Abstract
Tinnitus is a common sensorineural complication that can occur de novo or after cancer treatments involving cisplatin or radiotherapy. Considering the heterogeneous etiology and pathophysiology of tinnitus, the extent to which shared genetic risk factors contribute to de novo tinnitus and cancer treatment-induced tinnitus is not clear. Here we report a GWAS for de novo tinnitus using the UK Biobank cohort with nine loci showing significantly associated variants (p < 5 × 10–8). To our knowledge, significant associations in four of these loci are novel, represented by rs7336872, rs115125870, rs1532898 and rs2537, with UBAC2, NUDT9, TGM4 and MPP2 as their nearest protein coding genes, respectively. Through quantitative comparison of results from GWAS of de novo tinnitus with GWAS of radiation-induced tinnitus, two intronic variants (rs7023227 and rs3780395) from a locus within immunoregulatory gene PD-L1 (CD274) reached the replication threshold using comparison thresholds of 10–5 and 10–4, with no other shared genetic risk factors identified. We did not observe shared genetic risk factors between de novo and cisplatin-induced tinnitus. Our results suggest that genetic risk factors are mainly distinct based on etiology of tinnitus and future efforts to study, prevent or treat tinnitus are expected to benefit from strategies that allow for distinction of cases based on the primary environmental risk factor.
Similar content being viewed by others
Introduction
Tinnitus is the phantom perception of sound characterized by persistent ringing, buzzing, or hissing in the ears. The global prevalence of tinnitus and its severe form among adults are estimated at 14% and 2%, respectively1. Tinnitus is associated with anxiety, depression, and reduced quality of life2,3. Severe tinnitus is associated with higher suicide attempts in females4.
Tinnitus is a heterogenous condition5,6 with a broad array of potentially causal risk factors associated with it, including head and neck trauma, infections and cancers that affect the ear, otosclerosis, hearing loss7, ototoxic medications including cisplatin7,8, radiation therapy resulting in cochlear exposure9,10, neurological and endocrine conditions, and autoimmune or psychological conditions7. Tinnitus appears related to the neural activity of different cortical regions depending on whether it is accompanied by severe hearing loss11. Further, differences in brain connectivity have been reported based on tinnitus severity12.
Genome-wide association studies (GWAS) have investigated the genetic aspects of tinnitus. These include a pilot study of 167 cases and 794 controls13 and more recent studies that analyzed large populations in UK Biobank14, and demonstrated shared genetic components with psychiatric conditions15,16,17. In addition, there have been GWAS studies of tinnitus following cisplatin-based treatment18,19 and radiation20. Yet, to our knowledge, no investigation has examined the extent to which genetic risk factors for de novo tinnitus are shared with iatrogenic cancer treatment-related tinnitus. To address this knowledge gap, we performed a GWAS for tinnitus in UK Biobank and compared the results with our previously reported GWAS for cisplatin-induced tinnitus19 and radiation-induced tinnitus in cancer survivors20. The resulting degree of overlap in genetic risk factors would be indicative of the extent to which a shared genetic architecture exists and of the degree of similarity in the involved cellular and molecular pathways. This information could in turn guide decisions to develop predictive genetic models and to better treat tinnitus-affected individuals, including cancer survivors.
Methods
Access to UK Biobank
GWAS for de novo tinnitus was conducted using UK Biobank. UK Biobank contains phenotypic, genetic and, health related data for ~ 500,000 individuals21. These include questionnaire results and imputed genotype data22. Our study was approved through an agreement between the University of Chicago and UK Biobank (application: ID = 73962) and was conducted in accordance with the University of Chicago institutional guidelines. All study subjects in UK Biobank provided informed consents for participation. Sample and data collections were conducted according to the UK Biobank ethical framework.
