{{\prime\:}}\) was used as the calibration weight for the corresponding element-wise multiplication of congruent elements followed by additional calibration work based on \(\:{Y}_{2}^{{\prime\:}}={F}_{3}\left({X}_{2}\right)\cdot\:{X}_{2}^{{\prime\:}{\prime\:}}\). The final feature transformation of the calibrated data can be expressed as \(\:{Y}_{2}={F}_{4}\left({Y}_{2}^{{\prime\:}}\right)\). Additionally, the output features after calibration were further combined with the features of the original spatial context. The cascaded features Y’ then underwent coordinated attention operations, resulting in the final output features Y.
Coordinate attention module
The attention mechanism enhances or selects important information concerning the target using attention distribution coefficients or weight parameters. To leverage both spatial location and channel information, we propose a coordinate attention mechanism based on a ResNet architecture. This mechanism enables the efficient extraction of features. Several alternatives to traditional pooling methods have emerged to prevent the loss of spatial and channel information caused by simple and brute-force pooling (Fig. 4D). In this study, we propose the use of average pooling to retain long-range interactions between location and channel information. We also use the attention mechanism to embed location information into channel attention. The image was first transformed by downscaling; the 3D image was transformed into a one-dimensional (1D) feature encoding using averaging pooling across three directions. The transformation can be expressed as
where H, W, and L are the height, width, and length of the 3D image Y, respectively; i, j, and l are the arbitrary values of height, width, and length, respectively; yc is the input feature of Y with dimensionality reduction operation performed on the cth channel of Y, and zc is the output of the cth channel. By performing a 1D pooling operation on the image from 3D, the features are aggregated into three separate direction-aware feature maps along different directions of the input features. Specifically, the features are transformed by (H, 1, 1), (1, W, 1), and (1, 1, L) with three different pooling kernels in different dimensions as follows:
where \(\:{H}^{{\prime\:}}\), \(\:{W}^{{\prime\:}}\) and \(\:{L}^{{\prime\:}}\) represent the height, width, and length features after pooling, respectively. The features pooled in the \(\:{H}^{{\prime\:}}\), \(\:{W}^{{\prime\:}}\) and \(\:{L}^{{\prime\:}}\) dimensions generated by the above operation are concatenated according to \(\:M=cat({H}^{{\prime\:}},{W}^{{\prime\:}},{L}^{{\prime\:}})\), and the combined data are convolved as a whole: \(\:{M}^{{\prime\:}}=Conv3d\left(M\right)\). Additionally, the batch normalization is used to mitigate distribution bias in the data. In this study, after nonlinear activation, the features are separated and convolved again separately depending on the division of the three dimensions of the image. Finally, the overall features are reassigned after applying the sigmoidal activation function to preserve important feature information.
The coordinated attention mechanism employs three 1D global pooling operations to encode global information. This approach enables the model to capture long-term dependencies among spatial locations, which is crucial for tasks related to disease diagnosis. Specifically, GSCANet was used to integrate the input features and perform feature extraction with an expanded effective receptive field, thereby ensuring the accurate extraction of features. This approach enabled coordinated attention to capture remote dependencies between spatial locations, thus rendering it highly effective for analyzing medical images.
Performance evaluation
This study compares the performance of various neural network-based classification models in distinguishing between different datasets to assess the generalization capabilities of deep learning models. The AD vs. MCI vs. NC triple classification and AD vs. NC binary classification models were trained using the ADNI dataset and validated using the in-house dataset. The remaining experiments (including AD vs. LMCI vs. EMCI vs. NC quadruple classification, EMCI vs. LMCI, EMCI vs. NC dichotomization) were trained using the ADNI dataset and validated using the ADNI test set, which is distinct from the subjects in the training set. To balance the sample sizes across different categories within the ADNI dataset, we utilized multimodal data from the same individuals collected over multiple years. To mitigate bias arising from data similarity, we ensured that data from the same individual were not included in both the training and testing sets simultaneously.
All training and testing in this study were carried out on the same hardware and software platform. The hardware environment was Windows Server 2016 (64-bit) operating system, Intel® Xeon® Gold 6238R CPU, and V100S-PCIE-32GB GPU. The software programming environment for GSCANet was Python 3.8.18, Pytorch 1.10.1, and CUDA 10.2. The learning rate (lr) was set to 3e-4, and the number of training epochs was set to 100. Training was performed using cross-entropy loss with incremental gradient descent as the training method.
