Abstract
Osteosarcoma (OS) is a cancerous tumor, and its development is greatly influenced by long non-coding RNA (lncRNA). Endoplasmic reticulum stress (ERS) is an essential biological defense process in cells and contributes to the progression of tumors. However, the exact mechanisms remain elusive. This study aims to develop a signature of lncRNAs associated with ERS in OS. This signature will guide the prognosis prediction and the determination of appropriate treatment strategies. The UCSC Xena database collected transcriptional and clinical data of OS and muscle, after identifying ERS differentially expressed genes, we utilized correlation analysis to determine the endoplasmic reticulum stress lncRNAs (ERLs). The Least Absolute Shrinkage and Selection Operator (LASSO) and Cox regression analysis were utilized to develop an ERLs signature. To clarify the fundamental mechanisms controlling gene expression in low and high-risk groups, Gene Set Variation Analysis (GSVA) were conducted. In addition, the distinction between the two groups regarding drug sensitivity and immune-related activity was investigated to determine the immunotherapy effects. Utilizing RT-qPCR, the expression of model lncRNAs in OS cell lines was ascertained. The functional analysis of LINC02298 was carried out through in vitro experiments and pan-cancer analysis. This study successfully constructed an ERLs prognostic signature for OS, which comprised 5 lncRNAs (AC023157.3, AL031673.1, LINC02298, LINC02328, SNHG26). The risk signature predicted overall survival in patients with OS and was confirmed by assessing the validation and whole cohorts. Further, it was discovered that individuals classified as high-risk displayed suppressed immune activation, decreased infiltration of immune cells, and decreased responsiveness to immunotherapy. The RT-qPCR showed that the constructed risk prognosis model is reliable. Experimental validation has demonstrated that LINC02298 can promote OS cells’ invasion, migration, and proliferation. In addition, LINC02298 exhibited significant differential expression in many types of cancer. Moreover, LINC02298 is an important biomarker in a variety of tumors. This study established a novel ERLs signature, which successfully predicted the prognosis of OS. The function of LINC02298 in OS was elucidated via in vitro experiments. Therefore, it offers new opportunities for predicting the clinical prognosis of OS and establishes the basis for targeted therapy in OS.
Similar content being viewed by others
Background
Osteosarcoma (OS), a primary malignant bone tumor from mesenchymal cells, is the 2nd most prevalent type of bone cancer, with a prevalence rate of approximately 3 cases per 1 million people1,2,3,4. The overall survival has substantially increased with improvements in treatment and considerable advances in diagnostic imaging5,6,7. However, because of the high recurrence rate and distant metastasis, OS still has a dismal prognosis8,9,10. Moreover, high tumor heterogeneity, which results in chemoresistance in certain patients, continues to provide therapeutic challenges11. Therefore, finding new molecular targets for tumor therapy and prognostic indicators is essential.
LncRNAs are RNA that consist of more than 200 nucleotides. These are crucial in regulating transcription and translation processes and are particularly essential in the formation of tumors12,13,14. Growing evidence suggests that lncRNAs may regulate many biochemical reactions in OS and show a complicated and precise regulatory function in cancer initiation. Furthermore, lncRNAs modulate tumor metastasis and tumor microenvironment (TME)15. For example, Ji S et al. demonstrated that the growth of OS cells was significantly increased, and the programmed cell death was reduced by overexpressing NEAT116. Ye K et al. showed that upregulation of GAS5 can inhibit the migration, proliferation, and epithelial-mesenchymal transition of OS cells via miR-221/ARHI mediation17. Therefore, lncRNAs can be potential targets for OS therapy18,19.
The endoplasmic reticulum (ER) is the most extensive organelle in eukaryotic cells and is a crucial place for protein synthesis, processing, and transport20. Endoplasmic reticulum stress (ERS) results from an ER homeostasis imbalance induced by unfolded protein aggregation and an unbalanced Ca2+ concentration21,22. It has been reported that many physiological and pathological processes can activate ERS as a defensive system, such as chronic inflammation and oxidative stress23,24,25. Studies have shown that lncRNAs can influence tumor progression by affecting ER stress. Huang et al. Chen et al. indicated that lncRNA MEG3 triggers tumor cell apoptosis through ER stress in esophageal and liver cancers26,27. Ding et al. demonstrated that the lncRNA CASC2 enhances PERK mRNA’s stability, activating the PERK/eIF2α/CHOP endoplasmic reticulum stress pathway. This activation leads to cell death and increased sensitivity to radiation in non-small cell lung cancer28. Nevertheless, little is known about ERLs in OS, and more study is required to comprehend the underlying mechanisms.
This study developed a novel predictive model of ERLs to improve the prognosis of OS and explore changes in the TME, immunotherapy, and drug response. The impact of LINC02298 on OS was elucidated via in vitro experiments. These findings will present novel opportunities for predicting clinical prognosis and establish the basis for targeted therapy in OS.
Research methodology
The collection of data
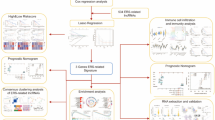
UCSC Xena database (https://xenabrowser.net/) provided the easy-to-use pre-compiled transcriptome data and clinical details of muscle and OS samples. The OS cases expression matrix were acquired from Therapeutically Applicable Research to Generate Effective Treatments (TARGET) OS database, and the expression data of normal muscle samples were obtained from the Genotype-Tissue Expression(GTEx) project. A total of 252 ERS-related genes (ERGs) were downloaded from MSigDB (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp) (Supplementary Table S1). The clinical information on the tumors and the pan-cancer gene expression patterns was collected from UCSC Xena database. Figure 1 illustrated the flowchart of the study.
