Abstract
Maternal use of e-cigarette (e-cig) aerosols poses significant risks to fetal brain development, potentially increasing susceptibility to neurodevelopmental disorders in later life. However, the underlying mechanisms remain incompletely understood. This study aimed to understand the effects of fetal e-cig exposure on DNA methylome and transcriptomic changes in the neonatal brain. Pregnant rats were exposed to e-cig aerosols, and neonatal brains (5 males and 5 females/group) from both control and e-cig-exposed groups were used for experimental analysis. Results indicated that prenatal e-cig exposure altered site-specific DNA methylation patterns at both CpG and CH (non-CpG) sites, predominantly in intergenic and intronic regions, with sex dimorphism in methylation and gene expression changes. Gene ontology analysis revealed that e-cig exposure not only affected neuron projection development and axonogenesis but also altered pathways related to neurodegeneration and long-term depression. These findings provide novel insights into the dynamic changes of CpG and CH methylation induced by e-cig exposure, underscoring the susceptibility of the developing brain to maternal e-cig exposure and its potential implications for developmental disorders and neurodegenerative diseases later in life.
Similar content being viewed by others
Introduction
Electronic cigarettes (e-cig) have emerged as devices designed to deliver a nicotine-containing aerosol by heating a solution typically composed of nicotine, propylene glycol or glycerol, and flavoring agents. Over the past decade, the prevalence of e-cig use has surged1,2,3, particularly among demographics such as young adults and reproductive women, surpassing conventional cigarette use during pregnancy4,5,6,7. This trend may arise from the perception among pregnant smokers that e-cigs are safer alternatives, often used as aids to quit smoking. However, the rapid increase in e-cig use during pregnancy poses significant risks to fetal development and increases the risk of neural and cardiovascular diseases later in life. Numerous studies have shed light on the adverse effects of nicotine on perinatal brain development and the autonomic nervous system, emphasizing a concerning shift in the balance of physiological processes8,9,10.
Research efforts, including investigations conducted by our group and others, have underscored the heightened vulnerability of the developing embryo and fetus to the deleterious effects of e-cigarette exposure11,12,13. E-cigarette exposure exhibits greater cytotoxicity in embryonic stem cells compared to adult cells, impacting early stage of embryogenesis and angiogenesis12. Moreover, e-cigarette exposure during early pregnancy has been linked to impaired embryo implantation, inhibited fetal growth13, resulting in fetal programming of neurobehavioral dysfunction14,15, and the induction of a brain hypoxic-ischemic sensitive phenotype in postnatal life11. Collectively, these findings further emphasize the urgent need to understand the underlying molecular mechanisms.
Building upon knowledge garnered from epidemiologic and animal studies on adverse stresses during pregnancy, particularly maternal cigarette smoking, recent studies have revealed the role of epigenetic mechanisms, notably DNA methylation, in mediating the long-term effects of perinatal stresses on health and disease later in life. Children exposed to maternal cigarette smoking exhibit alterations in DNA methylation patterns16,17,18, and interventions targeting DNA methylation have shown promise in mitigating the adverse effects19,20,21. Our recent study has revealed that prenatal exposure to e-cigarettes significantly disrupts the expression of autophagy-related gene 5 (ATG5) through CpG hypermethylation in its promoter region, leading to an elevated risk of neonatal brain ischemic injury11. However, whether prenatal e-cigarette exposure influences the genomic DNA methylome and transcriptomic profile in the neonatal brain remains unexplored.
In this study, we sought to investigate the impact of prenatal e-cigarette exposure on the DNA methylome and transcriptomic profiles in the developing brain using a pregnant rat model. Through RNA-sequencing analysis and reduced representation bisulfite sequencing (RRBS), we determined the effect of maternal e-cig exposure on (epi)transcriptomic biomarkers in the neonatal brain, implicated in e-cig-mediated developmental programming of neurodevelopmental disease. Our findings offer novel insights into the epigenetic mechanisms contributing to the adverse effects of fetal e-cigarette exposure, thereby suggesting potential strategies to address its impact on offspring health.
Results
Overview of data and study design
To investigate the DNA methylome and transcriptomic changes induced by maternal e-cig exposure in utero, we designed an experiment outlined in Fig. 1. Pregnant rats were randomly divided into two groups: the control group and the e-cigarette aerosol exposed group. Briefly, pregnant dams were exposed to either normal air (control) or e-cig aerosol vaporing through identical apparatuses during gestational day 4 through day 20 (E4-E20). A total of 32 offspring (13 males, 19 females) were born from the control group consisting of 3 litters, while 22 offspring (8 males, 14 females) were born from the e-cig exposed group of 2 litters.
Experimental design and data analysis workflow. Pregnant Sprague–Dawley rats were exposed to e-cigarette aerosol vaping (E-cig) or normal air (Control) from gestational day 4 (E4) to 20 (E20). At postnatal day 7 (P7), ten pups (5 males and 5 females) were randomly selected from each group of the control and e-cig-exposed offspring. Their brains were collected for RNA/DNA extraction and subsequent construction of RRBS methylation and RNA-seq libraries. The DNA methylome and transcriptomic profiles between offspring exposed to maternal e-cig aerosol and those from control dams were obtained and compared between these two groups.
At postnatal day 7 (P7), we randomly selected 10 pups (5 males and 5 females) from each group of the control and e-cig-exposed animals for the experimental analysis. Their brains were collected and frozen at − 80 °C until analysis for RNA/DNA extraction and subsequent construction of RRBS methylation and RNA-seq libraries. The study design enabled us to compare the DNA methylome and transcriptomic profiles between offspring exposed to maternal e-cigarette aerosol and those from control dams, providing insights into the molecular mechanisms underlying the effects of prenatal e-cigarette exposure on brain development.