Furthermore, we conducted ten-fold cross-validation for multiple classifications to assess the variability in the classification performance. Specifically, we randomly divided all participants into ten folds, using the data of participants in nine folds to build and train the model. The scans of participants in the remaining fold were used for model testing. We performed iterative testing using subjects in the remaining fold to prevent bias in the random assignment of data during cross-validation. Specifically, 15 replications per experiment were conducted using different combinations of training and test sets.
Finally, the diagnostic performances were evaluated using quantitative metrics, such as accuracy (ACC), sensitivity (SEN), and specificity (SPE), and the area under receiver operating characteristic curve (AUC), which are expressed as.
where TP, TN, FN, and FP represent true positive, true negative, false negative, and false positive, respectively. The classification results were averaged for ten-fold validation.
Results
Analysis of GSCANet’s early diagnostic results on ADNI data and its comparison with other models
First, we trained our model in the ADNI dataset and tested it using a completely non-overlapping ADNI dataset, conducting a tenfold cross-validation. Note that the in-house dataset was not used in this cross-validation process. As shown in Table 2, the final result for differentiating among the four categories of subjects (AD vs. LMCI vs. EMCI vs. NC) was 78.70% (95% CI: 77.5–81.5). For the binary classification of EMCI and LMCI, the final score was 84.67% (95% CI: 84.16–85.18), and the accuracy of the classification of EMCI and NC was 82% (95% CI: 81.79–82.21).
The GSCANet framework proposed in this study demonstrated improved performance compared to previous studies12,33,34,35,36,37,38,39. Table 2 shows that the accuracy of the four categories was significantly improved (by approximately 14%) compared to the adaptive sparse learning model34. In the EMCI vs. LMCI binary study, the SPE improved by 15% compared to that of the SWRLS dFC model37. The accuracy of EMCI and NC classification in our study improved by 2.75% compared to that of the GDCA model40. These results highlight the superior performance of our model in complex classification tasks.
Generalizability investigation of GSCANet model with independent database and comparison to other models
To further investigate generalizability and reproducibility of GSCANet model, training was performed on the ADNI dataset while testing was conducted on the in-house dataset. Subsequently, the classification performance of this study was explored in three classifications (AD, MCI, and NC) and two classifications (AD and NC). The results are presented in Table 3. The final result for the AD vs. MCI vs. NC triple classification was 83.33% (95% CI 77.5 to 81.5), which improved compared to those in 3D-CNN41, THAN42, STNet33 and LSTM-Robust43. The final result for the AD vs. NC dichotomy was 92.81% (95% CI 77.5 to 81.5), which was better than that of BB + pA-blocks (A) + Bili44, 3D-ResAttNet3445, THAN42, PT DCN26, and Dense CNN46. We observed that the method proposed in the present study had higher classification accuracy than that of other studies. Table 3 shows that the final results of the three classifications in this study improved by 7.37% compared to the LSTM-Robust model43. Our results in the AD vs. NC experiment improved by 8% in SEN compared to the DenseCNN2 model46. Based on these experiments, our proposed model has good generalizability and reproducibility in diagnosing AD and shows better classification performance than some previous models.
A comparative study of using the same features with other existing deep learning methods for diagnosing AD
As different datasets and features are used, it would be unfair to make direct comparisons between the different methods. We utilized the same dataset and experimental environment and only changed the parts of the experiment that required comparisons to validate fully the effectiveness of the proposed method in this study47. Our models were compared with classical models, such as VGG, ResNet, and ResNeSt networks for classification. All these models use the same features, including GLCM, FC, and neuropsychological scores. We compared the results of four-class and three-class classifications using these same features on different models. Specifically, the four-class classification (AD vs. LMCI vs. EMCI vs. NC) was trained on the ADNI dataset and tested using a completely non-overlapping ADNI dataset, whereas the three-class classification (AD vs. MCI vs. NC) was trained on the ADNI dataset and tested on the in-house dataset. As shown in Table 4, our model outperformed the VGG48 by 8.5% in the four-classification study. Furthermore, when compared with traditional models, such as the deep-residual network 50 (ResNet50)49 and split-attention networks (ResNeSt50)50, our model exhibited improved performances of 11.09% and 22.96%, respectively. In the three-classification experiment of AD vs. MCI vs. NC, our model exhibited improved performances of 5.45, 7.32, and 22.96%, compared to the VGG, ResNet50, and ResNeSt50 models, respectively. The results demonstrated that GSCANet has stronger feature extraction capabilities than other networks.
