Abstract
Explainability of convolutional neural networks (CNNs) is integral for their adoption into radiological practice. Commonly used attribution methods localize image areas important for CNN prediction but do not characterize relevant imaging features underlying these areas, acting as a barrier to the adoption of CNNs for clinical use. We therefore propose Semantic Exploration and Explainability using a Style-based Generative Adversarial Autoencoder Network (SEE-GAAN), an explainability framework that uses latent space manipulation to generate a sequence of synthetic images that semantically visualizes how clinical and CNN features manifest within medical images. Visual analysis of changes in these sequences then facilitates the interpretation of features, thereby improving explainability. SEE-GAAN was first developed on a cohort of 26,664 chest radiographs across 15,409 patients from our institution. SEE-GAAN sequences were then generated across several clinical features and CNN predictions of NT-pro B-type natriuretic peptide (BNPP) as a proxy for acute heart failure. Radiological interpretations indicated SEE-GAAN sequences captured relevant changes in anatomical and pathological morphology associated with clinical and CNN predictions and clarified ambiguous areas highlighted by commonly used attribution methods. Our study demonstrates SEE-GAAN can facilitate our understanding of clinical features for imaging biomarker exploration and improve CNN transparency over commonly used explainability methods.
Similar content being viewed by others
Introduction
Convolutional neural networks (CNNs), a class of artificial intelligence (AI) algorithms, have become increasingly popular in radiology for their ability to automate complex imaging tasks with high accuracy1. Their strong performance stems from their ability to approximate complex relationships between images and relevant outcomes using a hierarchy of imaging features learned through training2. However, their architectural complexity also makes it exceedingly difficult to explain their predictions, which can act as a barrier to the adoption of these algorithms for clinical practice3.
In response to the need for interpretable artificial intelligence algorithms, several explainable AI (XAI) methods have been proposed4,5, the majority of which are some form of attribution6,7,8,9,10,11,12,13. Attribution maps are static visualizations that highlight areas of an input image important for a CNN’s prediction. Although widely used for their ease of implementation, their trustworthiness has been an ongoing subject of debate14,15,16,17,18. Attribution maps localize areas of importance, but they do not describe the relevant imaging features underlying these highlighted areas (e.g., texture, intensity, morphology). This lack of detail can be detrimental to the explainability of CNNs, especially for diffuse disease processes, where large areas of the input image may be potentially relevant for a CNN’s prediction or where multiple focal observations are spatially colocalized. Therefore, an explainability method capable of both highlighting and characterizing imaging features would be beneficial as a supplement to existing XAI methods.
In recent years, generative adversarial networks (GANs) have emerged as the state-of-the-art approach for synthetic image generation and latent space manipulation for image editing19,20,21. We build on these prior works to propose Semantic Exploration and Explainability using a Style-based Generative Adversarial Autoencoder Network (SEE-GAAN), a GAN-based XAI framework that uses latent space manipulation to generate a sequence of synthetic images that semantically visualizes how clinical features and CNN predictions manifest within medical images. Visual analysis of changes in these synthetic image sequences then facilitates the interpretation of these features, thereby improving explainability.
In this feasibility study, we hypothesized that SEE-GAAN synthetic image sequences can overcome the limitations of commonly used attribution methods by providing dynamic visual explanations of the relationship between clinical features and relevant imaging patterns. We first assess SEE-GAAN’s ability to semantically visualize a variety of clinical features on chest radiographs. We then compare SEE-GAAN to commonly used attribution methods using a CNN developed to infer pulmonary edema severity from chest radiographs as a model for comparison22.
Methods
This retrospective study is Health Insurance Portability and Accountability Act-compliant and institutional review board-approved by the University of California, San Diego Human Research Protections Program with waived requirement for written informed consent; all research was performed in accordance with relevant guidelines/regulations.
SEE-GAAN framework
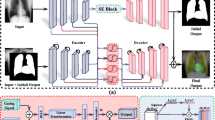
SEE-GAAN (Fig. 1) is a model-agnostic, local and global, generative XAI framework that visualizes how clinical features correlate with imaging patterns. Through autoencoder latent space manipulation, SEE-GAAN generates sequences of synthetic images correlated with these externally observed features. Subsequently, the analysis of changes in these sequences facilitates the interpretation of these features for improved explainability.
Semantic Exploration and Explainability using a Generative 1 Adversarial Autoencoder Network (SEE-GAAN), a framework for visualizing how clinical features and CNN predictions present within medical images. (a) SEE-GAAN autoencoder designed to reconstruct an image from its latent space representation. (b) SEE-GAAN latent space manipulation and synthetic image sequence generation using the SEE-GAAN autoencoder. (c) SEE-GAAN sequence of synthetic images and subtractions for global explanations of clinical and CNN features. Subtraction images visually highlight specific changes in augmented images and facilitate interpretation.
