Abstract
Plasmids, as important genetic elements apart from chromosomes, often carry multiple resistance genes and various mobile genetic elements, enabling them to acquire more exogenous genes and confer additional resistance phenotypes to bacteria. Various carbapenem resistance genes are often located on IncN plasmids, and several reports have linked fusion plasmids to IncN plasmids. Therefore, this study aims to explore the emergence, molecular structure characteristics, and resistance features mediated by IncN fusion plasmids carrying multiple carbapenem resistance genes. In this study, species identification was performed using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS). Polymerase chain reaction (PCR) was employed to detect the presence of carbapenem resistance genes in the strains. PCR-based replicon typing (PBRT) was used to identify IncN plasmids. Plasmids were analyzed through S1-nuclease pulsed-field gel electrophoresis (S1-PFGE), Southern blotting, conjugation experiments, and stability tests. Whole-genome sequencing (WGS) and antimicrobial susceptibility testing (AST) were conducted to characterize the target strains. Four strains containing IncN plasmids were identified: two Klebsiella pneumoniae, one Escherichia coli, and one Enterobacter cloacae, all harboring carbapenem resistance genes. Among them, two IncN plasmids (pFAHZZU7605-KPC-IMP and pFAHZZU7865-IMP) contained blaIMP−4 and exhibited similar molecular structure characteristics. Notably, the pFAHZZU7605-KPC-IMP plasmid harbored both IncN and IncR replicons. We hypothesize that the pFAHZZU7605-KPC-IMP fusion plasmid resulted from the recombination of a pFAHZZU7865-IMP-like plasmid and an IncR-like plasmid. Further analysis of the plasmid’s genetic elements revealed that insertion sequences ISKpn19 and ISKpn27 played crucial roles in the plasmid recombination and fusion process. In clinical settings, plasmids carrying different resistance genes can undergo fusion, mediated by genetic elements, thereby expanding the resistance spectrum of host bacteria. Hence, it is essential to enhance the monitoring and research of transposable elements to control the spread of multidrug-resistant bacteria.
Similar content being viewed by others
Introduction
In recent years, with the widespread use of carbapenem antibiotics, such as meropenem and imipenem in clinical settings, the detection rate of carbapenem-resistant strains has been increasing annually. These strains can cause high mortality rates in patients, posing severe challenges to global public health and clinical treatment1,2.
Genes encoding carbapenemases (blaNDM, blaKPC, blaGES, blaVIM, blaIMP, blaOXA−48−like) are frequently located on mobile genetic elements known as plasmids, which facilitate horizontal gene transfer through plasmid conjugation3. As a result, plasmids have become critical genetic elements in mediating bacterial resistance. Most carbapenemase-producing strains typically harbor a single type of carbapenemase-encoding gene. However, in cases where bacteria carry two or more carbapenemase-encoding genes, these genes are usually located on distinct plasmids4. There have been documented instances where both blaKPC−2 and blaIMP−4carbapenemase-encoding genes were found on the same plasmid. In one case, this plasmid was a fusion of IncF and IncHI plasmids. Unfortunately, the plasmid characteristics in the other case were not specified5,6. Additionally, there are reports of blaNDM−5 and blaKPC−2coexisting on a fusion plasmid composed of IncFII-IncR-IncN, alongside cases where different carbapenemase-encoding genes coexist on a single plasmid7,8,9,10.
The KPC-type carbapenemase was first identified in the United States in 1996 and subsequently led to outbreaks in Klebsiella pneumoniae11. Since then, KPC-type carbapenemase have become the most prevalent form of carbapenemase12. IMP-type carbapenemase were initially detected in Pseudomonas aeruginosa in 1988 and have subsequently emerged in other strains, including Acinetobacter baumannii, Stenotrophomonas maltophilia, and Klebsiella pneumoniae, contributing to their widespread dissemination globally13,14,15,16. Research indicates that IncN plasmids play a crucial role in the dissemination of KPC, VIM and IMP-type carbapenemase-encoding genes17,18,19. The reference plasmid R46, first identified in Escherichia coliin 1971, was the initial IncN plasmid characterized20. Over time, it underwent two deletions of resistance genes and genetic elements, evolving into the plasmid pKM101. Furthermore, in 2014, Yang et al. observed fusion events between IncN plasmids and IncF (F33: A-: B-) plasmids in Escherichia coliisolated from chickens21. IncN plasmids can enhance resistance by losing genes or merging with other structures, thereby providing their hosts with additional phenotypes and contributing to increased resistance.