We used confusion matrices to present the results from Tables 2 and 3 to assess the network’s performance on the validation data for each class (Fig. 5). A confusion matrix is a tool used to assess the performance of a classification model. It shows the relationship between the predicted categories and actual categories, helping to identify areas where the model performs well and where it is prone to errors. Figure 5A, B, and C illustrate the training results on the ADNI dataset. Figure 5D and E show the training results on the ADNI dataset and the testing results on the in-house dataset. In the four-class classification, NC was frequently misclassified into MCI(including EMCI) (Fig. 6A). This may be attributed to the subjective nature of diagnoses and the multi-site data sources, which can lead to similarities between NC and MCI. In the three-class classification, two AD subjects were misclassified as MCI, eleven MCI subjects were misclassified as AD, three MCI subjects were misclassified as NC, and sixteen NC subjects were misclassified as MCI. In the binary classification, subjects were more often misclassified into a more severe disease stage than into a less severe one. For example, in the EMCI and NC classification, 48 NC subjects were misclassified as EMCI, whereas no EMCI subjects were misclassified as NC. This indicates the effectiveness of the model, as in medical diagnostics, it is preferable to identify individuals with potential disease rather than incorrectly predicting negatives and missing those with the condition.
ROC curve of the multiclassification model in this study. (a) Binary classification. (b) Graph plots of the three classification results, including those for other model validation methods. (c) Graph plots of four classification results, including other model validation methods. Abbreviations: ROC, receiver operating characteristic; AUC, area under curve.
To better display our classification results, we plotted the time-dependent receiver operating characteristic (ROC) curves of the aforementioned binary classification, three classification, and four-classification as shown in Fig. 7. The AUC for the AD vs. MCI vs. NC three-category study was 0.92. The AUC for the AD vs. LMCI vs. EMCI vs. NC four-category was 0.95, and in the dichotomous category, the AUCs for the EMCI vs. LMCI and AD vs. NC were 0.89 and 0.82 respectively.
Ablation experiment
To thoroughly validate the effectiveness of the method proposed in this study, we conducted ablation tests on both the feature and model modules to assess the contributions of GLCM, FC, MMSE/CDR modalities, as well as the group self-calibrated convolution module and coordinate attention module to the overall performance. Specifically, we utilized the same dataset and experimental setup to conduct three-class classification (AD vs. MCI vs. NC), altering only the components under comparison in each experiment.
In the feature ablation experiments, we selected one or more from GLCM, FC, and MMSE&CDR on the basis of the original model to evaluate the contribution of each modality to the overall performance. The results of classification using features of a certain modality show that the accuracy of GLCM features alone was 73.39% (Table 5), which is higher than that of FC (58.49%) and CDR&MMSE (65.56%). By combining three modal features, we can enhance the model’s predictive power and achieve better classification results. This proves that multimodal data can provide supplementary information for AD diagnosis.
In the module ablation experiments, we chose VGG as the backbone network and incorporated either the group self-calibrated convolution module or the coordinate attention module, or both, to compare the impact of different configurations on the model’s parameters and recognition accuracy. The comparative results, as shown in Table 6, demonstrate how the addition of these components step-by-step contributes to the improvement of the model’s performance. The results indicate that the baseline accuracy of 77.88% was improved by both the group self-calibrated convolution module (80.38%) and the coordinate attention module (79.43%). Their combination exhibited the highest accuracy of 83.33%, which clearly indicate the effectiveness of both modules in enhancing feature extraction and facilitating interactions among channels and spaces.
Model interpretability
In computer-aided diagnosis, identifying specific brain regions closely related to the predictions of deep learning models is vital. Understanding these regions aids in explaining the model’s decision-making process, enhancing its interpretability and reliability. In our study, we utilized the Gradient Weighted Class Activation Mapping (Grad-CAM)51 technique to extract feature mappings from the proposed network. Before using concatenation to obtain the desired labels, Grad-CAM was applied to the final convolutional layer of the proposed GSCANet model. The attention maps generated serve as visual descriptions of the suggested network, highlighting the image regions essential for determining the target class. The attention maps and visualizations created using the Grad-CAM algorithm for GLCM images of NC and EMCI are shown in Fig. 7.
In our model, energy feature is used less frequently in classification which may imply that its discriminative power is weaker in the current dataset and model. While the Asm and entropy feature holds the highest proportion, which indicates that in the classification of AD, the Asm and entropy feature plays a more significant role in the model’s prediction, possibly because it captures texture information more relevant to the disease. Our findings indicate that the key regions, including the hippocampus, medial superior frontal gyrus, precuneus, middle temporal gyrus, posterior cingulate gyrus, lingual gyrus, and dorsolateral superior frontal gyrus—critical areas in AD pathology—provide the richest features for our model’s predictions. Changes in these regions are not only markers of the disease but also provide vital information to the model, aiding in improving classification accuracy and reliability. This visual evidence enhances our understanding of the model’s predictions. It also indicates that the self-calibration and coordination attention mechanisms enhance feature extraction and interactions between channels and spatial regions, enabling the model to focus on critical areas in brain images associated with AD.