Finding lncRNAs associated with ERS and establishing the ERLs signature in OS
The “limma” R package was used to screen for differentially expressed genes between OS and normal muscle tissue.The selection criteria were p adjusted < 0.05 and |FoldChange| > 4 (|log2 FoldChange|>2). Finally, 5179 genes were identified as differentially expressed genes. After intersecting with 252 ERGs, we obtained 78 differentially expressed ERGs in OS. The Pearson correlation analysis was utilized to find the correlation between LncRNAs and ERGs with |Cor|> 0.5, P value < 0.001, to identify ERLs. Analysis of univariate Cox proportional hazards was done at p < 0.05. LASSO regression analysis was done to determine the best panel of prognostic lncRNAs. After conducting a multivariate Cox analysis, the model genes were ultimately determined. The formula for the risk score determination is given below:
Validation of the ERLs signature
Using the “caret” R package, 88 OS patients were randomly categorized into training and testing groups at a 1:1 ratio. The samples were separated into high and low-risk groups based on median risk scores. The “survival” R package was used to perform survival analysis. The “survminer” R package was performed for data visualization, and the “pROC” R package was used to validate the model efficacy.
Principal component analysis (PCA) and nomogram construction
PCA was conducted using “limma” and “scatterplot3d” R packages to clarify the distribution of low- and high-risk samples. The calibration and nomogram curves were generated using “survival” and “rms” R packages. The nomogram used clinical criteria and risk scores to estimate the survival rates of 1-, 3-, and 5-year.
Gene ontology (GO), Kyoto encyclopedia of genes and genomes (KEGG) and gene set variation analysis (GSVA)
GO enrichment analysis includes biological processes (BP), molecular function (MF), and cellular components (CC)29. KEGG is a bioinformatics resource for mining metabolic pathways that are enriched in the gene list30. The R package “clusterProfiler” was used for GO and KEGG enrichment analysis. To explore the differentially enriched pathways between two groups, a GSVA was performed using the “GSEABase”, and “GSVA” R packages, with “h.all.v7.5.1.symbols.gmt” as the reference gene set. The “ggplot2” R package was used for visualization.
Immune-related functional analysis
The “estimate” R package was utilized to determine the stromal, immunological, and estimation scores for each patient with OS. The ssGSEA method was applied to evaluate the amounts of infiltration of 24 immune cells in each sample. These levels were then compared between the two risk groups. Subsequently, an immunological correlation study was performed to determine the association between the immune cells and risk score.
Sensitivity assessment of potential drugs and pancancer analysis
To evaluate the clinical potential of ERLs as a treatment for OS and assess its efficacy, the “pRRophetic” R package was employed to obtain the IC50 values of the selected 198 chemotherapeutic drugs. The data was graphically represented using the “ggplot2” R package. A screening criterion of p ≤ 0.001 was employed in the analysis. The pan-cancer analysis was performed in Xiantao (https://www.xiantaozi.com/).
Cell culture, transfection, lentivirus infection
The osteoblast cell lines (hFOB1.19) and human OS cell lines (HOS, MG63, and U2OS) were obtained from ATCC. HOS cells were cultured in minimal essential medium (MEM) from Gibco, USA, whereas hFOB1.19, MG63, and U2OS cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) from Gibco, USA. Based on a complete medium supplemented with fetal bovine serum (FBS, 10%; Gibco, USA) and 1% penicillin-streptomycin solution (NCM Biotech, China), all cell lines are cultured with 5% CO2 at 37 °C. A manufacturer, RiboBio, in Guangzhou, China, delivered the siRNAs for LINC02298 and a negative control known as NC. It was done with the riboFECT CP Transfection Kit from RiboBio in China. Genechem, based in Shanghai, China, provided the lentivirus.
RNA extraction and RT-qPCR
Total RNA was extracted from cells using TRIzol (Thermofisher, USA) followed by Chloroform (Thermofisher, USA). Spectrophotometric RNA purity and concentration analysis were performed using Nanodrop One (Thermofisher, USA). cDNA was synthesized from the extracted RNA utilizing the Prime Script RT reagent. An RT-qPCR assay was conducted using an ABI 7500 Real-Time PCR machine (Thermofisher, USA) and a reverse transcription reagent (TaKaRa, Japan). Supplementary Table S2 shows the primers that were used for the confirmation.
CCK-8 assay
Using cells in a viable growth state, a cell suspension was prepared. 1000 cells/100 µL were added to each well of a 96-well plate, which was then incubated at 37 °C for eight hours. At 24, 48, and 72 h, a volume of 10 µL of CCK-8 solution was introduced into each well of a 96-well plate and allowed to incubate for 3 h at a temperature of 37 °C. Subsequently, the absorbance of the solution was determined.
The wound healing assay
About 5 × 105 cells were put into a six-well plate. Once the cells were attached, the plate was scratched with a 10 µL pipette tip. The serum-free culture medium was added, and the plate was incubated for 24 h. Cell migration at specified locations was observed and documented using a microscope. Subsequently, the Image J software computed the average distance between the cells.
Transwell assay
The density of the suspended cells in the serum-free culture medium was adjusted to 5 × 105 cells/mL. A culture medium (600 µL) containing 10% FBS was introduced in the lower chamber, while the cell suspension (100 µL) was added in the upper chamber. Following this, the plate was incubated for 24 h. Following fixation with 4% paraformaldehyde, the cells were stained with 0.1% crystal violet. Counting and observing the cells was performed using a microscope.
Statistical analysis
R software version (v4.1.3) was used for all data collection and statistical analysis. A p-value of < 0.05 was considered statistically significant.
