Abstract
This study aimed to preliminarily develop machine learning (ML) models capable of predicting the risk of device-associated infection and 30-day outcomes following invasive device procedures in intensive care unit (ICU) patients. The study utilized data from 8574 ICU patients who underwent invasive procedures, sourced from the Medical Information Mart for Intensive Care (MIMIC)-IV version 2.2 database. Patients were allocated into training and validation datasets in a 7:3 ratio. Seven ML models were employed for predicting device-associated infections, while five models were used for predicting 30-day survival outcomes. Model performance was primarily evaluated using the receiver operating characteristic (ROC) curve for infection prediction and the survival model’s concordance index (C-index). Top-performing models progressively reduced the number of variables based on their importance, thereby optimizing practical utility. The inclusion of all variables demonstrated that extreme gradient boosting (XGBoost) and extra survival trees (EST) models yielded superior discriminatory performance. Notably, when restricted to the top 10 variables, both models maintained performance levels comparable to when all variables were included. In the validation cohort, the XGBoost model, with the top 10 variables, achieved an area under the curve (AUC) of 0.810 (95% CI 0.808–0.812), an area under the precision-recall curve (AUPRC) of 0.226 (95% CI 0.222–0.230), and a Brier score (BS) of 0.053 (95% CI 0.053–0.054). The EST model, with the top 10 variables, reported a C-index of 0.756 (95% CI 0.754–0.757), a time-dependent AUC of 0.769 (95% CI 0.763–0.775), and an integrated Brier score (IBS) of 0.087 (95% CI 0.087–0.087). Both models are accessible via a web application. The internally evaluated XGBoost and EST models demonstrated exceptional predictive accuracy for device-associated infection risks and 30-day survival outcomes post-invasive procedures in ICU patients. Further validation is required to confirm the clinical utility of these two models in future studies.
Similar content being viewed by others
Introduction
Healthcare-associated infections (HAIs) pose a significant threat to patient care and safety, affecting over 7% of patients globally and resulting in 37,000 deaths annually in Europe and 99,000 in the United States. They also impose substantial healthcare costs1. Among HAIs, device-associated infections represent the most severe risk to intensive care unit (ICU) patients’ life expectancy2. Common examples include ventilator-associated pneumonia (VAP), central line-associated bloodstream infections (CLABSIs), and catheter-associated urinary tract infections (CAUTIs). The incidence of such infections reaches up to 14.7% in countries like China, Korea, and other developing nations2,3,4. Device-associated infections correlate with prolonged hospital stays, increased costs, and higher mortality rates5. Reducing the incidence of these infections by up to 30% could significantly lower healthcare expenses6. Hence, preventing device-associated infections is crucial for reducing healthcare costs and improving patient outcomes.
Currently, several clinical strategies aim to mitigate device-associated infections, including primary education, infection surveillance, and continuous feedback. Effective surveillance is vital for implementing practical prevention measures. Continuous monitoring and management are essential to prevent these infections and enhance the prognosis for high-risk patients, particularly those in ICUs. A study involving 900 acute care hospitals in the United States demonstrated the efficacy of intervention practices in preventing device-associated infections7. Additionally, the International Nosocomial Infection Control Consortium (INICC) network, an online surveillance system, has proven effective in reducing infection rates8. Therefore, early risk assessment and identification of patients at risk for device-associated infections are critical to reducing incidence and improving care quality.
Recently, multivariate logistic analysis has been commonly used to identify risk factors and develop predictive models. However, this method is constrained by its reliance on the linearity assumption between risk factors and outcomes9. In contrast, machine learning (ML), a branch of artificial intelligence, can uncover hidden patterns in complex datasets by training algorithms. ML has significantly influenced clinical decision-making and shown promise in predicting clinical outcomes10,11. The trend of ML applications in HAI prediction is evident, with prior studies developing models to predict specific device-associated infections like VAP, CLABSI, and CAUTI12,13,14. However, these models are limited to predicting individual infection types, not encompassing all potential HAIs. ICU patients often undergo multiple invasive procedures, increasing their infection risk. Therefore, comprehensive predictive models are essential for mitigating device-associated infections and improving survival outcomes in ICU patients post-procedures.