Definition of tinnitus
Self-reported presence and degree of tinnitus is the clinical standard of care in otolaryngology and audiology practices. Therefore, tinnitus was defined based on subjects’ self-report of severe tinnitus, consistent with an earlier report using UK Biobank14. The UK Biobank data has been collected at four time points (iterations) thus far, with partial overlap of subjects across iterations. In each iteration, tinnitus groups were defined based on subjects’ answer to “Do you get or have you had noises (such as ringing or buzzing) in your head or in one or both ears that lasts for more than five minutes at a time?”. Subjects who answered “Yes, now most or all of the time” and “Yes, now a lot of the time” were grouped as cases, and subjects who answered “No, never” were grouped as controls. Subjects were excluded from controls if at any iteration they selected any of the following answers: “Yes, now some of the time”, “Yes, but not now, but have in the past”. Subjects answering “Prefer not to answer” or, “Do not know” were grouped as “no answer provided” for that iteration. Multi-iteration subjects were included in the tinnitus case group if they were defined with tinnitus at any iteration, with the age of the earliest report as a case used in our analyses (n = 19,907 cases: 11,634 males, 8273 females). Multi-iteration subjects were included in the control only if they were consistently designated as controls across all iterations for which an answer was provided (answered “No, never” each time they provided an answer), with the age at the last iteration used for our analyses (n = 114,930 controls: 51,397 males, 63,533 females).
Detailed methods for determining tinnitus groups in The Platinum Study (Pt-study) and the radiation-associated ototoxicity study in The Childhood Cancer Survivor Study (CCSS-Rad-study) were described previously19,20. Briefly, in Pt-study, subjects answered the question “Have you had in the last 4 weeks ringing or buzzing in the ears?” from the validated Scale for Chemotherapy-induced Long-term Neurotoxicity (SCIN) questionnaire23. Subject answering either “quite a bit” or “very much” to this question were defined as the tinnitus group (n = 238) with exclusion of any subject that replied “no” to a separate question of “Do you have ringing or buzzing in the ears?”. Subjects who answered “not at all” to the SCIN question were placed in the control group (n = 979). In CCSS-Rad-study, subjects who replied “yes” to “Have you ever been told by a doctor or other health care professional that you have, or have had: tinnitus or ringing in the ears?” were considered cases (n = 106) and those who replied “no” were placed in the control group (n = 846). Subjects replying “yes, no longer present” or “not sure” were excluded.
Genome wide association analysis for de novo tinnitus using UK Biobank
The summary of quality control steps for preparation of the UK Biobank subjects is shown in Supplemental Fig. 1. Imputed genotype data were filtered to remove subjects with sex discrepancy and of non-European genetically inferred ancestry. We took the intersection of genotype and phenotype data. Next, kinship cutoff of 0.0884 was used to remove related individuals, with the removal priority given to control subjects and subjects with higher number of presences in kinship pairs. Variants with imputation information scores below 0.7 and minor allele frequencies below 0.005 were excluded from the association analyses. Logistic regression via PLINK2 (v2.00a3.1LM)24 was used for the genome wide association analysis with age, sex, genotyping array and the first ten genetic principal components used as covariates.
Identification of the nearest protein coding genes
The VEP webtool25 (https://grch37.ensembl.org/Homo_sapiens/Tools/VEP) and Ensembl26 (https://grch37.ensembl.org) were used to obtain details of the nearest protein coding genes within 20 kb of the loci lead variants.
Preparation of GWAS summary statistics for pairwise comparison
GWAS summary statistics for tinnitus from Pt-study and CCSS-Rad-study were respectively downloaded from the data link in the publication and from GWAS Catalog (https://www.ebi.ac.uk/gwas/; Study: GCST90027272)19,20,27. An outline of the preparation steps used for clumping of GWAS summary statistics can be found in Supplemental Fig. 2. For harmonization of GWAS summary statistics between CCSS-Rad-study and UK Biobank, chromosome, position, and the minor allele information were used for matching variants across datasets and to infer reference and alternative allele information for the CCSS-Rad-study variants from the UK Biobank dataset.
Clumping of GWAS summary statistics
A genetic linkage disequilibrium (LD) reference panel for clumping was created from the imputed UK Biobank genotypes. One thousand randomly selected subjects of European ancestry without sex discrepancy and 3rd degree relatives were used as the reference panel. Variant identifiers were converted to chromosome position followed by the first 23 characters of reference and alternative alleles, respectively. Variants with duplicated IDs (n = 46) were excluded from the panel. Clumping was performed with PLINK1 (v1.90b6.26) using r2 threshold of 0.1 and distance of 500 kb for variants28.