Maternal e-cigarette exposure alters site-specific DNA methylation patterns in the neonatal brain
To elucidate the global and site-specific DNA methylation changes induced by e-cigarette exposure, we conducted an analysis of RRBS DNA-seq methylome. We obtained an average of 21 million raw reads (Table S1), resulting in an average of 1.6 million mapped CpG sites with minimum of 10 coverage. To ensure robust analysis, we analyzed only those CpG sites that were overlapping in females and males, yielding a total of 1,037,247 CpG sites in females and 1,108,515 CpG sites in males. To assess the extent of CpG methylation levels, we plotted the mean beta value (ranging between 0–1) across control and e-cigarette samples for males and females separately. This revealed a bimodal distribution, with the majority of CpGs either unmethylated (beta value < 0.1) or fully methylated (beta value > 0.9) (Fig. 2a). To understand the global differences between control and e-cigarette samples, we performed principal component analysis (PCA) on male and female samples separately (Supplementary Fig. 1a-b). We observed a global separation between control and e-cig samples, although two e-cig samples appeared closer to control samples. Genetic analysis revealed these two offspring were from different litter, indicating potential genetic differences.
CpG methylation landscape in control and e-cigarette (e-cig) exposed neonatal brains. (a) Distribution of methylation levels for each CpG site in male and female offspring. Methylation levels are represented by beta values between 0 to 1. Beta values between 0 to 1 are grouped into 10 bins. (b) Distribution of differentially methylated CpGs based on genomic regions in male and female offspring. (c-d) Heatmaps show methylation levels of differentially methylated CpGs on male rats (c) and female rats (d). (e–f) Bar plots show enriched gene ontology (biological processes) terms associated with genes overlapping differentially methylated CpGs in male rats (e) and female rats (f).
To understand the distribution of methylation in the genome, we plotted methylation values across genes and observed lower methylation levels at gene transcription start sites (TSSs), which gradually increased towards the 3’ end of genes (Supplementary Fig. 1c-d). Promoters overlapping with CpG islands (CGIs) exhibited lower methylation levels at TSSs compared to non-CGI promoters, consistent with the known correlation between DNA methylation and CG density. CpG islands within gene bodies (intragenic CGIs) displayed higher methylation levels compared to promoter CGIs (Supplementary Fig. 1e-f).
Given the similar global distribution of methylation between control and e-cig samples (Fig. 2a, Supplementary Fig. 1c-f), we focused on identifying site-specific methylation changes associated with key genes involved in brain development and function. To this end, we performed differential methylation analysis and identified 2573 differentially methylated CpGs (DmCs) in males (Table S2) and 2369 DmCs in females (Table S3), with minimal overlap (n = 25 CpGs) between sexes (Supplementary Fig. 1 g-h). These DmCs were predominantly located in intergenic and intronic regions (Fig. 2b). Equal proportions of DmCs were proportionally hyper and hypomethylated in both male and female samples (Fig. 2c-d). To understand which biological processes might be affected by DmCs, we extracted overlapping genes and identified many genes associated with neuronal development. Gene ontology (GO) analysis of overlapping genes associated with DmCs revealed enrichment of neurological terms such as axon guidance and regulation of neuron projection nervous system development in both male and female samples (Fig. 2e–f). Despite minimum overlap of methylated sites between male and female samples, the enrichment of dysregulated biological processes was similar between them. This comprehensive analysis highlights the complex and sex-specific alterations in DNA methylation patterns induced by maternal e-cig exposure, with potential implications for neurodevelopmental processes.
Maternal e-cig exposure changes site-specific CH methylation patterns in the neonatal brain
Recent studies have underscored the widespread presence of CH (non-CpG, representing CpA, CpC, and CpT) methylation during brain development22, prompting us to investigate the extent of CH methylation at P7 neonatal brains and its modulation by e-cig exposure. We identified 4.92 and 4.63 million consensus CH sites in male and female samples, respectively. Analysis of methylation levels revealed that the majority of CH sites were unmethylated (Fig. 3a), consistent with previous observations in other models22,23. As shown in Fig. 3b, the methylated CH (mCH) sites comprised less than 1% of CH methylation across all groups. Focusing on mCH sites, we performed differential analysis and identified 271 differentially methylated CH (DmCH) sites in male (Table S4) and 303 DmCH sites in female (Table S5) (Supplementary Fig. 2). Only six differentially CH methylated sites overlapped between the male and female offspring, which include chr2:237,157,925 (intronic: DKK2), chr3:82,426,834 (intergenic), chr4:171,983,593 (intronic: Slc15a5), chr4:55,384,644 (intronic: Grm8), chr5:76,888,018 (intronic: RGD1310951) and chr6:131,702,022 (intergenic). These sites were predominantly located in intergenic and intronic regions (Fig. 3c), with comparable distributions of hypermethylated and hypomethylated CH sites observed in both male (Fig. 3d) and female (Fig. 3e). Gene ontology analysis of genes associated with DmCH sites revealed enrichment of numerous neurological terms, including synapse assembly, dendrite morphogenesis, neuron projection morphogenesis, glutamate receptor signaling pathway, and axon guidance, which were also mainly conserved between male (Fig. 3f) and female (Fig. 3g) samples. These findings highlight the impact of maternal e-cig exposure on site-specific CH methylation patterns in the developing brain, with potential implications for neurodevelopmental processes.