SEE-GAAN autoencoder
The backbone of SEE-GAAN is an adversarially trained style-based autoencoder (Fig. 1a) inspired by the method of Han et al.23, comprising a residual encoder network \(\:E\) and style-based generator \(\:G\) (i.e. decoder network). For an image \(\:{\varvec{I}}_{\varvec{E}},\:E\) first computes a 512-dimension latent representation, \(\:{\varvec{w}}_{E}=E\left({\varvec{I}}_{E}\right)\). \(\:G\) then inverts this transformation by reconstructing the same image using only its latent representation, \(\:{\widehat{\varvec{I}}}_{E}=G\left({\varvec{w}}_{E}\right)\). The latent representation \(\:{\varvec{w}}_{E}\) contains high level semantic information (e.g., anatomy, morphology) about the input image useful for the reconstruction. However, we systematically manipulate the latent representation \(\:{\varvec{w}}_{E}\) to augment the reconstructed image according to an externally observed clinical feature.
SEE-GAAN latent space manipulation for global interpretation
Figure 1b provides an example of SEE-GAAN latent space manipulation to visualize the global relationship between a qualitative clinical feature (e.g., acute heart failure [AHF] status) and the appearance of chest radiographs. Radiographs are first stratified into AHF- and AHF + mutually exclusive groups. Each radiograph is then propagated through the encoder network to extract their corresponding 512-dimension latent representations. Latent representations are then averaged at the feature level, producing a single 512-dimension latent vector for AHF- (\(\:{\stackrel{-}{\varvec{w}}}_{E,AHF-}\)) and AHF+ (\(\:{\stackrel{-}{\varvec{w}}}_{E,\:AHF+}\)), respectively. We then traverse the latent space from AHF- to AHF+ using basic linear interpolation at each latent neuron, producing interpolated latent vectors \(\:{\stackrel{-}{\varvec{w}}}_{E}\). These latent vectors are then propagated through the generator network for reconstruction in image space [\(\:G\left({\stackrel{-}{\varvec{w}}}_{E}\right)\)]. The result is a smooth sequence of synthetic images that visualizes the transition from AHF- and AHF+ in image space.
For semantic visualization of quantitative clinical features, such as laboratory values, radiographs are first ordered by the clinical feature of interest and propagated individually through the encoder network to extract their corresponding latent representations. We then traverse the latent space using regression-based interpolation. Additional details on this procedure are provide in Appendices E1-E2 (supplement).
SEE-GAAN visualization and permutation testing
To facilitate interpretation, we subtract synthetic images from the first image in the sequence and visualize using the spectral color map24, as shown in Fig. 1c. Pixel-wise significance across each subtraction is then determined using permutation testing over 100 permutations, such that the latent vectors are randomly shuffled across images at each iteration, to reduce false positives. Pixel values that are not statistically significant at a prespecified significance level \(\:\alpha\:\) are given a value of zero for the final SEE-GAAN visualization. Details on the permutation testing procedure are included in Appendix E3 (supplement).
SEE-GAAN latent space manipulation for local interpretation
SEE-GAAN can also manipulate the latent space of individual images for local explainability (Fig. 2). Let \(\:{\varvec{w}}_{E}\) represent the SEE-GAAN encoder latent representation of an AHF- patient’s radiograph, and let \(\:{\stackrel{-}{\varvec{w}}}_{E,AHF-}\) and \(\:{\stackrel{-}{\varvec{w}}}_{E,AHF+}\) represent the average latent representations of images corresponding to AHF- and AHF+ status, respectively. We can then linearly shift \(\:{\varvec{w}}_{E}\) so that its corresponding reconstruction contains imaging features consistent with either AHF- or AHF+ using \(\left({\stackrel{-}{\varvec{w}}}_{E,AHF+}-{\stackrel{-}{\varvec{w}}}_{E,AHF-}\right)\), where \(\:{{\varvec{w}}_{E}}^{\left(\lambda\:\right)}\) is the shifted latent vector for a given value of \(\:\lambda\:\ge\:0\) (Fig. 2a). As we increase \(\:\lambda\:\), reconstructions \(\:G\left({{\varvec{w}}_{E}}^{\left(\lambda\:\right)}\right)\) gradually show the appearance of the patient’s radiograph as AHF+. We then visualize their subtractions using the spectral color map and determine pixel-wise significance across these subtractions using permutation testing (Fig. 2b). Generally, latent space manipulation is performed using \(\:{{\varvec{w}}_{E}}^{\left(\lambda\:\right)}={\varvec{w}}_{E}+\:\lambda\:\left({\stackrel{-}{\varvec{w}}}_{E,desired}-{\stackrel{-}{\varvec{w}}}_{E,current}\right)\), where \(\:{\stackrel{-}{\varvec{w}}}_{E,current}\) and \(\:{\stackrel{-}{\varvec{w}}}_{E,desired}\) are the average latent representations of the patients current representation (e.g., AHF-) and desired representation (AHF+), respectively.