Results
Identification of differentially expressed ERS genes
By conducting differential analysis on normal muscle tissue and OS tissue, we obtained 5179 differentially expressed genes. Among them, there are 4274 upregulated genes and 905 downregulated genes (Fig. 2A, B). After intersecting with 252 ERSs downloaded from MSigDB, we obtained 78 differentially expressed ERGs (Fig. 2C). The results of GO and KEGG enrichment analysis on these 78 intersecting genes included “response to endoplasmic reticulum stress”, “response to topologically incorrect protein”, “response to unfolded protein”, “endoplasmic reticulum lumen”,“endoplasmic reticulum protein-containing complex”, “endoplasmic reticulum quality control compartment” and “Protein processing in endoplasmic reticulum” (Fig. 2D–F). The functions of these intersecting genes are mainly associated with ERS. This further proves the reliability of our screened ERGs.
Identification of differentially expressed ERGs. (A,B) Identification of differentially expressed genes between OS and muscle (A) volcano plot, (B) heatmap (C) Intersecting differentially expressed genes with ERGs (D–F) Functional enrichment analysis of differentially expressed ERGs (D) bubble plot, (E) EMAP plot, (F) chord plot.
Identification of ERLs and construction of a predictive model for ERLs in the training group
Based on the TARGET database, lncRNAs were extracted using Perl-based methods. Then, we conducted correlation analysis on the 78 differentially expressed ERGs (Cor > 0.5, p< 0.001), and we obtained 420 ERLs (Fig. 3A). Overall, 88 OS samples were classified into two groups: testing and training groups. 64 of these lncRNAs were selected using univariate Cox regression analysis for their potential as prognostic lncRNAs associated with ERS in OS patients (Fig. 3B). The Least Absolute Shrinkage and Selection Operator(LASSO) analysis and multi Cox regression analysis of the training cohort was conducted to build the model and 5 lncRNAs were finally identified when constructing the risk signature (Fig. 3C–E). The ERLs- signature score for each patient was subsequently determined through the formula as given below: ERLs score (Risk Score) = (– 1.954919033×expr AC023157.3) + (0.840838939×exprAL031673.1) + (0.193350763×expr LINC02298) + (– 1.160820595×expr LINC02328) + (0.512805281×expr SNHG26). The patient cohort was categorized into low and high-risk groups based on the median risk score. The survival status map revealed that an increase in the risk score corresponded to a progressive decline in the patients’ survival time and an increase in their mortality rate (Fig. 3F, G). The relative expression levels of each gene were graphically represented (Fig. 3H). Moreover, the KM survival curves indicated that the prognosis for the high-risk group was inferior to that of the low-risk group (Fig. 3I). At 1, 3, and 5 years, the AUC values for the ROC curves were 0.893, 0.936, and 0.927 (Fig. 3J).
Identification of lncRNAs associated with ERS and construction of the prognostic signature. (A) The correlation analysis of the 78 ERGs and their related lncRNAs. (B) A graphical representation of a forest plot displays the results of the univariate Cox regression analysis (C) A cross-validation curve is generated to assess the paired likelihood of deviance (D) Elucidation of the LASSO coefficient profiles of predictive lncRNAs. (E) The results of the multivariate Cox regression analysis (F) Patients are classified into high-risk and low-risk groups based on riskscore. (G) Survival status map (H) Risk heatmap of gene expression (I) Kaplan–Meier survival curves (J) ROC curves were generated to assess the overall survival rates at 1, 3, and 5 years.
Evaluation of the predictive model and PCA
The accuracy of the ERLs signature was checked with both validation and total sets. The results were similar to what was seen in the training set: participants with a higher risk score showed a lower chance of survival and a higher death rate (Fig. 4A, B, F and G). Moreover, heatmaps were created to illustrate the distribution of expression for lncRNAs (Fig. 4C and H). Furthermore, the KM survival curves indicated a substandard prognosis for high-risk group patients (Fig. 4D and I). The model was excellent at predicting prognosis, as shown by the ROC curves of the validation set and the total set. The AUCs at 1, 3, and 5 years for the validation set were 0.716, 0.764, and 0.774, and they were 0.800, 0.867, and 0.873 for the total set (Fig. 4E and J). It was determined, through a PCA of all genes, ERGs, ERLs, and ERLs predictive model, the ERLs prognostic model could distinguish the patients between high and low risk groups more effectively (Fig. 5A–D).
The assessment of the prognostic signature of lncRNAs associated with the ERS in the validation and whole cohorts. (A,F) The distribution of riskscore (B,G) Plots indicating the state of survival (C,H) Risk gene expression heatmaps (D,I) The Kaplan-Meier survival curves (E,J) ROC curves were generated to evaluate the overall survival rate at 1, 3, and 5 years.
Nomogram construction and KM survival curve in different subgroups
A nomogram was generated by calculating the prognostic indicators related to clinical characteristics and risk scores (Fig. 6A). These results indicate a strong correlation between the observed and expected findings for the overall survival rates at 1, 3, and 5 years. The calibration curves of the nomogram suggested that the nomogram had a sufficient ability to predict outcomes (Fig. 6B). Survival analysis was further conducted to compare groups based on baseline features such as age, gender and metastasis. The findings demonstrated that OS patients divided as high-risk demonstrated significantly shorter survival durations compared to low-risk patients across many categories, including gender (male, female), age (age >12, age ≤ 12), and metastasis (metastasis, non-metastasis). These results provide strong evidence supporting the efficacy of the risk model in accurately predicting outcomes for OS patients in multiple subgroups (Fig. 6C–H).