Differential CH methylation landscape in control and e-cigarettes (e-cig) exposed neonatal brains. (a) Distribution of methylation levels of 4.92 million (in male) and 4.63 million (female) CH sites. Methylation levels range between 0 (unmethylated) to 1 (methylated). Methylation values between 0 to 1 are grouped into 5 bins. Most CH sites are unmethylated. (b) Zoomed view shows the top 1% of CH methylation levels. Zoomed view shows about 0.02% of CH sites are methylated. (c) Distribution of differentially methylated CH sites based on genomic regions in male and female offspring. (d-e) Heatmaps show the methylation levels of differentially methylated CHs on male (d) and female (e) offspring. (f-g) Enriched gene ontology (biological processes) associated with differentially methylated CHs overlapping genes in male (f) and female (g) offspring.
Effects of maternal e-cig exposure on gene expression in the neonatal brain
To gain a comprehensive understanding of gene expression changes induced by e-cig exposure, we conducted RNA-seq differential analysis using the DESeq2 approach. An average of 26 million raw reads were produced for each sample, where percentage of reads aligned to the transcriptome ranged from 71.1% to 76.1% (Table S1). We used DESeq2 and identified 67 differentially expressed genes (DEGs) in male (Table S6) and 271 DEGs in female samples (Table S7). The heatmaps top DEGs between the control and e-cig exposed groups are shown in Fig. 4a for male and Fig. 4b for female offspring. The e-cig-induced DEGs in both male and female offspring had only 7 overlapping genes (Hnrnpk, Hba-a2, Megf8, Fndc1, H2bc12, AABR07000398.1 and AY172581.9), where 6 genes (Hba-a2, Megf8, Fndc1, H2bc12, AABR07000398.1 and AY172581.9) were upregulated in both male and female samples. Hnrnpk gene was downregulated in male but upregulated in female samples. Gene ontology analysis revealed widespread enrichment of neurological terms, including regulation of synaptic transmission, axon guidance, synapse assembly, axonogenesis, and regulation of neuron projection development, among differentially expressed genes (Fig. 4c-d). Despite the minimal overlap of differentially expressed genes, enriched pathways showed similarity between male and female samples.
Differential gene expression due to e-cig exposure in male and female offspring. (a-b) Heatmaps show differentially expressed genes (DEGs) between control and e-cig exposure in male (a) and female (b) offspring. (c-d) Bar plot shows enriched gene ontology (biological processes) associated with DEGs between male (c) and female (d) offspring.
To explore the global relationships between promoter DNA methylation and gene expression changes in response to maternal e-cig exposure in utero, we focused the analyses only on differentially expressed genes. We correlated promoter DNA methayltion level (defined by average methayltion of all CGs in 1000 bases around TSSs) with their gene expression levels. The majority of correlation coefficient ranged between -0.5 to 0.5 suggesting no correlation (Fig. 5a-b). Only few genes had significant negative correlation (cor < -0.5 and p-value < 0.05) between promoter methylation and gene expression in male and female offspring (Fig. 5a-b). Visualization of gene expression and promtoer methylation of significant genes revealed strong negative coorealtion in some genes (Fig. 5c-d), while in some genes there was little change in promoter methylation. This is expected because we observed no abberant promoter methayltion induced by e-cig (Supplementary Fig. 1c-d), as in case of cancer. This revealed that e-cig induced promoter DNA methylation changes had minimum impact on gene expression.
Correlation of gene expression and DNA methylation. (a-b) Bar plots show correlation values between differentially expressed genes and promoter DNA methylation levels in male (a) and female (b) offspring. Genes that are negatively correlated and p-value < 0.05 are listed in the plots. (c-d) Scatter plots show the correlation between gene expression and promoter DNA methylation levels of individual genes in male (c) and female (d) offspring. Green colors (•) represent samples from control offspring and black colors (•) represent samples from e-cig-exposed offspring.
Differentially methylated and expressed genes converge towards neurological pathways
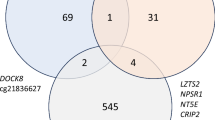
To explore the intersection between differentially methylated genes (DMGs) and differentially expressed genes (DEGs) in both male and female samples, we conducted gene intersection analysis. As depicted in Fig. 6a, we identified 1429 overlapped DEGs and DMGs in male offspring and 994 in female offspring, with 243 of these genes overlapping between male and female groups. Subsequently, gene ontology enrichment analysis for both male and female groups revealed an enrichment of neurological terms (Fig. 6b-c). Furthermore, upon comparing the enrichment of DEGs across the mouse tissue atlas, we made an intriguing observation: both DMGs and DEGs were significantly enriched among neuronal tissues, including the olfactory bulb, cerebral cortex prefrontal, cerebellum, hippocampus, and cerebral cortex regions (Fig. 6d-e). These findings suggest that in utero e-cigarette exposure dysregulates neurological processes, as evidenced by the convergence of DMGs and DEGs toward neuronal pathways in both male and female offspring.
Integrated analysis of differential DNA methylation and gene expression. (a) Overlap of differentially expressed genes, differentially methylated CG and CH sites in both male and female offspring. (b-c) Enrichment of gene ontology (GO) biological terms and KEGG pathways associated with enriched genes in male (b) and female (c) offspring. (d-e) Enrichment analysis of differentially expressed and methylated genes across mouse tissue atlas reveals enrichment of neuronal tissues in male (d) and female (e) rat offspring.