Discussion
This study employed a combination of local Haralick texture features based on sMRI, global FC features based on fMRI, and cognitive information based on neuropsychological score features to diagnose AD. Our distinctive approach lies in the proposed GSCANet model, which employs group self-calibrated convolution to extract meaningful features and expand our receptive field. We integrated coordinated attention to facilitate significant interactions among channels and spaces. The outcomes of our experiments consistently demonstrated the high performance of our model in classification tasks and when tested on external datasets.
The results presented demonstrate that the multimodal feature, encompassing Haralick texture features, functional connectivity, and neuropsychological scores combination method yields higher classification accuracy than traditional feature combination methods, for both tetra- and tri-classification tasks. This is because the proposed method extracts complementary information from multiple modalities and reduces the classification error, thus expanding the classification accuracy52,53. This approach provides a more comprehensive understanding of brain function by integrating the unique information of each modality54. The study used image intensity-derived GLCM to calculate the Haralick texture features and extract subtle structural changes that may be indicative of early AD55. This approach characterizes the local patterns and arrangement rules of brain images, providing subtle, high-resolution texture information of the brain surface56. Recent studies have shown that patients with early AD exhibit subtle structural changes before significant occurrence of brain tissue atrophy57,58. These changes may lead to mild brain atrophy manifested in the forms of various symptoms in AD patients, such as forgetfulness24. Therefore, the use of advanced features, such as subtle changes extracted from the original image, enables the diagnosis of AD57,59. Functional connectivity features can detect the functional changes in the brain that are related to the pathology of AD. It can enhance the diagnostic accuracy of brain disorders associated with AD-related cognitive impairments11. Moreover, neuropsychological scales, such as MMSE and CDR, each offer distinct advantages. These scales are among the most commonly used tools for assessing cognitive, emotional, and behavioral-related functions, providing valuable insights for diagnosis. In summary, the use of multimodal features, including Haralick texture, functional network, and neuropsychological score, provide a more comprehensive understanding of brain function and yields higher accuracy in AD diagnosis. This approach has the potential to facilitate early AD detection and intervention by characterizing subtle structural changes in the brain.
This study proposed a parameter-efficient GSCANet framework that exhibited improved performance compared to previous studies34,43,46. Tables 2 and 4 show that our results in the AD vs. NC experiment improved compared to those in LSTM-Robust43. The accuracy of the four categories was also significantly improved than that in adaptive sparse learning model34, and the accuracy of EMCI and NC classification in our study improved compared with that in GDCA model40. These results indicate that our model performs better in complex classification tasks. The improved performance of GSCANet can be attributed to several factors. First, the framework incorporates additional group self-calibrated convolution operations into the residual blocks after multiple sets of treatments, thereby enabling multiscale information interaction through heterogeneous convolutional kernels. This expands the receptive field and supplements the global information of features, facilitating the early identification of brain differences among different subjects. Second, GSCANet utilizes multiple group self-calibrated coordinate attention modules for feature extraction, wherein each block set operates independently. This approach effectively aggregates hierarchical and refined channel and spatial information using group self-calibrating features to coordinate attention. Using accurate location information to encode channel relationships and long-term dependencies, GSCANet facilitates effective interaction of channel and space information. This enhances the model’s sensitivity to semantic information among features, thereby improving the accuracy of diagnosis60. The proposed GSCANet framework incorporates group self-calibrated convolution and coordinates attention modules to improve performance in complex classification tasks. This approach enables effective multiscale information interaction and facilitates the aggregation of hierarchical and refined channel and spatial information. GSCANet improves the sensitivity of the model by utilizing accurate location information and encoding channel relationships and long-term dependencies to semantic information between features and enhances the accuracy of diagnosis.
The effectiveness of the method proposed in this study has been thoroughly validated. In a comparison with classic network models for classification, such as VGG, ResNet50, and ResNeSt50, our model demonstrated superior performance. It surpassed the VGG48 model in a four-category classification study. Furthermore, our model displayed exceptional performance improvement over traditional models, including ResNet49 and ResNeSt50, in the three-classification experiment of AD vs. MCI vs. NC. The results demonstrate that GSCANet has stronger feature extraction capabilities than other networks. We speculate that group self-calibrated coordinate attention in the residual block can identify potential features among data and possess robust recognition capabilities for networks with attention mechanisms or multiscale extraction60. Our model enables multiscale information interactions by expanding the receptive field and aggregating spatial channel information, thus enabling more efficient and faster detection of subtle predementia brain changes. The model framework proposed in this study ensures good classification performance and improves the flexibility and generalization performance, thereby indicating its potential to identify more effectively the early onset of AD compared with other methods. Our network model is based on a general framework that permits the flexible implementation and design of computer-aided diagnostic systems that can address disease diagnosis problems.