Currently, research on fusion plasmids, especially those harboring multiple resistance genes, remains limited. In our study, we identified a novel IncN-IncR fusion plasmid carrying blaTEM−40, blaIMP−4, and blaKPC−2 in a strain of ST1393 Klebsiella pneumoniae, which we named pFAHZZU7605-KPC-IMP. During our investigation, we observed that the IncN region of pFAHZZU7605-KPC-IMP bears striking similarity to another IncN plasmid, designated pFAHZZU7865-IMP, discovered in an Enterobacter cloacae strain carrying blaIMP−4. Consequently, our study provides a comprehensive characterization of the features of both plasmids.
Materials and methods
Collection of strains
During routine surveillance of carbapenem-resistant Enterobacteriaceae (CRE) conducted from April 2023 to September 2023 at a tertiary hospital in Zhengzhou, Henan Province, China, carbapenem-resistant Enterobacteriaceae strains were collected. Species identification was performed using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF/MS). The strains carrying carbapenem-resistance genes (blaKPC, blaNDM, blaVIM, blaIMP, blaOXA−48) were identified using PCR amplification. PCR-based replicon typing (PBRT) was employed to determine the types of IncN plasmids.
Antimicrobial susceptibility testing
The antimicrobial susceptibility testing was conducted using agar dilution and microbroth dilution methods, with Escherichia coliATCC 25,922 employed as the control strain. Interpretation of results for tigecycline and polymyxins followed the 2022 European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines22. For other antibiotics, interpretation was based on the 2022 Clinical and Laboratory Standards Institute (CLSI) guidelines.
Plasmid characterization and conjugation assay
The number and size of plasmids in Klebsiella pneumoniae FAHZZU7605 and Enterobacter cloacae FAHZZU7865 were determined using S1-PFGE. Southern blotting with digoxigenin-labeled specific probes was employed to identify the location of plasmids carrying blaKPC−2 and blaIMP−4. For the conjugation assay, rifampicin-resistant Escherichia coli EC600 was used as the recipient strain to verify the transferability of the target plasmid. Transconjugants were identified on MHA plates containing 300 µg/mL rifampicin and 2 µg/mL meropenem using MALDI-TOF/MS, and the conjugation frequency of the plasmids was calculated. PCR was performed to detect the presence of blaKPC−2, blaIMP−4, and IncN plasmids in the transconjugants. Antimicrobial susceptibility testing was also conducted on the transconjugants.
Whole genome sequencing analysis
To better understand the genetic characteristics of IncN plasmids, this study employed the SDS method for genomic DNA extraction, followed by DNA sequencing using the PacBio Sequel platform and Illumina NovaSeq PE150. Subsequently, the entire genome was annotated using Prokka, and multilocus sequence typing (MLST) was performed on the strains utilizing the pMLST database. The potential for strain transferability was predicted using the oriTfinder database. Furthermore, antibiotic resistance genes (ARGs) were detected with ResFinder 4.4.2, and plasmid replicon types were identified using PlasmidFinder 2.1. Insertion sequences, transposons, and integrons were detected using the ISFinder database and sequence alignment methods, while toxin-antitoxin system-related genes were queried through the TADB database23. Finally, multiple plasmid circular comparison images were generated using the BLAST Ring Image Generator (BRIG), and plasmid gene structure diagrams were illustrated using Easyfig 2.3.
Results
Distribution and antimicrobial susceptibility characteristics of carbapenem-resistant strains carrying IncN plasmids
From February 2023 to September 2023, a total of 1073 carbapenem-resistant strains were collected, predominantly comprising Acinetobacter baumannii (640), Klebsiella pneumoniae (273), Pseudomonas aeruginosa (101), Escherichia coli24, and Enterobacter cloacae17. Among these, 4 strains were found to carry IncN plasmids (0.37%). Specifically, these included 2 Klebsiella pneumoniae (FAHZZU7270 and FAHZZU7605), 1 Escherichia coli (FAHZZU8201), and 1 Enterobacter cloacae (FAHZZU7865). These strains were isolated from bronchoalveolar lavage fluid, tracheal aspirate, blood, and sputum samples. Their antimicrobial susceptibility testing results are detailed in Table 1. The aforementioned strains exhibited resistance to imipenem, meropenem, ceftriaxone, cefotaxime, ceftazidime, amoxicillin-clavulanic acid, and cefepime, but were sensitive to amikacin, gentamicin, tigecycline, and colistin. Additionally, due to the presence of the blaSHV gene in strains FAHZZU7270 and FAHZZU7605, these strains also showed resistance to aztreonam.