SEE-GAAN local explanations of clinical and CNN features. (a) Local explanations are created by linearly shifting the latent vector \(({\varvec{w}}_{E})\) of an image in the direction of the opposing class (e.g., \(\left({\stackrel{-}{\varvec{w}}}_{E,AHF+}-{\stackrel{-}{\varvec{w}}}_{E,AHF-}\right)\) by some weighting factor \(\:\lambda\:\). (b) Varying the weighting factor \(\:\lambda\:\) gradually augments the appearance of reconstructions\(\:G\left({{\varvec{w}}_{E}}^{\left(\lambda\:\right)}\right)\) as the opposing class (e.g., AHF+). The result is an image sequence that visualizes how a clinical or CNN feature presents on a specific patient’s image. Subtraction images visually highlight specific changes in augmented images and facilitate interpretation.
Dataset for feasibility testing
We demonstrate SEE-GAAN on a dataset comprising 26,644 frontal chest radiographs and clinical information across 15,409 deidentified patients from our institution (2017–2020). Radiographs were acquired on machines from several vendors. Laboratory and clinical measurements were collected within 24 h of each corresponding radiograph. Clinical and demographic information included sex, age (years), AHF status, and NT-pro B-type natriuretic peptide (BNPP [pg/mL]) laboratory measurements (Table 1).
SEE-GAAN autoencoder training
Radiographs were resized to 256 × 256 resolution using bicubic spline interpolation and then individually mapped to [-1, 1]. Images were then partitioned at the patient level into an 80/10/10 split, resulting in 21,351/2,602/2,691 images for training/validation/testing. We then trained the SEE-GAAN autoencoder from scratch for one million steps using an Adam optimizer with learning rate 0.001 and a batch size of four while saving weights every 10,000 training steps. During training, augmentations including random translations, rotations, zoom, and brightness adjustments were applied to images with a 0.2 probability. Fréchet Inception Distance (FID) was used to evaluate the quality of synthetic images across training steps25. The training step resulting in the smallest validation FID was selected for testing. Technical details on SEE-GAAN training are included in Appendix E4 (supplement) for reference. Python and Tensorflow-Keras code implementing SEE-GAAN is also made available at github.com/khasenst/see-gaan.
SEE-GAAN clinical feature exploration
Global interpretation
To assess SEE-GAAN’s ability to semantically visualize clinical features, we first generated SEE-GAAN sequences for sex, age, acute heart failure status, and device manufacturer (Carestream Health vs. General Electric [GE]). All visualizations were created using images from the testing set.
Local interpretation
To assess SEE-GAAN’s ability to augment an existing image by a preselected clinical feature for local interpretation, we selected a healthy 27-year-old female patient from the testing set and generated SEE-GAAN sequences for sex (female to male), age (younger to older), and AHF status (AHF- to AHF+).
SEE-GAAN for CNN explainability
We explore SEE-GAAN as a tool for CNN explainability using a CNN developed to infer pulmonary edema severity from chest radiographs (BNPP-CNN) as a test model22. The BNPP-CNN is a ResNet152 regression CNN developed to predict log BNPP as a biomarker for pulmonary edema using 256 × 256 chest radiographs as input.
Global explainability
For global analysis, we generated and compared SEE-GAAN sequences for ground-truth \(\:\text{B}\text{N}\text{P}\text{P}\) and predictions from the BNPP-CNN (\(\:\widehat{\text{B}\text{N}\text{P}\text{P}}\)).
Local explainability
We demonstrate a local analysis using four patients selected from the testing set designated as a true positive, true negative, false positive, and false negative for inflated BNPP (BNPP > 400 pg/mL) by the BNPP-CNN, respectively. For the true positive and true negative patients, we generated local SEE-GAAN sequences as shown in Fig. 2b across \(\:\widehat{\text{B}\text{N}\text{P}\text{P}}\) values. However, to illustrate how SEE-GAAN can be used to troubleshoot inaccurate predictions, we process the false positive and false negative patients in the following manner.
During latent space manipulation, we select \(\:\lambda\:\) such that the BNPP-CNN prediction of \(\:G\left({{\varvec{w}}_{E}}^{\left(\lambda\:\right)}\right)\) is approximately equal to the ground-truth BNPP value (log BNPP within 0.01). That is, we select \(\:\lambda\:\) that augments the image in such a way that the incorrect BNPP-CNN prediction is reversed, providing insight into the types of imaging features sought by the BNPP-CNN to make an accurate prediction. We then subtract this image from the reconstructed image \(\:G\left({\varvec{w}}_{E}\right)\) and visualize for interpretation.
Comparison with commonly used attribution maps
We computed vanilla gradient (VG)6 and gradient-weighted class activation mapping (Grad-CAM)9 attributions maps across the four patients for comparison with SEE-GAAN.