This study endeavors to develop ML models for predicting the risk of device-associated infections and 30-day survival outcomes in ICU patients following invasive device treatments, using data from the Medical Information Mart for Intensive Care (MIMIC)-IV cohort15. These ML models will be incorporated into a web-based application, facilitating clinicians in the early identification of high-risk patients and informed decision-making.
Materials and methods
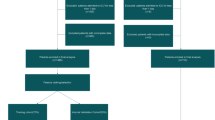
Study design and participants
Electronic inpatient records were retrieved from the MIMIC-IV database (version 2.2), which includes data on 180,733 inpatients and 50,920 ICU patients admitted to Beth Israel Deaconess Medical Center between 2008 and 2019. This database integrates information from hospital and ICU systems, along with external sources, capturing vital signs, laboratory and microbiological tests, admissions and discharges, medications, length of stay, survival data, and discharge or death records16. Access to this database was granted upon completion of the Collaborative Institutional Training Initiative (CITI Program) training course (certification ID: 12037493). Since the database is publicly available and anonymized, patient informed consent was not required. This study adhered to relevant guidelines to ensure patient privacy, and it was approved by the Ethics Committee of Tengzhou Central People’s Hospital in China (ethical review approval number: 2023-ethical reviews-45). All methodologies and protocols were rigorously conducted in full compliance with the principles set forth in the Declaration of Helsinki.
The study included patients over 18 years old who were first hospitalized in the ICU and underwent at least one invasive procedure, such as invasive mechanical ventilation (IMV), central venous catheter (CVC), or indwelling urinary catheter (IUC). Exclusion criteria were: (1) death within 48 h of ICU admission; (2) invasive procedures conducted in non-ICU departments; (3) failure to meet the Centers for Disease Control and Prevention (CDC) National Healthcare Safety Network (NHSN) guidelines17 for diagnosing VAP, CLABSI, or CAUTI.
Outcomes and definitions
This study targeted three types of infections: VAP, CLABSI, and CAUTI. The outcomes were defined as 30-day mortality and device-associated infections occurring in the ICU after more than 48 h following the specified operations. The predictive device-associated infections included VAP, CLABSI, and CAUTI. Invasive device procedures and diagnoses of device-associated infections were evaluated based on the CDC’s NHSN guidelines.
Data collection
Variables documented upon ICU admission were collected, encompassing general information, laboratory examinations, and vital signs. Basic data included age, gender, ethnicity, comorbidities, admission care unit, invasive interventions, device-associated infections, 30-day survival outcomes post-first invasive procedure, and hospitalization outcomes. Laboratory parameters and vital signs were collected within the first 24 h of ICU admission. These included white blood cell (WBC) count, platelet count, anion gap, bicarbonate levels, creatinine levels, chloride levels, glucose levels, hemoglobin levels, potassium levels, sodium levels, blood urea nitrogen (BUN) levels, calcium levels, activated partial thromboplastin time (APTT), partial thromboplastin time (PT), international normalized ratio (INR), systolic blood pressure (SBP), diastolic blood pressure (DBP), mean arterial pressure (MAP), temperature, peripheral blood oxygen saturation (SpO2), heart rate, respiratory rate, Glasgow Coma Scale (GCS), Sequential Organ Failure Assessment (SOFA), Acute Physiology Score III (APS III), and Simplified Acute Physiology Score II (SAPS II). The code for the study is available on GitHub (https://github.com/susu223344/Device-Associated-Infections-Hyperparametric-Search/blob/main/sql).