Characterization of antimicrobial resistance genes on IncN plasmids
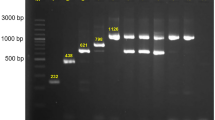
By PCR amplification, we detected the presence of carbapenemase-encoding genes in strains FAHZZU7270, FAHZZU7865, and FAHZZU8201, each containing a single carbapenemase-encoding gene: blaKPC, blaIMP, and blaNDM, respectively. However, strain FAHZZU7605 harbors both blaKPC and blaIMP resistance genes simultaneously. S1-PFGE and Southern Bolting results revealed that the KPC and IMP-type carbapenemase-encoding genes in Klebsiella pneumoniae FAHZZU7605 and the IMP-type carbapenemase-encoding gene in Enterobacter cloacae FAHZZU7865 are located on plasmids. Specifically, blaKPC−2 and blaIMP−4 in FAHZZU7605 are located on a plasmid approximately 160 kb in size, while blaIMP−4 in FAHZZU7865 is located on a plasmid of approximately 50 kb (Fig. 1). Whole-genome sequencing of both strains indicated that blaIMP−4 in FAHZZU7865 is located on an IncN plasmid, whereas blaKPC−2 and blaIMP−4 in FAHZZU7605 are situated on a fusion plasmid of IncN/IncR types (Table 2). Additionally, both plasmids contain a class 1 integron In823 associated with blaIMP−4 (Fig. 3), which is carried by a transposon-like structure IS26-xerC-blaIMP−4-itrA-IS610025.
Conjugation transfer experiments demonstrated that both plasmids, pFAHZZU7605-KPC-IMP and pFAHZZU7865-IMP, along with their associated resistance genes, were successfully transferred to the recipient strain EC600. This finding is consistent with the oriTfinder database results, which indicated that both plasmids possess the oriT region, Relaxase, T4CP, and T4SS gene clusters, predicting their capability for conjugative transfer. Specifically, the conjugation frequency of plasmid pFAHZZU7605-KPC-IMP is 9.6 × 10^-5, while that of plasmid pFAHZZU7865-IMP is 4.18 × 10^-4. Stability assays showed that, after 20 passages without antibiotic selection, both plasmids and their resistance genes remained stable in the transconjugants. Similarly, the antimicrobial susceptibility profiles of the transconjugants containing these plasmids are presented in Table 1.
Detection of fusion symbolism
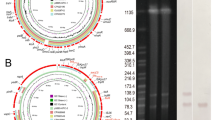
Due to the pFAHZZU7605-KPC-IMP plasmid harboring replication determinants of two plasmids, IncN and IncR, and carrying both carbapenemase-encoding genes, blaKPC and blaIMP, whereas the IncN plasmid pFAHZZU7865-IMP only carries the IMP carbapenemase-encoding gene, we hypothesize that the pFAHZZU7605-KPC-IMP plasmid may have arisen from recombination between an IncN plasmid similar to pFAHZZU7865-IMP and an IncR plasmid. BLASTN searches in the NCBI nucleotide database showed that pFAHZZU7605-KPC-IMP shares 63% query cover and 100% nucleotide identity with plasmid pSMBC50-107 K from Klebsiella pneumoniae subsp. rhinoscleromatis (GenBank Accession number: CP109830.1) (Fig. 2). Similarly, it exhibits 62% query cover and 100% nucleotide identity with plasmid p16055-KPC (GenBank Accession number: MN823985.1). Plasmid database analysis indicates that both pSMBC50-107 K and p16055-KPC are IncR plasmids.
Comparative circular maps of plasmids pFAHZZU7605-KPC-IMP, pFAHZZU7865-IMP, and pSMBC50-107 K. The innermost circle represents the reference plasmid pFAHZZU7605-KPC-IMP, shown in blue. The pFAHZZU7865-IMP plasmid is shown in yellow, and the pSMBC50-107 K plasmid is shown in pink. The light pink indicates the IncR region of the fusion plasmid, while the orange indicates the IncN region of the fusion plasmid.