Our analysis demonstrated that integrating features from all three modalities—GLCM from sMRI, FC from fMRI, and cognitive scales—achieves higher classification accuracy. To identify which feature contributes most significantly, we conducted ablation experiments. The results revealed that Gray-GLCM features alone provided superior classification accuracy compared to FC and cognitive scale features individually. This highlights the critical role of GLCM features as they offer deeper` insights into localized texture information and brain abnormalities associated with AD, beyond what functional connectivity can provide. Additionally, we explored which network module contributes most to classification improvement through further ablation experiments. Both group self-calibrated convolution and coordinated attention modules enhanced the network’s performance and the combination of these two modules resulted in significant improvement in classification accuracy. Furthermore, the visualizations result indicates that key regions, including the hippocampus, medial superior frontal gyrus, precuneus, middle temporal gyrus, posterior cingulate gyrus, lingual gyrus, and dorsolateral superior frontal gyrus, critical areas in AD pathology61,62, provide the richest features for our model’s predictions. This visual evidence not only enhances our understanding of the model’s predictions but also paves the way for verifying the diagnosis of AD.
However, a few limitations of the study exist, first, deep learning-based methods require large and diverse datasets to efficiently extract features and ensure robust performance. However, the amount of data used in this study is small, mainly consisting of Caucasian participants (ADNI dataset) and Asian participants (in-house dataset), which may limit the model’s generalizability. Future research should include more diverse datasets to improve the model’s robustness and applicability across different populations. Second, potential biases in the current datasets, stemming from demographic factors, imaging protocols, and clinical settings, could affect the generalization of our model. Future work will involve careful consideration and mitigation of these biases to enhance the model’s applicability across different populations and settings. Third, the interpretability of complex deep learning models remains a significant challenge in medical contexts. Ensuring transparency and trust in the model’s predictions is crucial. Future work will focus on developing methods to improve model interpretability, such as incorporating visualization techniques and explainable AI approaches, to provide clearer insights into the decision-making process and help clinicians understand and trust the model’s predictions. Lastly, the testing set we used did not include EMCI and LMCI subjects, which restricts the potential applicability of our model. Expanding the testing set to include these subjects in future studies will be essential to fully evaluate the model’s performance and utility in diagnosing different stages of AD.
Conclusion
In this study, we proposed a novel approach to achieve the early diagnosis of AD using GSCANet’s CNN model. Our approach involved the extraction of effective features through group self-calibration to expand our receptive field. We employed coordinated attention to enable characteristic interactions between channels and spaces. The experimental results demonstrated that our model consistently performed well in evaluation tasks and in the generalized testing of external datasets. Our approach effectively identifies subgroups of subjects with different patterns of AD progression and has potential clinical applications.
Data availability
Some of the data used in this study were from the third-party organization “Alzheimer’s Disease Neuroimaging Initiative” (ADNI) database, which is publicly available on the ADNI website (http://adni.loni.usc.edu/) after registration and compliance with the data use agreement. For detailed information, please refer to http://adni.loni.usc.edu/data-samples/access-data/. The other part of data used in this study is from the First Hospital of Jilin University. Due to privacy or ethical restrictions, the original neuroimaging data is not publicly available. The data that support the findings of this study are available from the corresponding author, Z.Z., upon reasonable request.
References
Alzheimer’s disease facts and figures. Alzheimer’s Dement. 19, 1598–1695 (2023).
Knopman, D. S. et al. Alzheimer disease. Nat. Rev. Dis. Primers 7, 1–21 (2021).
Xu, X., Xu, S., Han, L. & Yao, X. Coupling analysis between functional and structural brain networks in Alzheimer’s disease. Math. Biosci. Eng. 19, 8963–8974 (2022).
delEtoile, J. & Adeli, H. Graph theory and brain connectivity in Alzheimer’s disease. Neuroscientist 23, 616–626 (2017).
Ibrahim, B. et al. Diagnostic power of resting-state fMRI for detection of network connectivity in Alzheimer’s disease and mild cognitive impairment: a systematic review. Hum. Brain Mapp. 42, 2941–2968 (2021).