Development and visualization of ML models
Patients were randomly assigned to training and validation datasets in a 7:3 ratio. Missing data were imputed using the k-nearest neighbor (KNN) strategy, and continuous variables were normalized using a min-max scaler. To mitigate the risk of data leakage, the KNN imputation method and min-max scaling were separately applied to the training and validation datasets following data partitioning18. Two separate ML models were developed: one for device-associated infections and one for 30-day survival outcomes. Seven ML models were applied for predicting device-associated infections: random forest (RF), logistic regression (LR), support vector machine (SVM), extreme gradient boosting (XGBoost), Gaussian naive Bayesian (GNB), decision tree (DT), and recurrent neural networks with long short-term memory (LSTM). For 30-day survival outcomes, five models were used: Cox regression, extra survival trees (EST), survival tree (ST), gradient boosting survival tree (GBST), and deep learning survival neural network (DeepSurv)19,20. A grid search approach was employed to optimize each model, identifying the optimal hyperparameters through 10-fold cross-validation (Supplementary Table 1). The code for this grid search is accessible on GitHub (https://github.com/susu223344/Device-Associated-Infections-Hyperparametric-Search/blob/main/Code%201.ipynb). All models were trained and evaluated using 10-fold cross-validation with the identified optimal hyperparameters.
Assessment of the device-associated infection ML model was conducted using the area under the curve (AUC) of the receiver operating characteristic (ROC), the area under the precision-recall curve (AUPRC), and the Brier score (BS). Evaluation of the 30-day survival ML model employed concordance index (C-index), integrated Brier score (IBS), and time-dependent AUC. Both AUC and C-index measure discrimination; they are equivalent for binary classification ML models, but the C-index remains unaffected by survival time censoring, making it ideal for survival data analysis. C-index and AUC values below 0.60 indicate poor discrimination, between 0.60 and 0.75 indicate potentially helpful discrimination, and above 0.75 indicate clearly useful discrimination21. Additionally, AUPRC, similar to ROC-AUC, excels at evaluating discrimination in class-imbalanced scenarios, distinguishing positive and negative samples more effectively. The time-ROC curve, illustrating ROC curves at various time points with evolving AUC, captures the model’s discrimination variations over time, with a time-dependent AUC closer to 1 indicating higher predictive performance. BS and IBS are essential for assessing prediction accuracy; BS calculates the mean squared difference between predicted and actual values, while IBS integrates BS for continuous-time model assessment, with values closer to 0 signifying superior predictive accuracy.
Selection of the optimal ML algorithm for the device-associated infection model involved training seven ML models with all variables and comparing metrics such as AUC, AUPRC, and BS, alongside disparities in decision curve analysis (DCA) and calibration curves. This rigorous evaluation identified the most suitable algorithm. A similar process was followed for the 30-day survival model, training five ML models with all relevant variables and selecting the best algorithm based on C-index, IBS, and time-dependent AUC. The Shapley additive explanation (SHAP)22 method evaluated the variable importance of the optimal ML algorithms for both models. The performance of the optimal ML algorithms was tested on the full set of variables and the top 30, top 20, top 15, and top 10 variables, emphasizing fewer variables without compromising performance, thereby determining the final model.
The optimal ML models were converted and implemented into a web-based application. This application displays the predicted probability of device-associated infections and provides insights into the decision-making process using the SHAP method. Additionally, it includes the patients’ 30-day Kaplan-Meier survival curve.
Statistical analysis
Data were presented as medians with interquartile ranges for continuous variables and as counts with percentages for categorical variables. Differences between the two datasets were assessed using the Wilcoxon rank sum test for continuous data and Fisher’s exact test for categorical data. All statistical analyses were performed using R version 4.3.0 and Python version 3.7.0 (version 3.8.17 for the web-based application). A P-value of less than 0.05 was considered statistically significant.
Results
Baseline characteristics
A total of 8574 patients were included in this study, with 6001 in the training dataset and 2573 in the validation dataset. No significant differences were observed between the baseline parameters of the two cohorts (P > 0.05) (Table 1). The baseline characteristics are detailed in Table 1, with a median age of 69.03 years (interquartile range: 57.33, 80.04). Among the participants, 4774 (55.68%) were male. Following invasive device procedures, 491 patients (5.73%) developed device-associated infections, and 1329 patients (15.50%) died within 30 days.