Pairwise comparison of GWAS results
Clumped variants with p-values below specified cutoffs from the base dataset were tested for replication in the target dataset using the Bonferroni corrected thresholds and were plotted as paired dot plots. The distribution of the lead variants from the base dataset were also plotted against the target dataset using quantile–quantile (QQ) plots.
Colocalization analyses of GWAS results
Coloc package29 (V5.2.3) was used for the pairwise comparison of loci between the cancer cohorts and UK Biobank. The lead variants with p < 10–5 in the base dataset were selected and all variants within 250 kb of the lead variant (500 kb window) were used for colocalization analyses. The default prior probability values were used (p1 = p2 = 10–4 and p12 = 10–5). The posterior probability H4 > 0.5 was used as the threshold for colocalization at a locus.
LocusFocus colocalization of the shared risk locus and tissue eQTLs for PD-L1
LocusFocus webtool30 (V1.6.0, https://locusfocus.research.sickkids.ca) which implements the Simple Sum method was used for colocalization analyses of variants in the shared locus and eQTL associations for PD-L1 in tissues. GWAS variants with duplicated positions were removed and variant IDs were converted to chromosome, position, reference allele, alternative allele. All 48 preloaded GTEx (V7) eQTL datasets were selected as secondary datasets. Preloaded European ancestry from the 1000 Genomes was selected for population LD calculations. The default test distance of 100 kb (at each direction) from the lead GWAS variant was used.
Previously reported variants associated with tinnitus using UK Biobank
The eight lead variants with reported associations with tinnitus using UK Biobank14,15,16 and 47 variants from a recent meta-analysis (using UK Biobank and the Million Veteran Program) were used for comparison31. We compared the association p-values for these 55 variants in CCSS-Rad-study and Pt-study against the Bonferroni corrected cut offs adjusted for the number of variants present in the cancer cohort results (1.2 × 10–3 in Pt-study and 9.6 × 10–4 in CCSS-Rad-study). In cases that the lead variant was absent in the target GWAS, all variants within 250 kb (500 kb window) were tested for association p-values below the Bonferroni corrected cut off for multiple testing for the 55 reported variants (9.1 × 10–4) to rule out the presence of a significant proxy variant.
Software
PLINK2 was used for the genotype format conversion, removal of samples, and for the tinnitus GWAS in UK Biobank. The GTEx portal (https://gtexportal.org/home/) was used to determine expression quantitative trait loci (eQTLs) or splicing quantitative trait loci (sQTLs) status of variants.
The UK Biobank Research Analysis Platform (RAP) was used as the medium for quality control steps and genome wide association analysis of de novo tinnitus. In addition, analyses steps that used the UK Biobank genotypes and subject phenotypes as their immediate inputs were performed on RAP. LDSC package32 (https://github.com/bulik/ldsc) was used for estimation of SNP heritability from GWAS summary statistics.
Bash (https://www.gnu.org/software/bash/), R (http://www.r-project.org/), JupyterLab (https://jupyter.org/) and RStudio (http://www.rstudio.com/) provided the analysis medium and the ggplot2 package33 was used to generate plots.
Results
Genome-wide association analysis for tinnitus using UK Biobank
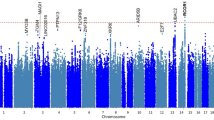
We defined severe cases of tinnitus using the same criteria as Wells et al.14. However, we combined samples across iterations to include as controls only those subjects who consistently reported no tinnitus (when the data were available) (n = 114,930), while pooling tinnitus cases across iterations (n = 19,907). After quality control steps, 10,865,531 variants were included in the genome wide association analysis of the UK Biobank dataset for tinnitus. The association results showed 375 variants with p-values below the suggestive association threshold (p < 10–6) from 28 independent loci, including 150 variants from nine loci with significant associations (p < 5 × 10–8) (Fig. 1, Supplemental Table 1). From these nine loci, eight contained a variant with p < 5 × 10–8 that was listed in the GTEx portal of which seven variants have demonstrated eQTL or sQTL roles for the nearest protein coding gene of the top variant in their locus (Table 1). A summary of the known eQTL and sQTL status of these variants is provided in Supplemental Table 2 (details provided in Supplemental Tables 3 and 4).