The development of a predicted nomogram and subgroup survival analysis. (A) This nomogram displays risk scores and clinicopathological variables that can be used to predict overall survival in OS patients at 1, 3, and 5 years. (B) Calibration curves (C,D) Survival analysis in different age subgroups. (E,F) Survival analysis in different sex subgroups. (G,H) Survival analysis in different metastasis subgroups.
GSVA
Then, we conducted GSVA and found that the high-risk group had lower scores than the low-risk group in KRAS signaling, Allograft rejection, IL2/STAT5 pathway, P53 pathway, Inflammatory response, PI3K/AKT/mTOR pathway, Complement, Apoptosis, IL6/JAK/STAT3 pathway, and TNFα via NFkβ signaling pathways. In contrast, the high-risk group had higher scores than the low-risk group in E2F targets, G2M checkpoint, Wnt/beta/catenin, and Cholesterol homeostasis (Fig. 7A). The heatmap of gene expression and hallmark pathway scores further demonstrates the relationship between gene expression and pathways (Fig. 7B). Patients in different risk groups may affect their respective prognosis through the above-mentioned pathways.
Immune infiltration analysis
Due to its dynamic and heterogeneous characteristics, the TME significantly influences cancer development, progression, and metastasis while also altering the behavior of tumor cells. Compared to patients in the low-risk group, the APC_co_inhibition, APC_co_stimulation, CCR, Check point, Cytolytic_activity, Inflammation-promoting, and T_cell_co-inhibition in patients in the high-risk group are significantly suppressed (Fig. 8A). ESTIMATE algorithm indicated that the low-risk group exhibited higher stromal, immune, and estimate scores than the high-risk group (Fig. 8B–D). Infiltration of stromal and immune cells is decreased in the high-risk group. Using the ssGSEA algorithm, we found that CD8 T cells, iDCs, Macrophages, Neutrophils, Tgd, and Th1 cells were significantly reduced in the high-risk group (Fig. 8E). Additionally, we also observed a significant correlation between Neutrophils, Macrophages, and iDCs (Fig. 8F). Furthermore, examining the relationship between the risk score and immune cells revealed that iDC, Macrophages, Neutrophils negatively correlated with the risk score and Tgd positively correlated with the risk score (Fig. 8G–K). The expression of common immune checkpoint genes between two groups was analysed and found the patients in high risk group had lower expression levels of LAG3,CTLA4,TNFRSF9,CD80,CD70,LAIR1,TNFSF15,CD274,HAVCR2,TMIGD2,CD28, CD48, CD200R1 (Fig. 8L).These findings suggest that the immune function of patients in the high-risk group is impaired, leading to an increased possibility of immune escape.
Immune infiltration analysis and immune checkpoints analysis. (A) Analysis of immune function between two groups (B) ESTIMATE score between two groups (C) Stromal score between two groups (D) Immune score between two groups (E) Infiltration of 24 types of immune cells between high-risk and low-risk groups (F) Heatmap of immune cell infiltration correlation (G) Heatmap of correlation between model genes and immune cell infiltration (H) High infiltration of Tgd is associated with high risk score (I–K) Low infiltration of cytotoxic cells, macrophages, and iDCs is associated with high risk score (L) The expression of immune checkpoints in high-risk and low-risk groups.
Drugs with potential efficacy in OS
This study utilized the “pRRophetic” R package to determine the IC50 values of various drugs. These values served as markers to estimate the effectiveness of a drug in inhibiting tumor growth. The results revealed 11 small molecule drugs with higher sensitivity towards the high-risk group: CDK9_5576, CDK9_5038, Acetalax, TAF1_5496, NVP-ADW742, Nilotinib, Linsitinib, KRAS(G12C)inhibitor-12, I-BRD9, GSK1904529A, Dihydrorotenone (Fig. 9).
Confirmation of risk ERLs expression in OS cell lines
The risk prognosis predictive model was further validated by RT-qPCR analysis performed in three human OS cell lines (HOS, MG63 and U2OS). Normal osteoblast cells (hFOB1.19) were used as the control group. It was found that AL031673.1, SNHG26 and LINC02298 had increased expression in all three OS cell lines. AC023157.3 and LINC02328 had reduced expression in the OS cell lines (Fig. 10A-E). This indicates that the constructed risk prognosis model is persuasive and reliable.
The most significant differential expression was LINC02298, which promoted OS cell proliferation, migration, and invasion
After analyzing the differential expression of the model ERLs in OS cell lines, it was observed that LINC02298 exhibited the most significant differential expression as a risk factor in OS. Therefore, further experimental validation was performed on LINC02298. To further investigate the function of LINC02298 in OS, lentiviral transduction was used to overexpress it in the HOS, MG63 and U2OS cell lines. RT-qPCR confirmed the overexpression of LINC02298 in OS cell lines (Fig. 11A, C and E). The CCK-8 assays demonstrated that LINC02298 overexpression promoted cell proliferation (Fig. 11B, D and F). Moreover, the migration of OS cells was promoted by LINC02298 overexpression, as determined by the wound healing assay (Fig. 11G–I). Moreover, the transwell assay demonstrated that LINC02298 overexpression significantly induced the migration and invasion of OS cells (Fig. 11J–L). In summary, the overexpression of LINC02298 promoted the growth, migration, and invasion of OS cells.