It is noteworthy that linear comparison analysis reveals high similarity between regions 1 and 3 of the pFAHZZU7605-KPC-IMP plasmid and the corresponding regions of the pFAHZZU7865-IMP plasmid. Moreover, gene element ISKpn19 is present at the end of region 1, the beginning of region 3, and on the pFAHZZU7865-IMP plasmid. Additionally, the flanking sequences 8 bp to the left and right of ISKpn19 on the pFAHZZU7865-IMP plasmid are AGGCAATG and GCTCACGC, respectively, matching those flanking ISKpn19 in regions 1 and 3. Furthermore, in region 2, high overlap is observed with the pSMBC50-107 K plasmid. Similarly, ISKpn27 is found at the beginning and end of region 2, as well as on the pSMBC50-107 K plasmid. The flanking sequences 8 bp to the left and right of ISKpn27 on the pSMBC50-107 K plasmid are ACATTAAC and CGAGTTGC, respectively, identical to the flanking sequences of ISKpn27 at the beginning and end of region 2 (Fig. 3).
Discussion
Among the collected carbapenem-resistant strains, IncN plasmids were identified in Klebsiella pneumoniae, Escherichia coli, and Enterobacter cloacae, but not in the abundant strains of Acinetobacter baumannii and Pseudomonas aeruginosa. This suggests that IncN plasmids may be more prevalent among Klebsiella pneumoniae, Escherichia coli, and Enterobacter cloacae. Klebsiella pneumoniae and Enterobacter cloacae are opportunistic pathogens and significant causes of hospital-acquired infections. Two clinical isolates, FAHZZU7605 and FAHZZU7865, from these species caused severe pulmonary infections. Despite treatment with carbapenem antibiotics and/or polymyxins like tigecycline, the therapeutic outcomes remained unsatisfactory.
Based on Multi Locus Sequence Typing (MLST) database analysis, Klebsiella pneumoniae strain FAHZZU7605 was identified as sequence type ST1393. Research indicates that in China, the predominant sequence type of KPC-2-producing carbapenemase Klebsiella pneumoniae responsible for clinical infections is ST11, followed by ST23, ST37, and ST15. In contrast, in the Lisbon region of Portugal, the most common sequence type of KPC-3-producing carbapenemase Klebsiella pneumoniaeis ST13, followed by ST17, ST348, and ST23124,26. These findings suggest that the prevalent subtypes of carbapenemase-encoding genes and their associated sequence types vary across regions. Notably, ST1393 has not been previously reported, highlighting the need for increased vigilance.
Plasmids are key genetic elements that facilitate the transfer of carbapenem resistance genes between different species. Studies have shown that IncN plasmids are the primary plasmid type carrying and spreading the blaIMP−4gene27. In this study, we identified two IncN plasmids carrying blaIMP−4, designated as pFAHZZU7865-IMP and pFAHZZU7605-KPC-IMP. Both plasmids possess conjugative transfer capabilities; however, the conjugative transfer frequency of plasmid pFAHZZU7605-KPC-IMP is slightly lower than that of plasmid pFAHZZU7865-IMP. This difference may be attributed to the larger molecular weight of plasmid pFAHZZU7605-KPC-IMP. However, the recipient strain that acquired this plasmid exhibited reduced resistance to carbapenem antibiotics compared to the original host strain, Enterobacter cloacaeFAHZZU7865. Notably, we observed that pFAHZZU7605-KPC-IMP-EC600 exhibits higher resistance to imipenem and meropenem compared to pFAHZZU7865-IMP-EC600. This increased resistance is likely due to the presence of two carbapenem resistance genes in pFAHZZU7605-KPC-IMP-EC600, resulting in higher levels of resistance in the host. Research indicates that an increase in plasmid copy number, elevated expression of plasmid genes, and reduced cell membrane permeability can enhance the antibiotic resistance of host bacteria28,29,30. Due to the interactions between plasmids and their host bacteria, variations in gene expression levels and copy numbers of plasmids exist among different strains, along with differences in the permeability of host bacterial cell membranes. Therefore, even when carrying the same plasmid, different host bacteria may exhibit varying degrees of antibiotic resistance. Both plasmids pFAHZZU7605-KPC-IMP and pFAHZZU7865-IMP can stably exist in the recipient strains. Querying the TADB database revealed that plasmid pFAHZZU7605-KPC-IMP contains genes related to stability, including toxin and antitoxin system genes vapC, vagC, and the parA-parMsystem31,32. Although these related genes were not found in plasmid pFAHZZU7865-IMP, both plasmids contain the ardBgene, which has been shown to potentially aid in the colonization of plasmids in new hosts33.