Development and assessment of ML models
Figures 1 and 2 compare the performance of seven ML models in predicting device-associated infections when all variables are included. In the validation dataset, XGBoost exhibited the highest discriminative performance with an AUC of 0.812 (95% CI 0.809–0.815), while SVM had the lowest AUC of 0.591 (95% CI 0.558–0.625). All seven ML models displayed low AUPRCs, with XGBoost and RF achieving the highest AUPRCs of 0.230 (95% CI 0.224–0.236) and 0.230 (95% CI 0.224–0.237), respectively, and SVM the lowest at 0.089 (95% CI 0.076–0.101). Among all models, the XGBoost model was most closely calibrated to the perfectly calibrated curve and had the lowest BS of 0.054 (95% CI 0.053–0.054). DCA indicated that XGBoost provided the highest net benefit when the threshold probability ranged between 0.1 and 0.3. At a threshold probability of 0.1, the XGBoost model identified 2 true positives per 100 patients with invasive devices, with no false positives. Based on these findings, XGBoost was selected as the ML model for predicting device-associated infections.
Table 2 and Fig. 3 depict the performance of five ML models in predicting 30-day survival. In the validation dataset, EST demonstrated the highest C-index of 0.777 (95% CI 0.775–0.779), while DeepSurv had the lowest value of 0.678 (95% CI 0.642–0.714). EST and Cox achieved the lowest IBS of 0.086 (95% CI 0.086–0.086), whereas DeepSurv had the highest IBS of 0.101 (95% CI 0.099–0.102). Additionally, EST displayed a higher time-dependent AUC of 0.795 (95% CI 0.789–0.801) compared to the other ML models, particularly beyond five days following invasive device procedures. Based on the performance of all models in the validation cohorts, EST was selected as the ML model for predicting 30-day survival outcomes.
Variables selection
Figure 4 depicts the feature importance of the two models when all variables are included. Tables 3 and 4 present model performances with different numbers of variables, sorted by importance. The XGBoost model incorporating the top 10 variables achieved an AUC of 0.810 (95% CI 0.808–0.812), AUPRC of 0.226 (95% CI 0.222–0.230), and BS of 0.053 (95% CI 0.053–0.054), comparable to the model using all variables, which had an AUC of 0.812 (95% CI 0.809–0.815), AUPRC of 0.230 (95% CI 0.224–0.236), and BS of 0.054 (95% CI 0.053–0.054) (Table 3). Similarly, the EST model with the top 10 variables exhibited a C-index of 0.756 (95% CI 0.754–0.757) and a time-dependent AUC of 0.769 (95% CI 0.763–0.775) compared to the C-index of 0.777 (95% CI 0.775–0.779) and time-dependent AUC of 0.795 (95% CI 0.789–0.801) observed in models incorporating all variables (Table 4). Both XGBoost and EST eventually selected the top 10 variables to minimize the number of variables included.
Interpretability analysis
The SHAP algorithm was utilized to calculate each variable’s contribution to every example, revealing how the data influences the probability and how the model processes this information. Figure 5 illustrates the interpretability analysis of the SHAP algorithm in the XGBoost and EST models. Feature importance was estimated based on the training dataset to assess the global impact of each feature. In the XGBoost model, the top variables were length of stay (LOS) in the ICU before using a central venous catheter (CVC), LOS before using invasive mechanical ventilation (IMV), tracheostomy, medical intensive care unit (MICU)/surgical intensive care unit (SICU), temperature, APS III, LOS before using an indwelling urinary catheter (IUC), MAP, PT, and renal replacement therapy (RRT). In the EST model, the key variables included APS III, MICU/SICU admission, age, cancer, liver disease, SAPS II, anion gap, cerebrovascular disease, myocardial infarction, and the use of two or more devices. SHAP summary plots demonstrated the impact of each feature on the model output in terms of overall distribution. The XGBoost model indicated an increased risk of device-associated infections with prolonged ICU stay before the utilization of devices like CVC and IMV, as well as heightened risks for patients who underwent tracheostomy and were admitted to the MICU/SICU. The EST model revealed an increased risk of mortality within 30 days following an invasive device procedure for patients with elevated APS III scores, MICU/SICU admission, advanced age, and a cancer diagnosis. Decision plots depicted the decision-making process for the top 100 patients, with the prediction line illustrating the accumulation of the ML output value from the base SHAP value to the final value at the top of the figure.