Knockdown of LINC02298 inhibited the proliferation, migration, and invasion of OS cells
After determining the biological functions of overexpressing LINC02298, LINC02298 was knocked down to further validate changes in its function. It was knocked down in the HOS, MG63 and U2OS cell lines with siRNA. RT-qPCR confirmed the LINC02298 knockdown in OS cell lines (Fig. 12A, C and E). CCK-8 assay indicated that the knockdown of LINC02298 inhibited cell proliferation (Fig. 12B, D and F).Moreover, the migration of OS cells was inhibited by LINC02298 suppression, as determined by the wound healing assay (Fig. 12G–I). Conversely, the transwell assays demonstrated that LINC02298 inhibition substantially inhibited the migration and invasion of OS cells (Fig. 12J–L). In summary, the inhibition of LINC02298 inhibited the proliferation, invasion and migration of OS cells.
Pan-cancer analysis of LINC02298
When comparing the expression levels in normal tissue and tumor tissue, we have observed that in the cases of BLCA, CHOL, COAD, ESCA, HNSC, KIRC, KIRP, LIHC, LUAD, LUSC, READ, STAD, THCA, the expression level of LINC02298 is higher in tumor tissue compared to normal tissue. While in many cancers, such as GBM, KICH, PRAD, the expression of LINC02298 is reduced compared to the normal tissue (Fig. 13A). The paired figure shows that the expression of BLCA, BRCA, CHOL, COAD, HNSC, KIRC, KIRP, LIHC, LUSC, STAD, and THCA in tumor tissue is more than that in normal tissue, while the expression in KICH and PRAD is lower than that in normal tissue (Fig. 13B). Subsequently, the Overall Survival (OS) analysis revealed that LINC02298 was significantly associated with poorer OS in ACC, DLBC, THCA, and UVM, while a LINC02298 in LGG was significantly associated with better OS (Fig. 13C). The Disease-Specific Survival (DSS) analysis showed that LINC02298 was significantly associated with poorer DSS in DLBC, READ, THCA, and UVM, while LINC02298 in LGG was significantly associated with better DSS (Fig. 13D). The Progression-Free Interval (PFI) analysis indicated that LINC02298 was significantly associated with better PFI in ACC, COAD, DLBC, and UVM, whereas a LINC02298 in LGG and UCEC was significantly associated with poorer PFI (Fig. E).The Cibersort ssGSEA and ESTIMATE algorithm suggests that LINC02298 is closely related to various immune cells in multiple cancers, indicating that LINC02298 may affect tumor prognosis by mediating multiple immune cells (Fig. 13F–H).
Pan-cancer Analysis of LINC02298. (A,B) The expression of LINC02298 in pan-cancer analysis (C) Overall survival analysis. (D) Disease specific survival analysis. (E) Progress free interval analysis. (F–H) The correlation between LINC02298 and immune cells using Cibersort (F), ssGSEA (G), ESTIMATE (H) algorithm.
Discussion
OS, which has an unfavorable outcome, is among the highly prevalent malignant bone tumors in toddlers and adolescents31,32. Through the application of surgical treatment, chemotherapy, and several other methods, the survival rate has risen substantially due to the progressive improvement of science and technology. On the other hand, the prognosis for certain patients continues to be below average.
Previous studies have shown that ERS is a eukaryotic cell’s response to ER dysfunction, which activates unfolded protein reactions33. The highly proliferative nature of cancer cells can activate the unfolded protein response by disrupting the folding of ER proteins, allowing the continuous growth of cancer cells even in nutrient-deprived environments34. Thus, ERGs are of great significance for targeted therapy in cancer35. In addition, ERLs play a significant role in diagnosing, treating, and estimating the prognosis of specific malignancies. In the case of hepatocellular carcinoma, for instance, Shen et al. discovered six ERLs connected to the prognosis and immune response36. Chen et al. indicated 8 ERLs that were significantly linked with the prognosis of uterine corpus endometrial carcinoma37. Cai et al. identified 9 ERLs with predictive value for the breast cancer prognosis38. However, ERLs have not yet been established as a prognostic model for OS patients.
This study constructed a new clinical prognostic model comprising five ERLs (AC023157.3, AL031673.1, LINC02298, LINC02328, SNHG26) closely related to tumor development and clinical prognosis. He et al. proved that AC023157.3 is a protective factor for OS39. Although the functions of SNHG26 and LINC02298 in OS have not been reported, their functions in other tumors have been explored. Wu et al. have proved that lncRNA SNHG26 can promote the progression and metastasis of gastric cancer by inducing c-Myc protein translation and energy metabolism positive feedback loop40. Jiang et al. have proved that lncRNA SNHG26 can promote the growth, metastasis, and cisplatin resistance of tongue squamous cell carcinoma through the PGK1/Akt/mTOR pathway41. Wang et al. demonstrated that LINC02298 promotes the progression of HCC by targeting the miR-28-5p/CCDC6 pathway42. Therefore, they may also have similar effects on OS. Further, the literature has not confirmed the roles of LINC02328 and AL031673.1. GSVA validated that the effect of ERLs in signature on OS patients’ prognosis might be attributed to many pathways.