Studies on the pSMBC50-107 K plasmid indicate that it belongs to the IncR plasmid group and carries the blaKPC−2gene, conferring resistance to meropenem34. The pFAHZZU7605-KPC-IMP plasmid is relatively large and is identified as a fusion plasmid of the IncN/IncR types, composed of two plasmids: a pFAHZZU7865-IMP-like plasmid and a pSMBC50-107 K-like plasmid. Typically, IncR plasmids are unable to undergo conjugative transfer35; however, we observed that plasmid pFAHZZU7605-KPC-IMP can successfully transfer to Escherichia coli EC600 and maintain stability. Furthermore, IncR plasmids can expand their host range through plasmid fusion. This plasmid fusion may provide a novel mechanism for IncR plasmids to overcome host restrictions and disseminate antibiotic resistance genes.
Bacteria can capture resistance genes through the action of insertion sequences (IS) or transposons, thereby increasing their resistance levels under high antibiotic pressure36. In this study, plasmid circular maps and linear comparison analyses reveal the presence of IS elements ISKpn19 and ISKpn27at the junctions between the pSMBC50-107 K-like and pFAHZZU7865-IMP plasmids. IS elements can transfer themselves, and even surrounding resistance genes, to other genomic locations via a cut-and-paste mechanism37. Consequently, the pFAHZZU7605-KPC-IMP plasmid likely evolved through the recombination of a pFAHZZU7865-IMP-like plasmid and a pSMBC50-107 K-like plasmid, facilitated by the actions of ISKpn19 and ISKpn27.
Studies on the pSMBC50-107 K plasmid indicate that it was also isolated from Zhengzhou, Henan Province, China, and can successfully undergo conjugative transfer. Additionally, related research has reported that IncR plasmids have the highest prevalence among carbapenem-resistant strains in the Henan region38. This provides both geographical and numerical bases for the recombination of pFAHZZU7865-IMP-like and pSMBC50-107 K-like plasmids. Although previous reports have described IncN/IncR fusion plasmids, we observed the fusion of two plasmids, each carrying carbapenem resistance genes, which further exacerbates the issue of antibiotic resistance. Additionally, we revealed the critical role of the gene elements ISKpn19 and ISKpn27 in the plasmid fusion process, indicating that these genetic elements may play a significant role in the formation of multidrug-resistant bacteria. This finding underscores the need to closely monitor the potential impact of gene elements on the evolution of multidrug-resistant strains. Similarly, a previous study reported the fusion of plasmids carrying blaNDM and blaIMP−4 genes mediated by the IS26element9. Therefore, transposable elements not only facilitate the transfer of gene segments, but also mediate the recombination between different plasmids, thereby enhancing the resistance of the host bacteria.
Conclusion
In this study, we characterized the transferrable fusion plasmid pFAHZZU7605-KPC-IMP, which carries both blaIMP−4 and blaKPC−2 genes simultaneously. Additionally, we described the transferrable plasmid pFAHZZU7865-IMP, which shares high homology in the IncN region with the fusion plasmid. The fusion of these two plasmids was mediated by ISKpn19 and ISKpn27 elements, each carrying distinct resistance genes. This resulted in the formation of a superplasmid. This phenomenon expands the resistance spectrum of the host bacteria, thereby facilitating the emergence of multidrug-resistant strains.
Data availability
Sequence data that support the findings of this study have been deposited in the National Center for Biotechnology Information with the primary accession code PRJNA1130433.
References
Debnath, A. et al. Clinical outcomes and treatment strategies in patients with Non-carbapenemase-producing Carbapenem-Resistant Versus Carbapenem-Susceptible enterobacterales infections. Ann. Pharmacother. 57 (7), 803–812 (2023).
Chen, J. et al. Carbapenem-resistant Enterobacter cloacae complex in a tertiary hospital in Northeast China, 2010–2019. BMC Infect. Dis. 21 (1), 611 (2021).
Rankin, D. A. et al. Concurrent transmission of multiple carbapenemases in a long-term acute-care hospital. Infect. Control Hosp. Epidemiol. 45 (3), 292–301 (2024).