Software and Hardware
SEE-GAAN was developed using Python v3.9.17 on Keras-Tensorflow v2.12.026. Regression-based interpolations were performed using pyGAM v0.8.027 and statistical testing was performed using statsmodels v0.14.128. SEE-GAAN was trained on a single NVIDIA Quadro RTX 8000 graphics processing unit (GPU) with 48 gigabytes of GPU memory. Training duration was ~ 8 days to complete the one million training steps.
Statistical analysis
Clinical information and demographics were summarized using mean ± standard deviation for quantitative variables and frequencies for qualitative variables. Statistical differences in clinical and demographic variables across the training/validation/testing sets were assessed using t-tests for quantitative variables and chi-squared tests for qualitative variables. Statistical significance was evaluated using a type I error rate of \(\:\alpha\:=\)5%. All SEE-GAAN sequences were reviewed by two board-certified radiologists for interpretation (LH, AH).
Results
Overview of dataset
A summary of clinical and demographic information is shown in Table 1. Average overall age was 63 years and 44% (6792/15409) of the cohort were female. Average BNPP measurements were 4904 pg/mL and approximately 16% (4193/26667) of radiographs were from patients with AHF. The majority of radiographs were acquired on Carestream Health (36% [9650/26667]) or GE Healthcare machines (9526/26667). There were no significant differences in clinical or demographic variables across the training/validation/testing sets (p-values > 0.05).
SEE-GAAN Clinical Feature Exploration
Global interpretation
SEE-GAAN sequences for several clinical features are shown in Fig. 3. The SEE-GAAN sequence for sex shows a decrease in breast soft tissue density in the lower chest and increased density elsewhere in the chest wall, which may reflect increased muscle and bone density in males. For age, as patients approach 55 years, SEE-GAAN captures increased attenuation in the bilateral flanks which could reflect a change in body habitus. Throughout the aging process, there is a decrease in attenuation of the chest wall, possibly due to reduced muscle/bone mass. For AHF status, SEE-GAAN sequences indicate increased size of the cardiomediastinal silhouette and central pulmonary vasculature for AHF+. SEE-GAAN also captures differences in device manufacturer, where we observe increased attenuation in the chest wall for GE devices, possibly due to differences in dynamic range post-processing or device settings.
Global SEE-GAAN sequences visualizing the overall 1 presentation of clinical features in chest radiographs. (a) The SEE-GAAN sequence for sex shows decreased breast soft tissue density but increased chest wall density elsewhere for males, possibly reflecting increased muscle or bone density. (b) Age captures increases and decreases in the attenuation of the flanks and decreases in chest wall soft tissue density. (c) Patients with AHF exhibit increased size of the cardiomediastinal silhouette and central pulmonary vasculature. (d) GE devices show increased attenuation throughout the chest wall for GE devices.
Local interpretation
A SEE-GAAN reconstruction from latent space and local SEE-GAAN sequences for a healthy 27-year-old female are shown in Fig. 4. As expected, the native image and reconstruction are largely identical with exception to minor differences in imaging features due to the 512-dimensional latent space compression. Sequence interpretations for sex, age, and acute heart failure are consistent with the global interpretations from Figs. 2and 3 but are shown on a specific patient’s radiograph to facilitate local interpretation.
Reconstruction and local SEE-GAAN sequences for a healthy 1 27-year-old female across several clinical features. (a) The reconstruction is largely identical to the native image with exception to minor differences in imaging features due to the 512-dimensional latent space compression. (b)–(d) Local sequences emphasize the same imaging features as the global sequences, except visualize the presentation of these features on a specific patient’s radiograph
SEE-GAAN for CNN explainability
Global explainability
Global sequences for BNPP and \(\:\widehat{\text{B}\text{N}\text{P}\text{P}}\) are shown in Fig. 5. For elevated BNPP patients, SEE-GAAN sequences indicate increased size of the cardiomediastinal silhouette and pulmonary vasculature and decreased chest wall soft tissue density. The SEE-GAAN sequence for \(\:\widehat{\text{B}\text{N}\text{P}\text{P}}\) suggests the BNPP-CNN heavily focuses on the size of the cardiomediastinal silhouette and the chest wall soft tissue density to make its predictions, with somewhat less emphasis on vascularity.
Comparison of global SEE-GAAN sequences for (a) ground-1 truth BNPP and (b) BNPP-CNN 2 predictions \(\:\widehat{\text{B}\text{N}\text{P}\text{P}}\) for CNN explainability. Increases in BNPP are associated with increased size of the cardiomediastinal silhouette and pulmonary vasculature and decreased chest wall density. The SEE GAAN sequence for \(\:\widehat{\text{B}\text{N}\text{P}\text{P}}\) suggests the BNPP-CNN heavily focuses on the size of the cardiomediastinal silhouette and chest wall soft tissue density to make its predictions, with somewhat less emphasis on vascularity.