The immune microenvironment of the tumor is essential in developing and treating cancers43. Immunotherapy has been the treatment of choice for tumors in recent years44. In addition to influencing the clinical prognosis of cancer patients, the abundance, diversity, and activation status of immune cell expansion in the tumor microenvironment have been demonstrated to be critical factors in tumorigenesis45. Immune function analysis showed that multiple immune pathways were closely related to the prognosis of OS. The Immune, Stromal, and Estimate score were increased in the low-risk group compared to the high-risk group, suggesting that immune and stromal cell infiltration increased considerably in the low-risk group. Most research indicates that an augmentation in immune cells is, on average, more advantageous for a patient’s prognosis46. More immune cell infiltration may mean that the immune system is more effective at recognizing and removing tumor cells, thereby inhibiting the growth and spread of the tumor47. Immune infiltration analysis showed a decrease in CD8 T cells in the high-risk group, which is considered to be the first-line defense cell type against cancer progression48. Studies have shown that OS cells secrete CXCL14 to stimulate fibroblasts to produce TGFβ and increase the invasion and migration of OS, and our study also found that CD8 T cell infiltration was reduced in the high-risk group, which was associated with a worse prognosis49. Th1 cells are one of the CD4 T cell subsets, and Th1 cells mainly produce IFN-γ, which promotes cell-mediated immune responses and plays an important role in the clearance of tumor cells50. The infiltration of Th1 cells in the high-risk group decreased, which led to a worse prognosis. Immune checkpoint genes play a crucial role in immune responses, affecting the intensity and direction of immune reactions by regulating the activity and function of T cells51. Decreased expression of CD80, CD70, CD28, CD48 in high-risk group may directly lead to inactivation and apoptosis of T cells, thereby reducing the attack on tumor cells52. It may also promote the proliferation of immunosuppressive cells (such as regulatory T cells and myeloid-derived suppressor cells) in the tumor microenvironment, further weakening the anti-tumor immune response. In summary, the decreased expression of these immune checkpoint genes in high-risk patients may reflect impaired immune system function and active immune escape in OS patients. This finding has important guiding significance for the prognosis assessment and the formulation of immunotherapy strategies for OS. Targeted therapy targeting these immune checkpoint molecules may help enhance patients’ anti-tumor immune response and improve prognosis. However, additional experimental investigation is required to ascertain the precise functions. CDK9_5576, CDK9_5038, Acetalax, TAF1_5496, NVP-ADW742, Nilotinib, Linsitinib, KRAS(G12C)inhibitor-12, I-BRD9, GSK1904529A, Dihydrorotenine showed higher sensitivity towards the high-risk group according to drug susceptibility analysis indicating that they may have a possibility to be utilized as a therapeutic agent for OS.
Here, the bioinformatics analysis and RT-qPCR validation revealed that LINC02298 was upregulated in OS cells and was negatively correlated with prognosis. This study investigated the function of LINC02298 in OS cells. Its overexpression significantly promoted the invasion, migration, and proliferation of OS cells. The knockdown experiment of LINC02298 provided more evidence of its biological function in OS. In summary, targeting LINC02298 could potentially be a therapeutic target in OS. The pan-cancer analysis showed that it was differentially expressed in a variety of cancers and may be a risk or protective factor for multiple cancers, and the immune infiltration analysis further suggested the complex immune mechanism of its role. This revealed that LINC02298 exhibits considerable prognostic potential for various cancers.
Nevertheless, this study is subject to some particular limitations. Initially, a more thorough examination of the mechanics behind ERLs is required. Similarly, more OS cell lines could prove advantageous for conducting a more comprehensive analysis of the results. Furthermore, confirming the authenticity of signatures is necessary in other external databases and perhaps in large-scale multicenter processes. Finally, a total of 5 ERLs were screened in this study, but the research focused on the function of LINC02298, and the functions of the other 4 lncRNAs would be further confirmed in future studies.
Conclusion
In conclusion, the current research involved the identification of a new signature using ERLs. The validity of this signature was confirmed via RT-qPCR on OS cell lines. It was also found that LINC02298 can promote cell development, movement, and invasion. The results of this study suggest that the ERL signature can successfully predict the prognosis of OS, and LINC02298 may be a potential therapeutic target.
Data availability
Data is provided within the manuscript or supplementary information files.
Abbreviations
- OS:
-
Osteosarcoma
- ER:
-
Endoplasmic reticulum
- ERS:
-
Endoplasmic reticulum stress
- ERGs:
-
Endoplasmic reticulum stress related genes
- ERLs:
-
Endoplasmic reticulum stress related lncRNAs
- LASSO:
-
Least absolute shrinkage and selection operator
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- GSVA:
-
Gene set variation analysis
- ATCC:
-
American type culture collection
- MEM:
-
Minimal essential medium
- DMEM:
-
Dulbecco’s modified eagle medium
- FBS:
-
Fetal bovine serum
- cDNA:
-
Complementary DNA
- TME:
-
Tumor microenvironment
- IC50:
-
Half-maximal inhibitory concentration
- SiRNA:
-
Small interfering RNA
- APC:
-
Antigen-presenting cells
- CCRs:
-
Chemokine receptors
- OS:
-
Overall survival
- DSS:
-
Disease-specific survival
- PFI:
-
Progression-free interval
- ACC:
-
Adrenocortical cancer
- BLCA:
-
Bladder cancer
- BRCA:
-
Breast cancer
- CESC:
-
Cervical cancer
- CHOL:
-
Bile duct cancer
- COAD:
-
Colon cancer
- DLBC:
-
Large B-cell lymphoma
- ESCA:
-
Esophageal cancer
- GBM:
-
Glioblastoma
- HNSC:
-
Head and neck cancer
- KICH:
-
Kidney chromophobe
- KIRC:
-
Kidney clear cell carcinoma
- KIRP:
-
Kidney papillary cell carcinoma
- LAML:
-
Acute myeloid leukemia
- LGG:
-
Lower grade glioma
- LIHC:
-
Liver cancer
- LUAD:
-
Lung adenocarcinoma
- LUSC:
-
Lung squamous cell carcinoma
- MESO:
-
Mesothelioma
- OV:
-
Ovarian cancer
- PAAD:
-
Pancreatic cancer
- PCPG:
-
Pheochromocytoma
- PRAD:
-
Prostate cancer
- READ:
-
Rectal cancer
- SARC:
-
Sarcoma
- SKCM:
-
Skin cutaneous melanoma
- STAD:
-
Stomach cancer
- TGCT:
-
Testicular cancer
- THCA:
-
Thyroid cancer
- THYM:
-
Thymoma
- UCEC:
-
Endometrioid cancer
- UCS:
-
Uterine carcinosarcoma
- UVM:
-
Uveal melanoma
References
Arndt, C. A., Rose, P. S., Folpe, A. L. & Laack, N. N. Common musculoskeletal tumors of childhood and adolescence. Mayo Clin. Proc. 87, 475–487 (2012).