Liu, L. et al. Carbapenem-resistant isolates of the Klebsiella pneumoniae Complex in Western China: the common ST11 and the Surprising Hospital-specific types. Clin. Infect. Dis. 67 (suppl_2), S263–s5 (2018).
Zheng, B. et al. Emergence of Raoultella ornithinolytica coproducing IMP-4 and KPC-2 carbapenemases in China. Antimicrob. Agents Chemother. 59 (11), 7086–7089 (2015).
Dong, H. et al. Characterization of a conjugative hybrid plasmid coharboring bla(KPC-2) and bla(IMP-4) in a Klebsiella quasipneumoniae clinical isolate. Microbiol. Spectr. 11 (1), e0261622 (2023).
Qiao, J. et al. Coexistence of bla(IMP-4), bla(NDM-1) and bla(OXA-1) in bla(KPC-2)-producing Citrobacter freundii of clinical origin in China. Front. Microbiol. 14, 1074612 (2023).
Sun, L. et al. Characterisation of a Novel Hybrid IncFII(pHN7A8):IncR:IncN plasmid co-harboring bla(NDM-5) and bla(KPC-2) from a clinical ST11 carbapenem-resistant Klebsiella pneumoniae strain. Infect. Drug Resist. 16, 7621–7628 (2023).
Shi, Q. et al. Chromosomal integration and plasmid fusion occurring in ST20 carbapenem-resistant Klebsiella pneumoniae isolates coharboring bla(NDM-1) and bla(IMP-4) induce resistance transmission and fitness variation. Emerg. Microbes Infect. 13 (1), 2339942 (2024).
Bocanegra-Ibarias, P. et al. Identification of Providencia spp. clinical isolates co-producing carbapenemases IMP-27, OXA-24, and OXA-58 in Mexico. Diagn. Microbiol. Infect. Dis. 109 (1), 116246 (2024).
Yigit, H. et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45 (4), 1151–1161 (2001).
Iovleva, A. & Doi, Y. Carbapenem-Resistant Enterobacteriaceae. Clin. Lab. Med. 37 (2), 303–315 (2017).
Watanabe, M., Iyobe, S., Inoue, M. & Mitsuhashi, S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 35 (1), 147–151 (1991).
Osano, E. et al. Molecular characterization of an enterobacterial metallo beta-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 38 (1), 71–78 (1994).
Zhao, W. H. & Hu, Z. Q. IMP-type metallo-β-lactamases in Gram-negative bacilli: distribution, phylogeny, and association with integrons. Crit. Rev. Microbiol. 37 (3), 214–226 (2011).
Yan, J. J., Ko, W. C. & Wu, J. J. Identification of a plasmid encoding SHV-12, TEM-1, and a variant of IMP-2 metallo-beta-lactamase, IMP-8, from a clinical isolate of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45 (8), 2368–2371 (2001).
Liu, C. et al. Phenotypic and genomic characteristics of clinical IMP-producing Klebsiella spp. Isolates in China. Commun. Med. (Lond). 4 (1), 25 (2024).
Lerminiaux, N. et al. Plasmid genomic epidemiology of Bla(KPC) carbapenemase-producing enterobacterales in Canada, 2010–2021. Antimicrob. Agents Chemother. 67 (12), e0086023 (2023).
Yao, Y. et al. Plasmid-mediated spread of Carbapenem Resistance in Enterobacterales: A three-year genome-based survey. Antibiot. (Basel) ;13(8). (2024).
Datta, N. & Hedges, R. W. Compatibility groups among fi - R factors. Nature. 234 (5326), 222–223 (1971).
Yang, X. et al. F33: A-: B-, IncHI2/ST3, and IncI1/ST71 plasmids drive the dissemination of fosA3 and bla CTX-M-55/-14/-65 in Escherichia coli from chickens in China. Front. Microbiol. 5, 688 (2014).
Ge, H. et al. First report of Klebsiella pneumoniae co-producing OXA-181, CTX-M-55, and MCR-8 isolated from the patient with bacteremia. Front. Microbiol. 13, 1020500 (2022).
Guan, J. et al. TADB 3.0: an updated database of bacterial toxin-antitoxin loci and associated mobile genetic elements. Nucleic Acids Res. 52 (D1), D784–d90 (2024).