Local explainability
Local SEE-GAAN sequences for the true positive and negative patients (Fig. 6a) are consistent with the global interpretations from Fig. 5, but are shown on specific patients’ radiographs to facilitate local interpretation. For the false positive and false negative patients (Fig. 6b), we observe the BNPP-CNN expects a smaller or larger cardiomediastinal silhouette and higher or lower chest wall soft tissue density, respectively, for a correct prediction.
Local SEE-GAAN sequences for CNN explainability and troubleshooting. (a) Sequence for a 53 year-old male correctly classified as having elevated BNPP (>400). (b) Sequence for a 51 year-old male correctly classified as not having elevated BNPP (<400). Both sequences are consistent with the global interpretations of cardiomediastinal silhouette, pulmonary vasculature, and decreased chest wall density. Sequences further characterize the ambiguous regions highlighted by commonly used attribution methods. (c)–(d) CNN troubleshooting using SEE-GAAN on a (c) false positive case (83 year old female) and (d) false negative case (51 year-old male). For both (c)–(d), we augment patients’ images until the BNPP-CNN correctly predicts their ground-truth BNPP values. We observe that the BNPP-CNN expects differences in the size of the cardiomediastinal silhouette and attenuation of the chest wall soft tissue to make correct predictions
Comparison commonly used attribution maps
VG and Grad-CAM attribution maps and local SEE-GAAN sequences for four patients are shown in Fig. 6. VG shows scattered activations centered on the heart and lungs but the interpretation of these activations remains unclear. In contrast, Grad-CAM shows more localized areas of attention spanning the heart, lungs, and some peripheral areas outside of the lungs. Although both methods highlight anatomical features known to be correlated with BNPP, the specific imaging characteristics underlying these areas of attention, such as heart size, remain ambiguous. From the local SEE-GAAN sequences, we determine the BNPP-CNN is focusing on heart size and body composition to make its predictions.
Discussion
In this feasibility study, we proposed a GAN-based XAI framework that uses autoencoder latent space manipulation to generate synthetic image sequences that semantically visualize clinical features. We applied our proposed SEE-GAAN framework to a variety of clinical features and showed that changes in their corresponding sequences provide an understanding of the presentation of these features within medical images. We further showed that SEE-GAAN produces both local and global visualizations for the analysis of clinical features and CNN predictions. Finally, we showed SEE-GAAN sequences also clarify ambiguities in the interpretation of popular explainability visualizations (i.e. VG maps and Grad-CAMs) by further characterizing the precise imaging patterns underlying their most salient areas through dynamic visualization.
As an XAI framework, SEE-GAAN has several favorable properties. SEE-GAAN uses dynamic visualization to elicit interpretation of clinical features and CNN predictions. Dynamic visualization enables feature interpretation through perceived changes in intensity, morphology, and texture within a synthetic image sequence. This is especially useful for interpreting features associated with diffuse disease processes or spatially colocalized focal observations, as we observed from several of the SEE-GAAN sequences. SEE-GAAN also does not depend on any specific type of model or algorithm (i.e. model-agnostic) and can be applied to any type of feature (e.g., CNN or clinical, quantitative or qualitative) for interpretation. In addition, SEE-GAAN provides both global and local interpretations of clinical features.
In comparison, attribution maps provide static visualizations that only localize regions of interest and therefore do not provide an explicit understanding of the imaging characteristics underlying their highlighted areas. Although attributions maps are model-agnostic, open-source implementations are often exclusive to neural networks and are not agnostic to other types of algorithms. Attribution maps provide both local and global visualizations, however, they typically require anatomical structures in images to be relatively coregistered for aggregation and global visualization. Nevertheless, in practice, we recommend using attribution maps in conjunction with the proposed SEE-GAAN sequences to synergize interpretation. Attribution maps can be used to first localize relevant areas of attention, while subsequently using SEE-GAAN to uncover the underlying anatomical or pathophysiological nature of this attention.
While we propose the use of GANs to improve CNN explainability, we acknowledge that other GAN-based alternatives have been proposed for this same purpose. Seah et al. used a GAN to synthesize chest radiographs with and without disease using permuted features from a CNN trained to predict B-type natriuretic peptide as a marker for congestive heart failure29. Another study by Hasenstab et al. proposed a GAN-based framework to interpret the features learned by a CNN trained for a similar task30. Both algorithms’ visual explanations were consistent with the interpretations provided by our SEE-GAAN sequences for BNPP. However, these methods are either not model-agnostic or do not provide both local and global explanations of clinical features. Moreover, they do not use style-based generators, which enable smoother latent space manipulation for dynamic visualization.