Discussion
Clinical and preclinical studies have shown that prenatal e-cig exposure can impair fetal development, inhibit growth, and lead to neurobehavioral dysfunction in postnatal life13,14,15. Our present study further provides novel epigenetic molecular mechanisms underlying prenatal e-cig aerosol vaping exposure-induced brain developmental disorder in offspring. In the present study, we demonstrate for the first time that prenatal e-cig exposure alters the pattern of DNA methylome in a sex-dependent manner in the neonatal brain, suggesting epigenetic programming of brain development in response to fetal stress. Specifically, our DNA methylome data show a bimodal distribution where a large majority of CpGs are either unmethylated or fully methylated. Both male and female neonatal brains showed similar global DNA methylation features, in which a low methylation level at gene transcription start sites (TSS) is observed and the methylation level gradually increases towards the 3’ end of genes. These findings are consistent with previous studies showing that DNA hypomethylation within the TSS region24. Hypomethylation of TSS regions provides an important mechanism in gene expression. The RRBS DNA-seq analysis shows notable alterations in site-specific DNA methylation patterns in response to prenatal e-cig exposure. While the global distribution of methylation appeared comparable between control and e-cig-exposed samples, the differential methylation analysis revealed thousands of CpG sites exhibiting changes in methylation levels. These sites are predominantly situated in intergenic and intronic regions, suggesting a potential impact on the regulatory elements governing gene expression. Gene body methylation has variable effects on gene expression leading to different functional outcomes25. It is well known that DNA methylation of gene promoters is associated with the silencing of gene expression by blocking the binding of transcription factors26. Intergenic methylation can impact gene expression and activity via the regulation of distal enhancers27. The dynamic changes of DNA methylation at enhancers and other regulatory regions play important regulatory roles in embryonic development and developmental disorders28. Consistent with these previous findings, our enriched gene ontology analysis indicates that prenatal e-cig exposure-mediated dynamic changes of DNA methylation at all regions, including intergenic, intronic, exonic, and promoter regions, are significantly involved in the aberrant development of offspring brains. We found that multiple neurological signaling pathways such as the regulation of neuron project development, axon guidance, synaptic tramisson, nervous system development, etc., are associated with the dynamic changes of CpGs methylation, which highlight the regulatory role of global CpG methylation in brain aberrant development in response to prenatal e-cig exposure.
The observed separation between control and e-cig-exposed offspring in PCA plots indicates a robust methylation response. Intriguingly, two e-cig-exposed samples exhibited similarities to control samples, prompting an investigation into genetic factors. Upon scrutinizing the genetic breeding information, it was discerned that these two offspring were from different litters, emphasizing the role of genetic variations in modulating the response to e-cigarette exposure. Despite this, our analysis identifies site-specific methylation changes associated with key genes implicated in brain development. The present finding that the overlap of differentially methylated CpGs was minimal between males and females, suggests a sex-dependent response to prenatal e-cig exposure. Interestingly, despite the minimum overlap of methylated sites, the enrichment of dysregulated biological processes is similar between male and female offspring. These findings suggest that prenatal e-cig exposure-induced sex-dependent changes of CpGs methylation can lead to brain developmental defects in both male and female offspring. Indeed, previous studies have provided substantial evidence that prenatal exposure to e-cig aerosols impairs neurodevelopment, resulting in adverse behavioral and neuroimmunological consequences in both male and female offspring29. However, our previous study has demonstrated that prenatal e-cig exposure causes a differential alteration of DNA methyltransferase expression in both male and female offspring neonatal brains but induces the development of a hypoxic-ischemic sensitive phenotype in male, not female neonatal offspring11. These findings raise an intriguing question of how much of the sex-dependent changes of DNA methylation contribute to the neurobiological variation and brain developmental disorder and present an opportunity for the future study of the sex difference in epigenetic programming of brain dysfunction later in life in response to prenatal e-cigarette exposure.
It is well-known that DNA methylation exists predominantly in cytosines followed by guanine residues (CpG). However, it is also found in cytosines followed by adenine, thymine, or another cytosine, which is referred to as non-CpG methylation (mCH). Most interestingly, recent reports suggest that, during brain development, highly conserved non-CpG methylation accumulates in neurons and becomes the dominant form of methylation in the human neuronal genome23. Accumulating studies show that non-CpG methylation (mCH) is an important epigenetic mark in developing neurons and plays a key role in the regulation of cognitive function22. Recognizing the growing importance of CH methylation in brain development, we extended our analysis beyond CpG sites and investigated the effects of prenatal e-cig exposure on the mCH sites. Our present data show a subset of CH sites exhibiting differential methylation that is predominately located in intergenic and intronic regions. Gene ontology analysis underscores the enrichment of neurological terms (such as synapse assembly, dendrite morphogenesis, and axon guidance) among CH-methylated genes, which are also conserved between male and female offspring. These findings clearly suggest that prenatal e-cig exposure significantly reshapes the global mCH landscapes in the developing brain, highlighting the potential significance of non-CpG methylation in the regulation of brain development in the context of prenatal e-cig exposure.
Consistent with our previous study of snRNA analysis30, our present bulk RNA-seq analysis illuminates the landscape of gene expression changes induced by prenatal e-cig exposure in the neonatal brain. The differentially expressed genes (DEGs) identified in our study, including NEUROD1, NRXN1, SHANK1, and SHANK3, play crucial roles in regulating various aspects of neuron cell development, differentiation, and synaptic transmission. NEUROD1 is a transcription factor involved in neuronal differentiation and maturation, playing a key role in the development of the nervous system. NRXN1 (Neurexin 1) is a synaptic adhesion molecule that plays a critical role in synaptic formation, axonal guidance, and synaptogenesis. It is essential for proper neuronal communication and synaptic transmission. SHANK1/3 are scaffolding proteins found in the postsynaptic density of excitatory synapses and are involved in the regulation of synaptic development, spine morphogenesis, and synaptic plasticity. These genes are implicated in vocal/imitative learning, autism spectrum disorder (ASD), axonal fasciculation, and synaptic transmission, highlighting their importance in neurodevelopment and brain function31. The dysregulation of these genes due to prenatal e-cig exposure underscores the potential impact on brain development and function, contributing to the observed neurobiological consequences later in life. The non-overlapping patterns of DEGs in male and female samples hint at a potential sex-specific impact. Despite this sex-specificity, gene ontology analysis reveals consistent enrichment of neurological terms, suggesting a shared dysregulation of critical pathways in the developing brain in response to e-cig exposure.