Model application
The XGBoost and EST models were deployed through a web-based application (https://susudaihai.streamlit.app/). Users can input variables into the application to predict the probability of device-associated infection. The application generates SHAP force plots, illustrating the contribution of individual features in influencing the model’s predictions. Additionally, it presents the patient’s 30-day Kaplan-Meier survival curve, enabling an assessment of their prognosis.
Discussion
This study preliminarily developed ML-based models to predict the risk of device-associated infections and 30-day survival outcomes in ICU patients following initial invasive device procedures. The XGBoost and EST models were developed for these predictions, focusing on the top 10 variables. In the validation cohort, the XGBoost model achieved an AUC of 0.810 (95% CI 0.808–0.812), and the EST model achieved a C-index of 0.756 (95% CI 0.754–0.757). These models enhance personalized surveillance by providing individualized probabilities of device-associated infections and 30-day survival outcomes.
Device-associated infections have garnered significant attention in preventing HAIs over recent decades. A survey of 183 hospitals in the USA indicated that HAIs accounted for 4%, with device-associated infections contributing 25.6%. These infections place a substantial burden on patients23. Previous studies have primarily focused on predicting specific device-associated infections through model construction. Various ML algorithms have been proposed to predict the risk of VAP in ICU patients. For instance, Giang et al.12 and Liang et al.24 demonstrated that XGBoost (AUC: 0.854) and RF (AUC: 0.84) as ensemble ML algorithms performed well in predicting VAP. Wu et al.25 used the LR algorithm to predict VAP in patients with acute respiratory distress syndrome, achieving an AUC of 0.744. Han et al.26 established an LR model with an AUC of 0.817 to predict mortality in ICU patients with VAP. Additionally, other studies13,27,28,29 have used LR, XGBoost, RF, and deep learning algorithms to build ML models for predicting CLABSI in hospitalized patients, with AUCs ranging from 0.722 to 0.993. Furthermore, several studies14,30,31 have utilized neural networks, ensemble learning models, and XGBoost algorithms to develop ML models for predicting CAUTI in hospitalized patients, with AUCs ranging from 0.758 to 0.904. However, these models only predicted one of the outcomes: VAP, CLABSI, or CAUTI. A study conducted in Poland32 among ICU patients found that the incidence density for CLABSI was 8.0 per 1000 catheter days, VAP was 15.2 per 1000 ventilator days, and CAUTI was 3.0 per 1000 catheter days. Predicting just one outcome is insufficient for reducing the risk of HAIs in ICU patients. Using models from three different studies to predict the risk of device-related infections would present clinicians with numerous variables and inconsistent inclusion criteria, complicating clinical decision-making. Moreover, these models did not integrate patient mortality outcomes. In the ICU, both patient survival probability and device-associated infections are crucial for clinicians, as some ICU patients face poor prognoses following invasive device procedures. A model that can predict both outcomes would significantly enhance patient prognosis. The XGBoost model in this study achieved an AUC of 0.810, closely approximating the AUCs of the aforementioned models specific to device-related infections. Requiring only 10 variables, XGBoost facilitates easier data acquisition compared to similar studies. Additionally, it provides comprehensive predictions for VAP, CLABSI, and CAUTI, enhancing both the breadth and efficiency of predictions. The model includes variables such as patients’ ICU length of stay before employing IMV, CVC, and IUC, enabling clinicians to input diverse device usage times. This aids in predicting potential risks of device-related infections and offers insights into optimal device utilization timings and necessity. This particular aspect distinguishes our study from others.