Faria, N. A. et al. Genomic insights into the expansion of carbapenem-resistant Klebsiella pneumoniae within Portuguese hospitals. J. Hosp. Infect. 148, 62–76 (2024).
Lo, W. U. et al. Complete sequence of an IncN plasmid, pIMP-HZ1, carrying blaIMP-4 in a Klebsiella pneumoniae strain associated with medical travel to China. Antimicrob. Agents Chemother. 57 (3), 1561–1562 (2013).
Xiang, Y. et al. Clinical and molecular characteristics of Klebsiella pneumoniae infection in a tertiary general hospital of Wuhan, China. Eur. J. Clin. Microbiol. Infect. Dis. 43 (2), 269–278 (2024).
Wang, X. et al. Nosocomial dissemination of blaIMP-4 among Klebsiella pneumoniae by horizontal gene transfer and clonal spread: the epidemic IncN plasmids and the emerging high-risk IMP-4-producing ST101 clone. J. Antimicrob. Chemother. 78 (12), 2890–2894 (2023).
Buckner, M. M. C. et al. Clinically relevant plasmid-host interactions indicate that transcriptional and not genomic modifications ameliorate fitness costs of Klebsiella pneumoniae carbapenemase-carrying plasmids. mBio 9 (2), e02303–17 (2018).
Tellapragada, C. et al. Resistance to aztreonam-avibactam among clinical isolates of Escherichia coli is primarily mediated by altered penicillin-binding protein 3 and impermeability. Int. J. Antimicrob. Agents. 64 (3), 107256 (2024).
Xiang, G. et al. Porin deficiency or plasmid copy number increase mediated carbapenem-resistant Escherichia coli resistance evolution. Emerg. Microbes Infect. 13 (1), 2352432 (2024).
Unterholzner, S. J., Poppenberger, B. & Rozhon, W. Toxin-antitoxin systems: Biology, identification, and application. Mob. Genet. Elem. 3 (5), e26219 (2013).
Qiu, J., Zhai, Y., Wei, M., Zheng, C. & Jiao, X. Toxin-antitoxin systems: classification, biological roles, and applications. Microbiol. Res. 264, 127159 (2022).
Delver, E. P. & Belogurov, A. A. Organization of the leading region of IncN plasmid pKM101 (R46): a regulation controlled by CUP sequence elements. J. Mol. Biol. 271 (1), 13–30 (1997).
Tang, B. et al. Detection of clinical Serratia marcescens isolates carrying bla(KPC-2) in a hospital in China. Heliyon. 10 (8), e29702 (2024).
Papagiannitsis, C. C., Miriagou, V., Giakkoupi, P., Tzouvelekis, L. S. & Vatopoulos, A. C. Characterization of pKP1780, a novel IncR plasmid from the emerging Klebsiella pneumoniae ST147, encoding the VIM-1 metallo-β-lactamase. J. Antimicrob. Chemother. 68 (10), 2259–2262 (2013).
Tang, N. et al. Understanding the rapid spread of antimicrobial resistance genes mediated by IS26. mLife. 3 (1), 101–109 (2024).
Partridge, S. R., Kwong, S. M., Firth, N. & Jensen, S. O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31 (4), e00088–17 (2018).
Chen, R. et al. Difference analysis and characteristics of incompatibility group plasmid replicons in gram-negative bacteria with different antimicrobial phenotypes in Henan, China. BMC Microbiol. 24 (1), 64 (2024).
Funding
Henan Provincial Science and Technology Research Project (232102310176).
Author information
Authors and Affiliations
Contributions
The experiments were conceived and designed by XG. The samples and experiments were collected and performed by LF, YS, YJ, and SQ. The data was analyzed by RL, RC and CL. The manuscript was written by LF and revised by XG. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
This study was approved by the Ethics Committee of the first affiliated hospital of Zhengzhou University, Henan, China (2024-KY-0318). The First Affiliated Hospital of Zhengzhou University Ethics Committee also approved the waiver of informed consent to participate in this study owing to its retrospective design. All patient data were anonymised prior to analysis. Study procedures were conducted in accordance with the ethical standards of the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Fang, L., Shen, Y., Chen, R. et al. The characterization of an IncN-IncR fusion plasmid co-harboring blaTEM−40, blaKPC−2, and blaIMP−4 derived from ST1393 Klebsiella pneumoniae. Sci Rep 14, 26723 (2024). https://doi.org/10.1038/s41598-024-78205-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-78205-9