The integration of DNA methylation and gene expression data sheds light on the intricate relationship between these two layers of epigenetic regulation. A consistent trend of negative correlation is observed, particularly at the TSS CpG sites, suggesting a complex interplay between DNA methylation and transcriptional regulation in response to prenatal e-cig exposure. This negative correlation between DNA methylation and gene expression is further emphasized at the individual gene level. In addition, our previous studies of the single-gene approach also demonstrated that prenatal e-cig exposure increased promoter methylation and decreased gene expression of Atg5 in the developing brain11. These findings underscore the dynamic nature of epigenetic regulation in shaping gene expression profiles. The intersection analysis of differentially methylated and expressed genes reveals a substantial overlap, particularly in the context of neurological pathways. Approximately 25% of the genes are common between male and female groups, indicating a convergence of molecular responses to e-cigarette exposure.
Gene ontology enrichment analysis further supports the notion that e-cig exposure in utero dysregulates critical neurological processes. Our present findings from GO biological processes and KEGG pathways analysis show that prenatal e-cig exposure not only regulates neuron project development and axonogenesis but also alters the pathways of neurodegeneration and long-term depression. These findings suggest that the e-cig exposure-induced methylation/expression changes in the brain are likely to have a significant impact on both neurogenesis and neurodegeneration, which may result in an increased risk of brain developmental disorders and potential long-term brain degenerative diseases, such as Parkinson, Alzheimer’s Disease (AD), and dementia. Indeed, we demonstrated previously in the same animal model that prenatal e-cig exposure induces fetal programming of neonatal brain hypoxic-ischemia sensitive phenotype via DNA methylation epigenetic regulatory signaling pathway11. Our current findings provide a basis and present an opportunity for our future study to investigate whether prenatal e-cig exposure induces fetal programming of age-related diseases, such as AD and demenia, later in life.
To contextualize our findings, we compare the enrichment of differentially methylated and expressed genes across a tissue atlas. Strikingly, we observe a significant enrichment of these genes in neuronal tissues, including the olfactory bulb, cerebral cortex prefrontal, cerebellum, hippocampus, and cerebral cortex regions, emphasizing the specificity of the impact on different regions of the brain. This observation aligns with the broader implications of maternal e-cigarette exposure on neurodevelopment, raising concerns about the potential long-term consequences for cognitive and behavioral functions.
Exposure to maternal cigarette smoking during pregnancy has been associated with alterations in DNA methylation, as determined by both gene-candidate and genome-wide approaches32. However, investigations into DNA methylation profiles in neonatal offspring have primarily focused on cord blood and placental tissues. Previous studies have reported that maternal tobacco smoking during pregnancy led to over 6000 CpGs being differentially methylated in cord blood samples33. Moreover, infants exposed to maternal cigarette smoking exhibited significant DNA hypomethylation and identified 50 CpGs differentially methylated in placental tissues and increased placental CYP1A1 expression compared to non-smoking mothers17,34. Notably, prior research has predominantly concentrated on the effects of maternal cigarette smoking on CpG methylation, overlooking non-CpG methylation profiles. The present study utilized neonatal brain tissues to measure DNA methylation, encompassing both CpG and non-CpG profiles. This approach provided direct evidence that alterations in DNA methylation, spanning both CpG and non-CpG profiles in the neonatal brain, constitute an epigenetic mechanism underlying prenatal e-cigarette exposure-induced neurodevelopmental disorders in exposed offspring.
In conclusion, our study offers valuable insights into the molecular consequences of maternal e-cig exposure on DNA methylation and gene expression in the offspring neonatal brain. The observed dysregulation of neurological pathways raises pertinent questions about the potential long-term impact on cognitive and behavioral outcomes. Future research endeavors should focus on unraveling the functional consequences of these molecular changes, exploring potential interventions to mitigate the adverse effects of maternal e-cigarette exposure, and elucidating the temporal dynamics of these epigenetic and transcriptomic alterations. Our study underscores the critical importance of understanding the complex interplay between genetic, epigenetic, and transcriptional responses to perinatal stresses, particularly in the context of neurodevelopment. The findings highlight the need for targeted strategies to safeguard fetal brain development from the deleterious effects of e-cigarette use during pregnancy.
Limitations of study
In our current study, we explored the impact of maternal e-cig exposure on DNA methylome and transcriptomic signatures in neonatal brain tissue. While we randomly assigned offspring from each dam to experimental groups, a key limitation of this study lies in our use of offspring count as the experimental parameter (n). It’s important to note that genetic backgrounds may exhibit varying susceptibilities to e-cig exposure. Based on the current data obtained from bulk tissue, neonatal brain cell type specific DNA methylation profile in response to e-cig exposure is not addressed. In our future study, we plan to investigate whether prenatal e-cig exposure induces alterations of gene expression and DNA methylation in cell type-dependent manner in the neonatal brain tissue.
Materials and methods
Experimental animals and chronic intermittent e-cigarette (CIEC) exposure system
All procedures and protocols for animal experiments were approved by the Institutional Animal Care and Use Committee of Loma Linda University and followed the guidelines of the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The present study is reported in accordance with ARRIVE guidelines (https://arrivequidelines.org). Pregnant Sprague–Dawley rats were purchased from Charles River Laboratories (Portage, MI) and were housed under a control temperature (22 ºC) and photoperiod (12-h light and 12-h dark cycle) with food and water ad libitum.