The XGBoost algorithm achieved the highest AUC among the seven classification models included in the study. For unbalanced data, AUPRC is typically more discriminative than AUC33, and XGBoost also achieved the highest AUPRC. Predictive models often struggle to show high net benefits across all risk thresholds, our DCA curve demonstrated significant net benefit for XGBoost within the 0.1–0.3 risk threshold. Considering all these factors, XGBoost was ultimately selected as the optimal classification model, aligning with findings from previous studies. Among the five survival models examined, EST was chosen for its highest C-index and time-dependent AUC.
The SHAP algorithm revealed that LOS before using CVC and IMV were the most significant variables in the device-associated infection model. As an intravenous catheter, CVC is inserted into centrally-located veins to administer medications, fluids, and nutrition, and to draw blood. Most ICU patients require CVC due to advanced care needs. However, infection is a common complication associated with CVC, primarily due to bacteria infiltrating the catheter and forming biofilm colonies resistant to antibiotics. These biofilms can quickly spread to the bloodstream, leading to CRBSI34,35,36. The delayed use of CVCs and IMVs in the ICU correlates with an increased risk predicted by the ML model. This heightened risk is likely due to prolonged ICU stays, which elevate the chances of bacterial colonization and infection. Additionally, the use of invasive devices, such as CVCs and IMVs, compromises the patient’s protective barriers.
The 30-day survival model identified APS III, MICU/SICU admission, advanced age, and a cancer diagnosis as key variables increasing mortality risk within 30 days after invasive device procedures. Consistent with previous analyses, APS III and age are significant factors affecting ICU mortality37. This study demonstrates that patients admitted to the MICU/SICU with higher APS III scores not only face a higher risk of device-associated infections but also exhibit increased 30-day mortality rates. It is important to note that predictive models and causal studies differ, and identifying significant variables in interpretable findings does not always indicate causal associations38.
The ML model assists ICU physicians in making informed decisions on optimal device insertion timing to reduce the risk of device-associated infections. This assistance includes: (1) For patients newly admitted to the ICU without prior device usage, physicians can input varying durations of CVC and IMV usage based on the patient’s current fixed variables. This predicts the infection risk at different intervals, facilitating the determination of the latest suitable time for device implementation and the appropriate equipment category. (2) For patients with existing ICU tenure and prior device usage, physicians can input different device usage durations and adjust tracheostomy status to forecast infection risks. This helps in assessing the necessity of additional equipment and tracheostomy. (3) In scenarios where device usage duration is fixed, integrating forecasts of infection risk and 30-day mortality risk enables comprehensive evaluation of the patient’s prognosis, allowing timely intervention.
Conclusions
This study preliminarily developed an integrated ML model that predicts device-associated infections and 30-day survival outcomes following invasive device procedures in ICU patients. The ML model is accessible through a user-friendly web-based application. The clinical applicability of this integrated tool requires further validation in future studies.
Limitations
This study has several limitations. Firstly, while the MIMIC database contains extensive patient data and has facilitated the development of numerous ML algorithms, it is important to note that it is a single-center database, unlike multi-center databases such as eICU. Some of the data in the MIMIC database originate from two distinct systems, MetaVision and CareVue, which focus solely on ICU data and lack information about patients once they transition from the ICU to general wards. Moreover, the racial distribution within the MIMIC database is significantly imbalanced, posing substantial challenges in applying the findings across diverse global populations, particularly among Asians. Secondly, difficulties in collecting sufficient external validation data hindered the generalizability of our model to different settings. Thirdly, the incidence of device-associated infections was only 5.73%, resulting in substantial data imbalance. While the AUC reached 0.810, the AUPRC was merely 0.226, indicating a limitation in XGBoost’s capacity to accurately identify true positive cases. Thus, the practical effectiveness of the model warrants further investigation. Finally, this ML model predicts only 30-day survival outcomes rather than long-term prognosis and is specifically designed for ICU patients following invasive device procedures. It cannot be applied to predict infection risk and survival outcomes for non-ICU patients or those undergoing other types of operations.