Rojas, G. A., Hubbard, A. K., Diessner, B. J., Ribeiro, K. B. & Spector, L. G. International trends in incidence of osteosarcoma (1988–2012). Int. J. Cancer. 149, 1044–1053 (2021).
Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 72, 7–33 (2022).
Sadykova, L. R. et al. Epidemiology and risk factors of osteosarcoma. Cancer Invest. 38, 259–269 (2020).
Wakamatsu, T. et al. Prognostic implication of adjuvant/neoadjuvant chemotherapy consisting of doxorubicin and ifosfamide in patients with extraskeletal osteosarcoma. Int. J. Clin. Oncol. 24, 1311–1319 (2019).
Hao, Y. et al. Prognostic value of tumoral and peritumoral magnetic resonance parameters in osteosarcoma patients for monitoring chemotherapy response. Eur. Radiol. 31, 3518–3529 (2021).
Harrison, D. J., Geller, D. S., Gill, J. D., Lewis, V. O. & Gorlick, R. Current and future therapeutic approaches for osteosarcoma. Expert Rev. Anticancer Ther. 18, 39–50 (2018).
Kager, L. et al. Primary metastatic osteosarcoma: presentation and outcome of patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J. Clin. Oncol. 21, 2011–2018 (2003).
Simpson, S. et al. Comparative review of human and canine osteosarcoma: morphology, epidemiology, prognosis, treatment and genetics. Acta Vet. Scand. 59, 71 (2017).
Sayles, L. C. et al. Sweet-Cordero, Genome-informed targeted therapy for osteosarcoma. Cancer Discov. 9, 46–63 (2019).
Xiao, X. et al. Individualized chemotherapy for osteosarcoma and identification of gene mutations in osteosarcoma. Tumour Biol. 36, 2427–2435 (2015).
Zhang, X. et al. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res.70, 2350–2358 (2010).
Han, G. et al. LncRNA BCRT1 facilitates osteosarcoma progression via regulating miR-1303/FGF7 axis. Aging (Albany NY) 13, 15501–15510 (2021).
Xiong, J. et al. LncRNA FOXP4-AS1 promotes progression of Ewing sarcoma and is associated with Immune infiltrates. Front. Oncol.11, 718876 (2021).
Yang, P. et al. Long non-coding RNAs and cancer mechanisms: Immune cells and inflammatory cytokines in the tumor microenvironment. Med. Oncol. 39, 108 (2022).
Ji, S., Wang, S., Zhao, X. & Lv, L. Long noncoding RNA NEAT1 regulates the development of osteosarcoma through sponging miR-34a-5p to mediate HOXA13 expression as a competitive endogenous RNA. Mol. Genet. Genomic Med.7, e673 (2019).
Ye, K. et al. Long noncoding RNA GAS5 suppresses cell growth and epithelial-mesenchymal transition in Osteosarcoma by regulating the miR-221/ARHI pathway. J. Cell. Biochem.118, 4772–4781 (2017).
Ni, S., Hong, J., Li, W., Ye, M. & Li, J. Construction of a cuproptosis-related lncRNA signature for predicting prognosis and immune landscape in osteosarcoma patients. Cancer Med. 12, 5009–5024 (2023).
Ming, H., Li, B., Zhou, L., Goel, A. & Huang, C. Long non-coding RNAs and cancer metastasis: molecular basis and therapeutic implications. Biochim. Biophys. Acta Rev. Cancer. 1875, 188519 (2021).
Fagone, P. & Jackowski, S. Membrane phospholipid synthesis and endoplasmic reticulum function. J. Lipid Res.50 Suppl, S311–S316 (2009).
Yao, J. et al. The attenuating effect of the intraovarian bone morphogenetic protein 4 on age-related endoplasmic reticulum stress in chicken follicular cells. Oxid. Med. Cell Longev. 2020 4175613. (2020).
Li, Z. et al. ER stress-related molecules induced by Hantaan virus infection in differentiated THP-1 cells. Cell. Stress Chaperones. 26, 41–50 (2021).
Song, J. et al. Non-structural protein 2B of human rhinovirus 16 activates both PERK and ATF6 rather than IRE1 to trigger ER stress.Viruse 11 (2019).
Urra, H., Dufey, E., Avril, T., Chevet, E. & Hetz, C. Endoplasmic reticulum stress and the hallmarks of cancer. Trends Cancer. 2, 252–262 (2016).
Markouli, M., Strepkos, D., Papavassiliou, A. G. & Piperi, C. Targeting of endoplasmic reticulum (ER) stress in gliomas. Pharmacol. Res. 157, 104823 (2020).
Huang, Z. L. et al. Long non-coding RNA MEG3 induces cell apoptosis in esophageal cancer through endoplasmic reticulum stress. Oncol. Rep. 37, 3093–3099 (2017).
Chen, R. P. et al. Involvement of endoplasmic reticulum stress and p53 in lncRNA MEG3-induced human hepatoma HepG2 cell apoptosis. Oncol. Rep. 36, 1649–1657 (2016).