To better understand the effects of e-cigarette (e-cig) aerosol exposure during pregnancy on fetal development, we developed a pregnant rat model of chronic intermittent e-cigarette (CIEC) exposure11,30, which is slightly modified from previous rodent model35,36. Briefly, pregnant Sprague–Dawley rats were randomly divided into two groups: the first group was exposed to e-cigarette aerosol; and the 2nd group was exposed to air in the same chamber. Commercial (BluPlus Cig) E-cigarettes (2.4% nicotine) were used for this project to reflect what real-world E-cig users are experiencing. The e-cigarette rodent exposure system was obtained from AutoMate Scientific, Inc. (Berkeley, CA). To mimic the phenomenon of chronic intermittent E-cig exposure in human vapers, we have optimized the system to deliver E-cig aerosol with a puff duration of 4 s, 3 puffs in an inter-puff interval of 30 s per vaping episode, and one episode per hour in the dark phase of 12 h each day, which generates similar nicotine and cotinine blood pharmacokinetics in the pregnant rats (Supplementary Fig. 3) to those observed in human e-cigarette users37,38. The dams were exposed to e-cigarettes for a total of 17 days from gestational day 4 (E4) to E20, with a vaping cycle of 12 h per day. The rationale for starting on E4 is that this is the period just before embryo implantation on the uterine wall in rats. After CIEC exposure, dams were transferred to their home cages for natural delivery. After birth, the neonatal offspring at the age of 7 days (P7) were used for this project study. The rats were euthanized with an infusion of 5% isoflurane followed by heart removal after the experiments.
RNA and DNA extractions
Neonatal rat pups were divided into four groups, including control male (n = 5), control female (n = 5), maternal E-cig-exposed male (n = 5) and maternal E-cig-exposed female (n = 5), respectively. All pups were euthanized on postnatal day 7 (P7). Whole brains were freshly isolated and frozen in liquid nitrogen and stored in a − 80 °C freezer. The genomic DNA and total RNA extraction from the neonatal brain tissues were isolated with the AllPrep DNA/RNA/miRNA Universal Kit (Cat. No. 80224, Qiagen, USA) following the manufacturer’s instructions. This kit enables simultaneous purification of genomic DNA and total RNA from all types of tissue, including brain tissue. Since there is no need to divide the brain sample into two for separate purification procedures, maximum yields of DNA and RNA can be achieved. DNA and RNA quantity and quality were assessed using NanoDrop 2000 (Cat. No.: ND2000, Thermo Scientific, USA). 100 ng of total RNA and 100 ng of gDNA for each sample were used for the following sequence analysis.
RNA-seq library construction and sequencing
RNA quality was evaluated using the Agilent 2200 TapeStation and RNA ScreenTape (Santa Clara, CA). RNA samples had RNA integrity numbers (RIN) between 8.8 to 10. The Ovation® RNA-Seq System for rat (NuGEN Technologies, San Carlos, CA) was used to construct RNA-seq libraries according to the manufacturer’s instructions. Briefly, 100 ng of total RNA spiked with 1 µl of 1:500 diluted ERCC ExFold RNA Spike-In Mix 1 (Life Technologies, Carlsbad, CA) was used to start cDNA synthesis. Following primer annealing and cDNA synthesis. The cDNA was then subject to end-repair, adaptor index ligation, and strand selection. A custom InDA-C primer mixture SS5 V8 for rat was used to allow strand selection. Finally, libraries were amplified on a PCR thermocycler for 13 cycles (Mastercycler® pro, Eppendorf, Hamburg, Germany), and purified with Agencourt AMPure beads (Beckman Coulter, Indianapolis, IN). Library was quantified using Qubit dsDNA HS Kit on Qubit 3.0 Fluorometer (Life Technologies, Carlsbad, CA) and the peak size was determined using the D1000 ScreenTape on Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, CA). RNA-seq libraries were sequenced on Illumina NextSeq 550 with 75 bpx2, PE, approximately 25 M reads/each, at the Center for Genomics, Loma Linda University.
Reduced representation bisulfite sequencing (RRBS) library construction and sequencing
DNA was quantitated using Qubit dsDNA HS Kit on Qubit 3.0 Fluorometer (Life Technologies, Carlsbad, CA). 100 ng of gDNA was used to construct RRBS DNA library using the Ovation® RRBS Methyl-Seq System (NuGEN Technologies, San Carlos, CA) according to the manufacturer’s protocol. Briefly, the methylation insensitive MspI enzyme, which cuts the DNA at CCGG sites, was used to digest gDNA into fragments. The fragments were directly subject to end blunting and phosphorylation in preparation for ligation to a methylated adapter with a single-base T overhang. A unique index, out of 16 indices, was used per each sample for multiplexing. The ligation products were finally repaired in a thermal cycler under the program (60 °C–10 min, 70 °C–10 min, hold at 4 °C). The product of the final repair reaction was used for bisulfite conversion using QIAGEN EpiTect Fast DNA Bisulfite Kit according to Qiagen’s protocol. Bisulfite converted library was purified using Agencourt AMPure beads (Beckman Coulter, Indianapolis, IN), amplified for 12 PCR cycles (Mastercycler® pro, Eppendorf, Hamburg, Germany), and then bead-purified. RRBS libraries were sequenced using the Illumina Nextseq 550 (Illumina, San Diego, CA, USA) in a single-ended, 75 bp, with approximately 25 M reads/each at the Center for Genomics, Loma Linda University.
RNA-seq data processing and analysis
Raw RNA-seq fastq data was assessed for sequencing quality using FastQC (v0.11.5, http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Adapters and primers were trimmed using Trimmomatic (v0.3.7). Reads were aligned to the rat reference genome NCBI Rnor6.0, downloaded from iGenome, using Kallisto (v.0.44.0)39 with the default parameter settings. The read counts matrix from Kallisto was input to DESeq2 (v.1.38.0) to identify differentially expressed genes (DEG). The DEGs were identified by DESeq2 with the cutoff of log2FC > + /−0.2, p-value < 0.005 and padj < 0.1.