Other studies have proposed style-based generators to synthesize images across other modalities and anatomical regions. Fetty et al. explored the idea of latent space manipulation on CT and MR pelvic images31. Schutte et al. and Ren et al. proposed using latent space manipulation of style-based generators to explore musculoskeletal radiographs, pathology slides, and mammography32,33. Hong et al. extended this idea to 3D convolutions to study brain MR images34. We add to these works by proposing a framework that facilitates local and global interpretation of clinical features, which can be used in combination with commonly used XAI methods.
Outside of generative and commonly used gradient- or perturbation-based attribution methods, there are several alternative approaches for explainable AI in medical imaging35,36,37, including attention networks38 and feature analytic methods37,39,40. In contrast to attribution maps, which are model agnostic, post-hoc visualizations for explainability, attention networks use modules directly incorporated into the architecture, which offer a level of explainability. Attention modules act as feature selectors during training, preserving features important for a given CNN task and producing attention maps that are visualized for interpretation. However, similar to attribution, attention maps only localize areas of importance and do not describe the relevant imaging features underlying these highlighted areas. Moreover, CNN architectures typically contain thousands of feature maps throughout the network, making visual exploration of this feature space less tractable. SEE-GAAN addresses these limitations by generating a single dynamic visualization to elicit feature interpretation and applies to both CNN predictions and standalone clinical features.
Similar to SEE-GAAN’s latent space, low-dimensional embeddings have been used to visualize the feature space of CNNs to explain their predictions. Feature embeddings are often visualized across the distribution of output classes to identify challenging cases or clusters for automated assessment. Common techniques for feature analysis include principal components analysis, t-SNE, and UMAP37. Concept methods39,40 incorporate user-defined concepts during training in a supervised manner to encourage networks to learn high-level human-friendly features. The incorporation of these high-level features during training have achieved competitive accuracy with conventional end-to-end models while enabling human-friendly interpretation of the model features. In contrast, SEE-GAAN uses the StyleGAN framework for its autoencoder, which learns a disentangled representation of high-level image features for its image reconstruction without supervision. Despite its unsupervised training procedure, SEE-GAAN is still able to capture the relationship between human-friendly features and their presentation on medical images through latent feature interpolation, which maps human-friendly features (e.g., heart failure) to SEE-GAAN latent space and subsequently image space.
Our study has limitations. Due to computational restrictions during training, chest radiographs were downsampled to 256 × 256 resolution, thus reducing the visibility of finer imaging features (e.g., Kerley b lines). SEE-GAAN implementations at native resolution to study more granular imaging features are reserved for future research. Clinical observations in our dataset were also limited. Future studies should investigate SEE-GAAN’s ability to capture a variety of other conditions, such as fibrosis and pneumothoraces. In addition, although SEE-GAAN can be applied to features from other CNNs for interpretation and explainability, the current iteration of SEE-GAAN only applies to the modality on which it was trained (i.e. chest radiographs). We reserve the development of a conditional version of SEE-GAAN applicable to multiple modalities and anatomical locations as a direction for future research. More broadly, although our study emphasizes the importance of CNN explainability to facilitate the adoption of these algorithms in clinical practice, we note that many other factors contribute to the underuse of AI for clinical purposes. Therefore, in addition to explainability, future studies aiming to produce algorithms for clinical use should holistically consider legal requirements, algorithm traceability, and rigorous evaluation by domain experts to ensure their adoption41,42,43.
In conclusion, our proposed SEE-GAAN framework facilitates the interpretation of clinical features and improves CNN explainability by providing an understanding of the specific imaging characteristics highlighted by popular attribution methods. As a result, SEE-GAAN visualizations can help facilitate the adoption of AI algorithms in clinical practice and can be used as a tool for clinical data mining and biomarker discovery.
Data availability
The datasets generated and/or analyzed during the current study are not publicly available due to the possibility of patients’ identification from clinical and imaging patterns but are available from the corresponding author on reasonable request.
Code availability
Python and Tensorflow-Keras code implementing SEE-GAAN available at github.com/khasenst/see-gaan.
References
Najjar, R. & Redefining Radiology A review of artificial intelligence integration in medical imaging. Diagnostics. 13(17), 2760. https://doi.org/10.3390/diagnostics13172760 (2023).
Hasenstab, K. Convolutional neural networks and their applications in medical imaging: a primer for mathematicians. AMS Notices. 70 https://doi.org/10.1090/noti2598 (2023).
Reyes, M. et al. On the interpretability of artificial intelligence in radiology: challenges and opportunities. Radiol. Artif. Intell. 2(3), e190043. https://doi.org/10.1148/ryai.2020190043 (2020).
de Vries, B. M. et al. Explainable artificial intelligence (XAI) in radiology and nuclear medicine: a literature review. Front. Med. 10, 1180773. https://doi.org/10.3389/fmed.2023.1180773 (2023).
Borys, K. et al. Explainable AI in medical imaging: an overview for clinical practitioners – beyond saliency-based XAI approaches. Eur. J. Radiol. 162, 110786. https://doi.org/10.1016/j.ejrad.2023.110786 (2023).