Data availability
The dataset utilized in this paper was acquired from the MIMIC-IV (version 2.2) database (https://physionet.org/content/mimiciv/2.2/) with granted permission. The code for extracting variables is available on GitHub (https://github.com/susu223344/Device-Associated-Infections-Hyperparametric-Search/blob/main/sql). The datasets used and/or analyzed during the current study can be obtained from the corresponding author upon reasonable request.
Change history
13 June 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41598-025-02017-8
References
Mazzeffi, M., Galvagno, S. & Rock, C. Prevention of healthcare-associated infections in intensive care unit patients. Anesthesiology 135(6), 1122–1131 (2021).
Rosenthal, V. D. et al. Device-associated nosocomial infections in 55 intensive care units of 8 developing countries. Ann. Intern. Med. 145(8), 582–591 (2006).
Chen, Y. et al. A point-prevalence survey of healthcare-associated infection in fifty-two Chinese hospitals. J. Hosp. Infect. 95(1), 105–111 (2017).
Choi, J. et al. Trends in the incidence rate of device-associated infections in intensive care units after the establishment of the Korean nosocomial infections Surveillance System. J. Hosp. Infect. 91(1), 28–34 (2015).
Al-Tawfiq, J. A. et al. Surveillance of device associated infections in intensive care units at a Saudi Arabian Hospital, 2017–2020. J. Infect. Public. Health 16(6), 917–921 (2023).
Negm, E. M. et al. Impact of a comprehensive care bundle educational program on device-associated infections in an emergency intensive care unit. Germs 11(3), 381–390 (2021).
Saint, S. et al. What US hospitals are currently doing to prevent common device-associated infections: Results from a national survey. BMJ Qual. Saf. 28(9), 741–749 (2019).
Duszynska, W. et al. Device associated-health care associated infections monitoring, prevention and cost assessment at intensive care unit of University Hospital in Poland (2015–2017). BMC Infect. Dis. 20(1), 761 (2020).
Minami, T. et al. Machine learning for individualized prediction of hepatocellular carcinoma development after the eradication of hepatitis C virus with antivirals. J. Hepatol. 24 (2023).
Shung, D. L. et al. Validation of a machine learning model that outperforms clinical risk scoring systems for upper gastrointestinal bleeding. Gastroenterology 158, 160–167 (2020).
Spann, A. et al. Applying machine learning in liver disease and transplantation: A comprehensive review. Hepatology 71, 1093–1105 (2020).
Giang, C. et al. Predicting ventilator-associated pneumonia with machine learning. Medicine 100(23) (2021).
Parreco, J. P., Hidalgo, A. E., Badilla, A. D., Ilyas, O. & Rattan, R. Predicting central line-associated bloodstream infections and mortality using supervised machine learning. J. Crit. Care 45, 156–162 (2018).
Jakobsen, R. S., Nielsen, T. D., Leutscher, P. & Koch, K. Clinical explainable machine learning models for early identification of patients at risk of hospital-acquired urinary tract infection. J. Hosp. Infect. (2023).
Johnson, A. E. W. et al. MIMIC-IV, a freely accessible electronic health record dataset. Sci. Data. 10 (1), 1 (2023).
Liu, J. et al. Impact of stress hyperglycemia ratio on mortality in patients with critical acute myocardial infarction: Insight from American MIMIC-IV and the Chinese CIN-II study. Cardiovasc. Diabetol. 22(1), 281 (2023).
National Healthcare Safety. Network (NHSN) Patient Safety Component Manual (Centers for Disease Control and Prevention website). https://www.cdc.gov/nhsn/psc/index.html
Huang, R. et al. Stroke mortality prediction based on ensemble learning and the combination of structured and textual data. Comput. Biol. Med. 155, 106176 (2023).
Yue, S. et al. Machine learning for the prediction of acute kidney injury in patients with sepsis. J. Transl. Med. 20 (1), 1–12 (2022).
Katzman, J. L. et al. DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med. Res. Methodol. 18(1), 1–12 (2018).
Alba, A. C. et al. Discrimination and calibration of clinical prediction models: Users’ guides to the Medical Literature. JAMA 318(14), 1377–1384 (2017).