Yan, T. et al. Early-stage identification and pathological development of Alzheimer’s disease using multimodal MRI. J. Alzheimer’s Disease 68, 1013–1027 (2019).
Dai, Z. et al. Disrupted structural and functional brain networks in Alzheimer’s disease. Neurobiol. Aging. 75, 71–82 (2019).
Khatri, U. & Kwon, G. R. Alzheimer’s disease diagnosis and biomarker analysis using resting-state functional MRI functional brain network with multi-measures features and hippocampal subfield and amygdala volume of structural MRI. Front. Aging Neurosci. 14, 818871 (2022).
Gill, S. et al. Using machine learning to predict Dementia from neuropsychiatric symptom and neuroimaging data. J. Alzheimer’s Disease 75, 277–288 (2020).
Logothetis, N. K. The underpinnings of the BOLD functional magnetic resonance imaging signal. J. Neurosci. 23, 3963–3971 (2003).
Dennis, E. L. & Thompson, P. M. Functional brain connectivity using fMRI in aging and Alzheimer’s disease. Neuropsychol. Rev. 24, 49–62 (2014).
Mohammadian, F. et al. Quantitative assessment of resting-state functional connectivity MRI to differentiate amnestic mild cognitive impairment, late-onset Alzheimer’s disease from normal subjects. J. Magn. Reson. Imaging 57, 1702–1712 (2023).
Gonuguntla, V., Yang, E., Guan, Y., Koo, B. B. & Kim, J. H. Brain signatures based on structural MRI: classification for MCI, PMCI, and AD. Hum. Brain. Mapp. 43, 2845–2860 (2022).
Martinez-Murcia, F. J., Górriz, J. M., Ramírez, J. & Ortiz, A. A structural parametrization of the brain using hidden Markov models-based paths in Alzheimer’s Disease. Int. J. Neural Syst. 26, 1650024 (2016).
Cai, J. H. et al. Magnetic Resonance texture analysis in Alzheimer’s disease. Acad. Radiol. 27, 1774–1783 (2020).
Lee, S., Kim, K. W., Alzheimer’s Disease Neuroimaging Initiative. Associations between texture of T1-weighted magnetic resonance imaging and radiographic pathologies in Alzheimer’s disease. Eur. J. Neurol. 28, 735–744 (2021).
Folego, G., Weiler, M., Casseb, R. F., Pires, R. & Rocha, A. Alzheimer’s disease detection through whole-brain 3D-CNN MRI. Front. Bioeng. Biotechnol. 8, 534592 (2020).
Lao, H. & Zhang, X. Regression and classification of Alzheimer’s disease diagnosis using NMF-TDNet features from 3D brain MR image. IEEE J. Biomedical Health Inf. 26, 1103–1115 (2022).
Kang, W., Lin, L., Sun, S. & Wu, S. Three-round learning strategy based on 3D deep convolutional GANs for Alzheimer’s disease staging. Sci. Rep. 13, 5750 (2023).
Baker, N., Lu, H., Erlikhman, G. & Kellman, P. J. Deep convolutional networks do not classify based on global object shape. PLoS Comput. Biol. 14, e1006613 (2018).
Jo, T., Nho, K. & Saykin, A. J. Deep learning in Alzheimer’s disease: diagnostic classification and prognostic prediction using neuroimaging data. Front. Aging Neurosci. 11, 220 (2019).
Liu, J.-J., Hou, Q., Cheng, M.-M., Wang, C. & Feng, J. Improving convolutional networks with self-calibrated convolutions. In 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 10093–10102 (Seattle, WA, USA, 2020).
Hou, Q., Zhou, D. & Feng, J. Coordinate attention for efficient mobile network design. In 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 13708–13717 (Nashville, TN, USA, 2021).
Lian, C., Liu, M., Zhang, J. & Shen, D. Hierarchical fully convolutional network for joint atrophy localization and Alzheimer’s disease diagnosis using structural MRI. IEEE Trans. Pattern Anal. Mach. Intell. 42, 880–893 (2020).
Yee, E. et al. Construction of MRI-Based Alzheimer’s disease score based on efficient 3D convolutional neural network: comprehensive Validation on 7,902 images from a Multi-center dataset. J. Alzheimers Dis. 79, 47–58 (2021).
Gao, X., Shi, F., Shen, D. & Liu, M. Task-Induced pyramid and attention GAN for multimodal brain image imputation and classification in Alzheimer’s disease. IEEE J. Biomed. Health Inf. 26, 36–43 (2022).