Ding, Z., Kang, J. & Yang, Y. Long non-coding RNA CASC2 enhances irradiation-induced endoplasmic reticulum stress in NSCLC cells through PERK signaling. 3 Biotech. 10, 449 (2020).
Gene Ontology Consortium. Going forward. Nucleic Acids Res. 43, D1049–D1056 (2015).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Jo, V. Y. & Fletcher, C. D. WHO classification of soft tissue tumours: an update based on the 2013 (4th) edition. Pathology 46, 95–104 (2014).
Zhang, C. et al. Diagnostic and prognostic significance of dysregulated expression of circular RNAs in osteosarcoma. Expert Rev. Mol. Diagn. 21, 235–244 (2021).
Moreno, J. A. & Tiffany-Castiglioni, E. The chaperone Grp78 in protein folding disorders of the nervous system. Neurochem Res. 40, 329–335 (2015).
Lin, Y., Jiang, M., Chen, W., Zhao, T. & Wei, Y. Cancer and ER stress: mutual crosstalk between autophagy, oxidative stress and inflammatory response 118109249 (Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie, 2019).
Li, S. et al. The stability of FKBP9 maintained by BiP is crucial for glioma progression. Genes Dis. 11, 101123 (2023).
Shen, X., Wu, S., Yang, Z. & Zhu, C. Establishment of an endoplasmic reticulum stress-associated lncRNAs model to predict prognosis and immunological characteristics in hepatocellular carcinoma. PLoS One 18, e0287724 (2023).
Chen, J., Shen, L. & Yang, Y. Endoplasmic reticulum stress related IncRNA signature predicts the prognosis and immune response evaluation of uterine corpus endometrial carcinoma. Front. Oncol. 12, 1064223 (2022).
Cai, J. et al. Development and validation of a novel endoplasmic reticulum stress-related lncRNA prognostic signature and candidate drugs in breast cancer. Front. Genet. 13, 949314 (2022).
He, Y., Zhou, H., Xu, H., You, H. & Cheng, H. Construction of an immune-related lncRNA signature that predicts prognosis and immune microenvironment in osteosarcoma patients. Front. Oncol. 12, 769202 (2022).
Wu, Z. H. et al. LncRNA SNHG26 promotes gastric cancer progression and metastasis by inducing c-Myc protein translation and an energy metabolism positive feedback loop. Cell. Death Dis. 15, 236 (2024).
Jiang, Q. et al. lncRNA SNHG26 promoted the growth, metastasis, and cisplatin resistance of tongue squamous cell carcinoma through PGK1/Akt/mTOR signal pathway. Mol. Ther. Oncolytics. 24, 355–370 (2022).
Wang, J., Xu, B., Liang, L. & Chen, Q. Long non-coding RNA 02298 promotes the malignancy of HCC by targeting the miR-28-5p/CCDC6 pathway. Biochem. Genet. (2024).
Smyth, M. J., Ngiow, S. F., Ribas, A. & Teng, M. W. Combination cancer immunotherapies tailored to the tumour microenvironment. Nat. Rev. Clin. Oncol. 13, 143–158 (2016).
Kaymak, I., Williams, K. S., Cantor, J. R. & Jones, R. G. Immunometabolic interplay in the tumor microenvironment. Cancer Cell 39, 28–37 (2021).
Dyson, K. A. et al. Emerging trends in immunotherapy for pediatric sarcomas. J. Hematol. Oncol. 12, 78 (2019).
Zhang, Y. & Zhang, Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell. Mol. Immunol. 17, 807–821 (2020).
Fridman, W. H., Zitvogel, L., Sautès-Fridman, C. & Kroemer, G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 14, 717–734 (2017).
Kato, T. et al. Cancer-associated fibroblasts affect Intratumoral CD8(+) and FoxP3(+) T cells Via IL6 in the tumor microenvironment. Clin. Cancer Res. 24, 4820–4833 (2018).
Xu, Y. et al. Osteosarcoma cells secrete CXCL14 that activates integrin α11β1 on fibroblasts to form a lung metastatic niche. Cancer Res. 84, 994–1012 (2024).
Künzli, M. & Masopust, D. CD4(+) T cell memory. Nat. Immunol. 24, 903–914 (2023).
Wang, J. et al. Acetate reprogrammes tumour metabolism and promotes PD-L1 expression and immune evasion by upregulating c-Myc. Nat. Metab. 6, 914–932 (2024).
Shibru, B. et al. Detection of immune checkpoint receptors - a current challenge in clinical flow cytometry. Front. Immunol. 12, 694055 (2021).
Acknowledgements
The authors express their sincere gratitude to all the reviewers who contributed to the manuscript review process, as well as to MJEditor (www.mjeditor.com) for providing invaluable linguistic support during the preparation of this work.
Funding
This study was supported by the International Scientific and Technological Cooperation Project of Jiangxi Province (No.20232BBH80001) and National Natural Science Foundation of China (No.82060403).
Author information
Authors and Affiliations
Contributions
Study concept and design: Xu PC, Cheng XG and Yuan JH . Collection and analysis of the data: Wu ZW and Li KH. Administrative, technical or material assistance: He DW, Zhao JMH and Wang YM . Manuscript written: Wu TL, Li T and Xu PC. All authors participated in the article’s development and endorsed the final version for submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xu, P., Yuan, J., Li, K. et al. Development and validation of a novel endoplasmic reticulum stress-related lncRNAs signature in osteosarcoma. Sci Rep 14, 25590 (2024). https://doi.org/10.1038/s41598-024-76841-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-76841-9