Lundberg, S. M. & Lee, S-I. A unified approach to interpreting model predictions. Adv. Neural. Inf. Process. Syst. 30 (2017).
Magill, S. S. et al. Multistate point-prevalence survey of health care–associated infections. N. Engl. J. Med. 370(13), 1198–1208 (2014).
Liang, Y. et al. Early prediction of ventilator-associated pneumonia in critical care patients: A machine learning model. BMC Pulm. Med. 22(1), 1–9 (2022).
Wu, Z. et al. A ventilator-associated pneumonia prediction model in patients with acute respiratory distress syndrome. Clin. Infect. Dis. 71 (Supplement_4), S400–S408 (2020).
Han, X., Wu, W., Zhao, H. & Wang, S. Developing and validating a prediction model for in-hospital mortality in patients with ventilator-associated pneumonia in the ICU. Ann. Palliat. Med. 11(5), 1799–1810 (2022).
Rahmani, K. et al. Early prediction of central line associated bloodstream infection using machine learning. Am. J. Infect. Control 50(4), 440–445 (2022).
Bonello, K. et al. Prediction of impending central-line-associated bloodstream infections in hospitalized cardiac patients: Development and testing of a machine-learning model. J. Hosp. Infect. 127, 44–50 (2022).
Tabaie, A. et al. Deep learning model to predict serious infection among children with central venous lines. Front. Pead. 9, 726870 (2021).
Zhu, C. et al. Prediction of post-stroke urinary tract infection risk in immobile patients using machine learning: An observational cohort study. J. Hosp. Infect. 122, 96–107 (2022).
Taylor, R. A., Moore, C. L., Cheung, K-H. & Brandt, C. Predicting urinary tract infections in the emergency department with machine learning. PLoS ONE 13(3), e0194085 (2018).
Kolpa, M. et al. Incidence, microbiological profile and risk factors of healthcare-associated infections in intensive care units: A 10-year observation in a district hospital in southern Poland. Int. J. Environ. Res. Public. Health 15, 112 (2018).
Pinker, E. Reporting accuracy of rare event classifiers. NPJ Digit. Med. 1, 56 (2018).
Kolikof, J., Peterson, K. & Baker, A. Central Venous Catheter (StatPearls Publishing, 2021).
Ornowska, M., Smithman, J. & Reynolds, S. Locking solutions for prevention of central venous access device complications in the adult critical care population: A systematic review. PLoS ONE 18(10), e0289938 (2023).
Ziegler, M. J., Pellegrini, D. C. & Safdar, N. Attributable mortality of central line associated bloodstream infection: Systematic review and meta-analysis. Infection 43(1), 29–36 (2015).
Xie, W., Li, Y., Meng, X. & Zhao, M. Machine learning prediction models and nomogram to predict the risk of in-hospital death for severe DKA: A clinical study based on MIMIC-IV, eICU databases, and a college hospital ICU. Int. J. Med. Inform. 174, 105049 (2023).
Martin, S. A., Townend, F. J., Barkhof, F. & Cole, J. H. Interpretable Machine Learning for Dementia: A Systematic Review (Alzheimer’s & Dementia, 2023).
Acknowledgements
We express our heartfelt thanks to the Massachusetts Institute of Technology and the Beth Israel Deaconess Medical Center for their significant contributions to the MIMIC project.
Funding
This study was supported by the National Institution of Hospital Administration’s Infection Prevention and Control Research Project in Medical Institutions in China (GY2023055).
Author information
Authors and Affiliations
Contributions
Xiang Su and Quanguo Zhao conceived and designed the study. Xiang Su extracted the data from the MIMIC-IV database (version 2.2). Xiang Su, Xiaogang Sun, and Ling Sun contributed to data analysis and drafted the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Su, X., Sun, L., Sun, X. et al. Machine learning for predicting device-associated infection and 30-day survival outcomes after invasive device procedure in intensive care unit patients. Sci Rep 14, 23726 (2024). https://doi.org/10.1038/s41598-024-74585-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-74585-0