Gutman, B., Wang, Y., Morra, J., Toga, A. W. & Thompson, P. M. Disease classification with hippocampal shape invariants. Hippocampus 19, 572–578 (2009).
Guan, H., Wang, C., Cheng, J., Jing, J. & Liu, T. A parallel attention-augmented bilinear network for early magnetic resonance imaging-based diagnosis of Alzheimer’s disease. Hum. Brain Mapp. 43, 760–772 (2022).
Penny, W. D., Friston, K. J., Ashburner, J. T., Kiebel, S. J. & Nichols, T. E. Statistical Parametric Mapping: The Analysis of Functional Brain Images (Elsevier, 2011).
Haralick, R. M., Shanmugam, K. & Dinstein, I. Textural features for image classification. IEEE Trans. Syst. Man. Cybern. SMC-3, 610–621 (1973).
Zhang, T. et al. Predicting MCI to AD conversation using integrated sMRI and rs-fMRI: machine learning and graph theory approach. Front. Aging Neurosci. 13, 688926 (2021).
Ioffe, S. & Szegedy, C. Batch normalization: accelerating deep network training by reducing internal covariate shift. in Proceedings of the 32nd International Conference on Machine Learning 448–456PMLR, (2015).
Wang, M. et al. Spatial-temporal dependency modeling and network hub detection for functional MRI analysis via convolutional-recurrent network. IEEE Trans. Biomed. Eng. 67, 2241–2252 (2020).
Lei, B. et al. Adaptive sparse learning using multi-template for neurodegenerative disease diagnosis. Med. Image Anal. 61, 101632 (2020).
Wee, C. Y. et al. Cortical graph neural network for AD and MCI diagnosis and transfer learning across populations. Neuroimage Clin. 23, 101929 (2019).
Hao, X. et al. Multi-modal neuroimaging feature selection with consistent metric constraint for diagnosis of Alzheimer’s disease. Med. Image Anal. 60, 101625 (2020).
Li, Y., Liu, J., Jiang, Y., Liu, Y. & Lei, B. Virtual adversarial training-based deep feature aggregation network from dynamic effective connectivity for MCI Identification. IEEE Trans. Med. Imaging 41, 237–251 (2022).
Forouzannezhad, P. et al. A gaussian-based model for early detection of mild cognitive impairment using multimodal neuroimaging. J. Neurosci. Methods 333, 108544 (2020).
Lee, J., Ko, W., Kang, E. & Suk, H. I. & and the Alzheimer’s disease neuroimaging initiative. A unified framework for personalized regions selection and functional relation modeling for early MCI identification. Neuroimage 236, 118048 (2021).
Fang, C. et al. Gaussian discriminative component analysis for early detection of Alzheimer’s disease: a supervised dimensionality reduction algorithm. J. Neurosci. Methods 344, 108856 (2020).
Tufail, A. B. et al. Early-stage Alzheimer’s disease categorization using PET neuroimaging modality and convolutional neural networks in the 2D and 3D domains. Sens. (Basel). 22, 4609 (2022).
Zhang, Z. et al. THAN: task-driven hierarchical attention network for the diagnosis of mild cognitive impairment and Alzheimer’s disease. Quant. Imaging Med. Surg. 11, 3338–3354 (2021).
Mehdipour Ghazi, M. et al. Training recurrent neural networks robust to incomplete data: application to Alzheimer’s disease progression modeling. Med. Image Anal. 53, 39–46 (2019).
Wyman, B. T. et al. Standardization of analysis sets for reporting results from ADNI MRI data. Alzheimers Dement. 9, 332–337 (2013).
Zhang, X., Han, L., Zhu, W., Sun, L. & Zhang, D. An explainable 3D residual self-attention deep neural network for joint atrophy localization and Alzheimer’s Disease diagnosis using structural MRI. IEEE J. Biomed. Health Inf. 26, 5289–5297 (2022).
Katabathula, S., Wang, Q. & Xu, R. Predict Alzheimer’s disease using hippocampus MRI data: a lightweight 3D deep convolutional network model with visual and global shape representations. Alzheimers Res. Ther. 13, 104 (2021).
Liu, Y. et al. MPC-STANet: Alzheimer’s disease recognition method based on multiple phantom convolution and spatial transformation attention mechanism. Front. Aging Neurosci. 14, 918462 (2022).
Simonyan, K. & Zisserman, A. Very deep convolutional networks for large-scale image recognition. Preprint at (2015). https://doi.org/10.48550/arXiv.1409.1556
He, K., Zhang, X., Ren, S. & Sun, J., Deep residual learning for Image recognition. In 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 770–778 (Las Vegas, NV, USA, 2016).