RRBS data processing and analysis
Raw RRBS fastq data was trimmed using Trim Galore (v0.3.7) and NuGEN diversity trimming and N6 de-duplicate scripts, as described previously40. Reads were aligned to the rat reference genome NCBI Rnor6.0, using Bismark v0.16.334 with the default parameter settings. Methylated CG and CH sites were extracted using bismark_methylation_extractor (v0.16.334) with the following command bismark_methylation_extractor bamFile –multicore 8 –ignore 2 –ignore_3prime 2 –comprehensive –bedGraph –merge_non_CpG –CX_context. The output from bismark_methylation_extractor resulted in two files, one for CG methylation and another one for CH (merged CHH and CHG) methylation. CG and CH methylation files were merged with coverage file to combine coverage and methylation percentage for each CG and CH. Only CpGs with at least 10 × coverage in all samples were selected for subsequent analysis. We obtained an average of 21 million raw reads for each sample (Table S1), ranging from 18,987,532 to 23,256,101. The identified reads were annotated, resulting in an average of 1,625,772 CpG sites and 1,659,542 CpG sites with > 10 coverage in females and males, respectively. To identify differentially methyled CG probes, we performed t-test using matrixTests package. Methylation percentage change between control and e-cigarette group was > 10% and p value of 0.0001. To identify differentially methyled CH probes, methylation percentage change between control and e-cigarette group was > 5% and p value of 0.0001.
Enrichment analysis
Enrichment analysis of gene ontology, KEGG pathway and mouse tissue enrichment was performed using EnrichR41,42. For enrichment analysis, we used differential mCG and mCH sites and differentially expressed genes. Enrichment analysis was done separately for male and female samples.
Statistical analysis
All statistical analysis was prformed uing the R software. Principle component analysis (PCA) was performed using prcom function from stats package within R software. Experimental number (n) represents neonates. Five animals were used in each group.
Data availability
Data is provided within the manuscript or supplementary information files. Raw sequencing data generated from this study will be deposited on GEO database (GEO accession numbers: GSE278792). The datasets used and/or analyzed during the current study are available from corresponding author on reasonable request.
References
Ayers, J. W., Ribisl, K. M. & Brownstein, J. S. Tracking the rise in popularity of electronic nicotine delivery systems (electronic cigarettes) using search query surveillance. Am. J. Prev. Med. 40, 448–453 (2011).
Farsalinos, K. E., Romagna, G., Dimitris, T., Kyrzopoulos, S. & Voudris, V. Evaluation of electronic cigarette use (vaping) topography and estimation of liquid consumption: implications for research protocol standards definition and for public health authorities’ regulation. Int. J. Environ. Res. Public Health. 10, 2500–2514 (2013).
Noel, J. K., Rees, V. W. & Connolly, G. N. Electronic cigarettes: a new ‘tobacco’ industry?. Tobacco Control. 20, 81 (2011).
Jiang, N., Lee, L., Zelikoff, J. T. & Weitzman, M. E-cigarettes: Effects on the fetus. Pediatr. Review. 39, 156–158 (2018).
Liu, B. et al. National estimates of e-cigarette use among pregnant and nonpregnant women of reproductive age in the United States, 2014–2017. JAMA Pediatr. https://doi.org/10.1001/jamapediatrics.2019.0658 (2019).
Wanner, N. J., Camerota, M. & Propper, C. Prevalence and perceptions of electronic cigarette use during pregnancy. Matern. Child Health J. 21, 1655–1661 (2017).
Whittington, J. R. et al. The use of electronic cigarettes in pregnancy: a review of the literature. Obstet. Gynecol. Surv. 73(9), 544–549 (2018).
Li, Y. et al. Perinatal nicotine exposure increases vulnerability of hypoxic-ischemic brain injury in neonatal rats: role of angiotensin II receptors. Stroke 43(9), 2483–2490 (2012).
Pauly, J. R. & Slotkin, T. A. Maternal tobacco smoking, nicotine replacement and neurobehavioural development. Acta Paediatr. 97, 1331–1337 (2008).
Schweitzer, R. J., Wills, T. A. & Behner, D. E-cigarette use and indicators of cardiovascular disease risk. Curr. Epid. Rep. 4, 248–257 (2017).
Walayat, A. et al. Fetal e-cigarette exposure programs a neonatal brain hypoxic-ischemic sensitive phenotype via altering DNA methylation patterns and autophagy signaling pathway. Am. J. Physiol. Regul. Integr. Comp. Physiol. 321(5), R791–R801 (2021).
Ashour, A. et al. E-cigarette liquid provokes significant embryotoxicity and inhibits angiogenesis. Toxics. 8, 38 (2020).
Wetendorf, M. et al. E-cigarette exposure delays implantation and causes reduced weight gain in female offspring exposed in utero. J. Endocr. Soc. 3, 1907–1916 (2019).
Nguyen, T. et al. Maternal e-cigarette exposure results in cognitive and epigenetic alterations in offspring in a mouse model. Chem. Res. Toxicol. 31, 601–611 (2018).
Sifa, A. E. et al. Prenatal electronic cigarette exposure decreases brain glucose utilization and worsens outcome in offspring hypoxic-ischemic brain injury. J. Neurochem. 153, 63–79 (2020).
Joubert, B. R. et al. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ. Health Perspect 120, 1425–1431 (2012).
Suter, M. et al. Maternal tobacco use modestly alters correlated epigenome-wide placental DNA methylation and gene expression. Epigenetics. 6, 1284–1294 (2011).