Simonyan, K., Vedaldi, A. & Zisserman, A. Deep inside convolutional networks: Visualising image classification models and saliency maps. Preprint at (2014). https://arxiv.org/abs/1312.6034
Zeiler, M. D. & Fergus, R. Visualizing and understanding convolutional networks. ECCV Lecture Notes Comput. Sci. https://doi.org/10.1007/978-3-319-10590-1_53 (2014).
Springenberg, J. T., Dosovitskiy, A., Brox, T. & Riedmiller, M. Striving for simplicity: The all convolutional net. Preprint at (2015). https://arxiv.org/abs/1412.6806
Selvaraju, R. R. et al. Grad-CAM: Visual explanations from deep networks via gradient-based localization. IEEE International Conference on Computer Vision (ICCV). (2017). https://doi.org/10.1109/ICCV.2017.74
Smilkov, D., Thorat, N., Kim, B., Viégas, F. & Wattenberg, M. SmoothGrad: Removing noise by adding noise. Preprint at (2017). https://arxiv.org/abs/1706.03825
Shrikumar, A., Greenside, P. & Kundaje, A. Learning important features through propagating activation differences. ICML’17: Proceedings of the 34th International Conference on Machine Learning. 70, 3145–3153; (2017). https://doi.org/10.5555/3305890.3306006
Sundararajan, M., Taly, A. & Yan, Q. Axiomatic attribution for deep networks. ICML’17: Proc. 34th Int. Conf. Mach. Learn. 70, 3319–3328. https://doi.org/10.5555/3305890.3306024 (2017).
Kapishnikov, A., Bolukbasi, T., Viegas, F. & Terry, M. XRAI: better attributions through regions. IEEE/CVF Int. Conf. Comput. Vis. (ICCV). 4947-4956 https://doi.org/10.1109/ICCV.2019.00505 (2019).
Kindermans, P. J. et al. The (un)reliability of saliency methods in explainable AI: interpreting, explaining and visualizing deep learning. 11700, 267–280 ; (2019). https://doi.org/10.1007/978-3-030-28954-6_14
Boccignone, G., Cuculo, V. & D’Amelio, A. Problems with saliency maps. Image Anal. Processing – ICIAP. 11752 https://doi.org/10.1007/978-3-030-30645-8_4 (2019).
Saporta, A. et al. Benchmarking saliency methods for chest X-ray interpretation. Nat. Mach. Intell. 4, 867–878. https://doi.org/10.1038/s42256-022-00536-x (2022).
Arun, N. et al. Assessing the trustworthiness of saliency maps for localizing abnormalities in medical imaging. Radiol. Artif. Intell. 3(6), e200267. https://doi.org/10.1148/ryai.2021200267 (2021).
Zhang, J. et al. Revisiting the trustworthiness of saliency methods in radiology AI. Radiol. Artif. Intell. 6(1), e220221. https://doi.org/10.1148/ryai.220221 (2023).
Karras, T., Laine, S. & Aila, T. A style-based generator architecture for generative adversarial networks. IEEE Trans. Pattern Anal. Mach. Intell. 43, 4217–4228. https://doi.org/10.1109/TPAMI.2020.2970919 (2021).
Karras, T. et al. Analyzing and improving the image quality of StyleGAN. IEEE/CVF Conf. Comput. Vis. Pattern Recognit. (CVPR). 8107-8116 https://doi.org/10.1109/CVPR42600.2020.00813 (2020).
Karras, T. et al. Alias-free generative adversarial networks. Advances Neural Inform. Process. Systems. 34, 852–863. https://doi.org/10.48550/arXiv.2106.12423 (2021).
Huynh, J. et al. Deep learning radiographic assessment of pulmonary edema: optimizing clinical performance, training with serum biomarkers. IEEE Access. 10, 48577–48588. https://doi.org/10.1109/ACCESS.2022.3172706 (2022).
Han, L. et al. AE-StyleGAN: Improved training of style-based auto-encoders. IEEE/CVF Winter Conference on Applications of Computer Vision (WACV). 955–964; (2022). https://doi.org/10.1109/WACV51458.2022.00103
Hunter, J. T. & Matplotlib A 2D graphics environment. Computing Science Engineering. 9(3), 90–95. https://doi.org/10.1109/MCSE.2007.55 (2007).
Heusel, M., Ramsauer, H., Unterthiner, T., Nessler, B. & Hochreiter, S. GANs trained by a two time-scale update rule converge to a local nash equilibrium. Advances in Neural Information Processing Systems. Preprint at (2017). https://arxiv.org/abs/1706.08500
Abadi, M. et al. TensorFlow: A system for large-scale machine learning. Proc. 12th USENIX Conf. Operating Syst. Des. Implement. 12, 265–283. https://doi.org/10.5555/3026877.3026899 (2016).