Zhang, H. et al. ResNeSt: Split-attention networks. In 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW) 2735–2745 (New Orleans, LA, USA, 2022).
Selvaraju, R. R. et al. Grad-CAM: visual explanations from deep networks via gradient-based localization. in. IEEE International Conference on Computer Vision (ICCV) 618–626. https://doi.org/10.1109/ICCV.2017.74 (2017)
Liu, X. et al. Use of multimodality imaging and artificial intelligence for diagnosis and prognosis of early stages of Alzheimer’s disease. Translational Res. 194, 56–67 (2018).
El-Sappagh, S., Alonso, J. M., Islam, S. M. R., Sultan, A. M. & Kwak, K. S. A multilayer multimodal detection and prediction model based on explainable artificial intelligence for Alzheimer’s disease. Sci. Rep. 11, 2660 (2021).
Goel, A. et al. Integration of multimodal neuroimaging data to facilitate advanced brain research. J. Alzheimers Dis. 83, 305–317 (2021).
Chekouo, T., Mohammed, S. & Rao, A. A. Bayesian 2D functional linear model for gray-level co-occurrence matrices in texture analysis of lower grade gliomas. Neuroimage Clin. 28, 102437 (2020).
Lin, E., Lin, C. H. & Lane, H. Y. Deep learning with neuroimaging and genomics in Alzheimer’s disease. Int. J. Mol. Sci. 22, 7911 (2021).
Liao, W. et al. Discerning mild cognitive impairment and Alzheimer disease from normal aging: morphologic characterization based on univariate and multivariate models. Acad. Radiol. 21, 597–604 (2014).
Ferrari, C. & Sorbi, S. The complexity of Alzheimer’s disease: an evolving puzzle. Physiol. Rev. 101, 1047–1081 (2021).
Qiao, H., Chen, L., Ye, Z. & Zhu, F. Early Alzheimer’s disease diagnosis with the contrastive loss using paired structural MRIs. Comput. Methods Programs Biomed. 208, 106282 (2021).
Wu, Y., Zhou, Y., Zeng, W., Qian, Q. & Song, M. An attention-based 3D CNN with multi-scale integration block for Alzheimer’s disease classification. IEEE J. Biomed. Health Inf. 26, 5665–5673 (2022).
Pini, L. et al. Brain atrophy in Alzheimer’s disease and aging. Ageing Res. Rev. 30, 25–48 (2016).
Wu, B. S. et al. Cortical structure and the risk for Alzheimer’s disease: a bidirectional mendelian randomization study. Transl Psychiatry 11, 1–7 (2021).
Acknowledgements
We gratefully acknowledge all the participants, clinical doctors and researchers at the First Hospital of Jilin University. Part of the data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). ADNI investigators complete list can be found at:
https://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Funding
This work was supported by the National Natural Science Foundation of China under Grant 62103404, in part by the Shenzhen Overseas Innovation Team Project under Grant KQTD20180413181834876, in part by the Shenzhen Basic Research Program under Grant JCYJ20210324101402008, in part by the Shenzhen Science and Technology Program under Grant GJHZ20210705141405016, in part by the International Cooperation Projects of Science and Technology of Guangdong Province under Grant 2023A0505050162, in part by the Changchun Municipal Science and Technology Innovation Cooperation Special Project of the Chinese Academy of Sciences under Grant CC202205010111044454, and in part by the Japan Society for the Promotion of Science under Grant 21K15614.
Author information
Authors and Affiliations
Contributions
X.Y., J.L.,Y.L.,S.F.,T.M.,J.W.,Q.L.,Z.Z.Conceptualization: X.Y., J.L.,Y.L.,Z.Z. Data curation: X.Y. Formal analysis: X.Y., J.L. Investigation: X.Y., J.L.,S.F.,T.M.,J.W. Methodology: X.Y., S.F.,T.M. Project administration: Z.Z.,Q.L. Supervision: J.W., Q.L., Z.Z. Validation: X.Y. ,Y.L Visualization: X.Y., Y.L. Writing—original draft: X.Y. Preparation writing—review and editing: J.L., Y.L., Z.Z.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yu, X., Liu, J., Lu, Y. et al. Early diagnosis of Alzheimer’s disease using a group self-calibrated coordinate attention network based on multimodal MRI. Sci Rep 14, 24210 (2024). https://doi.org/10.1038/s41598-024-74508-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-74508-z
Keywords
This article is cited by
-
Enhancing early Alzheimer's diagnosis through machine learning on multimodal neuroimaging and cognitive data
Iran Journal of Computer Science (2025)