Chen, H. et al. Maternal e-cigarette exposure in mice alters DNA methylation and lung cytokine expression in offspring. Am. J. Respir. Cell Mol. Biol. 58, 366–377 (2018).
Mckee, S. E. & Reyes, T. M. Effect of supplementation with methyl-donor nutrients on neurodevelopment and cognition: considerations for future research. Nutr. Rev. 76, 497–511 (2018).
Bakker, R., Timmermans, S., Steegers, E. A., Hofman, A. & Jaddoe, V. W. Folic acid supplements modify the adverse effects of maternal smoking on fetal growth and neonatal complications. J. Nutr. 141, 2172–2179 (2011).
Ke, J. et al. Role of DNA methylation in perinatal nicotine-induced development of heart ischemia-sensitive phenotype in rat offspring. Oncotarget 8(44), 76865–76880 (2017).
de Mendoza, A. et al. The emergence of the brain non-CpG methylation system in vertebrates. Nat. Ecol. Evol. 5(3), 369–378 (2021).
Lister, R. et al. Global epigenomic reconfiguration during mammalian brain development. Science. 341(6146), 1237905 (2013).
Sun, M. et al. Mammalian brain development is accompanied by a dramatic increase in bipolar DNA methylation. Sci. Rep. 2(6), 32298. https://doi.org/10.1038/srep32298 (2016).
Yang, X. J. et al. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 26, 577–590 (2014).
Busslinger, M., Hurst, J. & Flavell, R. A. DNA methylation and the regulation of globin gene expression. Cell. 34, 197–206 (1983).
Stadler, M. B. et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 480, 490–495 (2011).
Greenberg, M. V. C. & Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 20, 590–607 (2019).
Church, J. S. et al. Neuroinflammatory and behavioral outcomes measured in adult offspring of mice exposed prenatally to E-cigarette aerosols. Environ. Health Perspect. 128(4), 47006 (2020).
Chen, Z. et al. Single-nucleus chromatin accessibility and RNA sequencing reveal impaired brain development in prenatally e-cigarette exposed neonatal rats. iScience. 25(8), 104686 (2022).
Sungur, A. O., Schwarding, R. K. W. & Wohr, M. Behavioral phenotypes and neurobiological mechanisms in the Shank1 mouse model for autism spectrum disorder: A translational perspective. Behav Brain Res. 15(352), 46–61. https://doi.org/10.1016/j.bbr.2017.09.038 (2018).
Nakamura, A., Francois, O. & Lepeule, J. Epigenetic alterations of maternal tobacco smoking during pregnancy: a narrative review. Int. J. Environ. Res. Public Health. 18(10), 5083. https://doi.org/10.3390/ijerph18105083 (2021).
Joubert, B. R. et al. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am. J. Hum. Genet. 98(4), 680–696. https://doi.org/10.1016/j.ajhg.2016.02.019 (2016).
Morales, E. et al. Genome-wide DNA methylation study in human placenta identifies novel loci associated with maternal smoking during pregnancy. Int. J. Epidemiol. 45(5), 1644–1655. https://doi.org/10.1093/ije/dyw196 (2016).
Espinoza-Derout, J. et al. Chronic intermitten electronic cigarette exposure induces cardiac dysfunction and atherosclerosis in apolipoprotein-E knockout mice. Am. J. Physiol. Heart Circ. Physiol. 317, H445–H459 (2019).
Shao, X. M. et al. A mouse model for chronic intermitten electronic cigarette exposure exhibits nicotine pharmacokinetics resembling human vapers. J. Neurosci. Methods. 326, 108376 (2019).
Lopez, A. A. et al. Effects of electronic cigarette liquid nicotine concentration on plasma nicotine and puff topography in tobacco cigarette smokers: A preliminary report. Nicotine Tob. Res. 18, 720–723 (2016).
St Helen, G., Havel, C., Dempsey, D. A., Jacob, P. 3rd. & Benowitz, N. L. Nicotine delivery, retention and pharmacokinetics from various electronic cigarettes. Addiction. 111, 535–544 (2016).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34(5), 525–527. https://doi.org/10.1038/nbt.3519 (2016).
Huang, L. et al. Foetal hypoxia impacts methylome and transcriptome in developmental programming of heart disease. Cardiovasc. Res. 115, 1306–1319 (2019).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 14, 128. https://doi.org/10.1186/1471-2105-14-128 (2013).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28(1), 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Funding
This work was supported by National Institutes of Health Grants U01DA058278 (CW, DX), HL135623 (DX, XMS), DA041492 (DX), and HD088039 (DX). This project was partially supported by funds provided by The Regents of the University of California, Research Grants Program Office, Tobacco Related Disease Research Program (TRDRP) grant # T29IR0437 and T34IR8076. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
D.X. conceived study and designed the experiments. A.W., C.N., M.H., Y.L., W.C., Z.C., and X.H. performed experiments including animal in vivo model, RNA and DNA isolation, library constructions and sequencing. A.W., C.N., M.H., C.W. and D.X. drafted the manuscript and performed bioinformatics data analyses. C.W., L.Z., M.S., and D.X. edited and revised the manuscript. A.W., Z.C. and Y.L. prepared the methods for the manuscript. All authors reviewed and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Walayat, A., Hosseini, M., Nepal, C. et al. Maternal e-cigarette exposure alters DNA methylome, site-specific CpG and CH methylation, and transcriptomic signatures in the neonatal brain. Sci Rep 14, 24263 (2024). https://doi.org/10.1038/s41598-024-75986-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-75986-x
Keywords
This article is cited by
-
MethPy: a new software for analyzing non-CpG methylation after bisulfite assay and Sanger sequencing
Scientific Reports (2025)