Serven, D. & Brummitt, C. pyGAM: generalized additive models in Python. Zenodo. https://doi.org/10.5281/zenodo.1208723 (2018).
Seabold, S. & Perktold, J. Statsmodels: Econometric and statistical modeling with python. Proc. 9th Python Sci. Conf. 9, 57–61. https://doi.org/10.25080/Majora-92bf1922-011 (2010).
Seah, J. C. Y., Tang, J. S. N., Kitchen, A. & Gaillard, F. D. A. F. chest radiographs in congestive heart failure: visualizing neural network learning. Radiology. 290 (2), 514–522. https://doi.org/10.1148/radiol.2018180887 (2019).
Hasenstab, K. A. et al. Feature interpretation using generative adversarial networks (FIGAN): a framework for visualizing a CNN’s learned features. IEEE Access. 11, 5144–5160. https://doi.org/10.1109/ACCESS.2023.3236575 (2022).
Fetty, L. et al. Latent space manipulation for high-resolution medical image synthesis via the StyleGAN. Z. Med. Phys. 30 (4), 305–314. https://doi.org/10.1016/j.zemedi.2020.05.001 (2020).
Schutte, K., Moindrot, O., Hérent, P., Schiratti, J. B. & Jégou, S. Using StyleGAN for visual interpretability of deep learning models on medical images. Preprint at (2021). https://arxiv.org/abs/2101.07563
Ren, Z., Yu, S. X., Whitney, D. & Controllable medical image generation via GAN. IS&T Int. Symp. Electron. Imaging. 33, art00003. https://doi.org/10.2352/issn.2470-1173.2021.11.hvei-112 (2021).
Hong, S. et al. 3D-StyleGAN: A style-based generative adversarial network for generative modeling of three-dimensional medical images. DGM4MICCAI DALI 2021 Lecture Notes in Computer Science. 13003; (2021). https://doi.org/10.1007/978-3-030-88210-5_3
Singh, A., Sengupta, S. & Lakshminarayanan, V. Explainable deep learning models in medical image analysis. J. Imaging. 6(6), 52. https://doi.org/10.3390/jimaging6060052 (2020).
Tjoa, E. & Guan, C. A survey on explainable artificial intelligence (XAI): toward medical XAI. IEEE Trans. Neural Networks Learn. Syst. 32(11), 4793–4813. https://doi.org/10.1109/TNNLS.2020.3027314 (2021).
Fuhrman, J. D. et al. A review of explainable and interpretable AI with applications in COVID-19 imaging. Med. Phys. 49(1), 1–14. https://doi.org/10.1002/mp.15359 (2022).
Cheng, J. et al. ResGANet: residual group attention network for medical image classification and segmentation. Med. Image. Anal. 76, 102313. https://doi.org/10.1016/j.media.2021.102313 (2022).
Kim, B. et al. Interpretability beyond feature attribution: Quantitative testing with concept activation vectors (tcav). International Conference on Machine Learning. 2673–2682; (2018). https://doi.org/10.48550/arXiv.1711.11279
Koh, P. W. et al. Concept bottleneck models. International Conference on Machine Learning. 5338–5348; (2020). https://doi.org/10.48550/arXiv.2007.04612
Pagallo, U. et al. The underuse of AI in the health sector: opportunity costs, success stories, risks and recommendations. Health Technol. 14, 1–14. https://doi.org/10.1007/s12553-023-00806-7 (2024).
Stoger, K., Schneeberger, D. & Holzinger, A. Medical artificial intelligence: the European legal perspective. Commun. ACM. 64(11), 34–36. https://doi.org/10.1145/3458652 (2021).
Sorantin, E. et al. The augmented radiologist: artificial intelligence in the practice of radiology. Pediatr. Radiol. 52(11), 2074–2086. https://doi.org/10.1007/s00247-021-05177-7 (2022).
Author information
Authors and Affiliations
Contributions
KH – developed SEE-GAAN algorithm, performed data analysis, created tables and figures, drafted and revised manuscript; LH – provided radiological interpretations and revised manuscript; NC – performed data analysis and implemented interpolation procedures; AH – clinical and imaging data collection, provided radiological interpretations, and revised manuscript.
Corresponding author
Ethics declarations
Competing interests
Dr. Hasenstab, Dr. Hahn, and Mr. Chao declare no potential conflicts of interest. Dr. Hsiao receives research grant support from GE healthcare, Bayer AG, and Bracco. He is a consultant for Canon and was a cofounder of Arterys Inc, which has been acquired by Tempus AI.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hasenstab, K.A., Hahn, L., Chao, N. et al. Simulating clinical features on chest radiographs for medical image exploration and CNN explainability using a style-based generative adversarial autoencoder. Sci Rep 14, 24427 (2024). https://doi.org/10.1038/s41598-024-75886-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-75886-0