Comparison of tinnitus GWAS results between CCSS-Rad-study and UK Biobank
There were 9,239,074 variants that aligned between the GWAS summary statistics from the CCSS-Rad-study20 and the UK Biobank datasets and also aligned with the genetic reference panel. LD clumping resulted in 388,998 variants from the CCSS-Rad-study and 390,697 variants from the UK Biobank GWAS summary statistics. For pairwise comparison of the clumped GWAS results, a p-value cutoff of 10–5 was used for the selection of variants in the base dataset. When UK Biobank was used as the base dataset and CCSS-Rad-study as the target dataset, from the 101 lead variants in UK Biobank only rs3780395 reached the Bonferroni corrected threshold for replication in CCSS-Rad-study (Fig. 2A, top panel). rs3780395 is an intronic variant within the coding sequence of PD-L1 (CD274) and GTEx analysis indicates that it is also an eQTL for PD-L1 expression in multiple brain tissues, skeletal muscle, pancreas and whole blood samples (Supplemental Table 5). When CCSS-Rad-study was used as the base dataset and UK Biobank as the target dataset, none of the 36 lead variants from CCSS-Rad-study reached the Bonferroni corrected threshold for replication significance in the UK Biobank dataset (Fig. 2B, top panel). Comparison of distribution of the lead variants from the base datasets in the target datasets (Fig. 2, middle panel) do not indicate noticeable enrichment in the target datasets, with only rs3780395 above the 99th percentile. Consistent with the pairwise comparison, colocalization test across GWAS using the association p-value threshold of 10–5 in the base dataset (Fig. 2, bottom panel) provides support for colocalization at the locus represented by rs3780395 from UK Biobank (PP.H4 > 0.5).
Pairwise comparison of tinnitus GWAS results across UK Biobank and CCSS-Rad-study using cutoff of 10–5. (A) UK Biobank as the base GWAS. (B) CCSS-Rad-study as the base GWAS. Top panel shows − log10(p) of the lead variants from the base GWAS in the target GWAS. Blue color indicates matching direction of effect size in the significant variants. Solid lines show the 10–5 threshold and dashed lines show the Bonferroni corrected thresholds. Middle panel shows position of the lead variants in the base GWAS (red) in the QQ plot of the target GWAS results (grey). Dotted lines show the 50th, 99th and 99.9th percentiles of the target dataset and values next to them show percent of the lead variants of the base dataset above each line. Bottom panel shows colocalization results for loci consisting of variants within 250 kb of the lead variants in the base dataset with p-values below the 10–5 cutoff. Dashed lines show 0.5 colocalization cutoffs for the posterior probabilities. PP.H4 is the posterior probability that the GWAS signals are colocalized and PP.H3 is the posterior probability that there are two independent signals in the locus. Rad-study: CCSS-Rad-study.
We performed a follow up comparison using alternative p-value cutoffs of 10–4 and 10–3 (Supplemental Fig. 3). When a cutoff of 10–4 was applied, rs3780395 from the UK Biobank dataset and rs7023227 from CCSS-Rad-study were replicated across studies. Both rs7023227 and rs3780395 are within the same locus and in high LD with each other (Fig. 3). Only 0.63% (cutoffs 10–4) and 1.23% (cutoffs 10–3) of the lead UK Biobank variants were within the 99th percentile of the CCSS-Rad-study GWAS results. Likewise, only 1.07% (cutoffs 10–4) and 0.72% (cutoffs 10–3) of the lead CCSS-Rad-study variants were within the 99th percentile of the UK Biobank GWAS results.
LocusFocus Manhattan plots for rs3780395 and rs7023227 in (A) the UK Biobank and (B) the CCSS-Rad-study GWAS results for tinnitus. Trendlines represent the most significant regional variant associations in eQTL datasets for PD-L1 expression. Only tissues with significant colocalization are included. Details of the colocalization analyses are provided in Supplemental Figs. 4 and 5. GWAS variants with − log10(p) < 1 are excluded from the plot. For plotting B, reference and alternative allele information were inferred from UK Biobank. Gray areas highlight the 100 kb distance from the GWAS lead variants that were used for the colocalization analyses.
Colocalization of the shared risk locus and tissue eQTLs for PDL1
We used LocusFocus to evaluate the colocalization of variants in the shared risk locus and associations for PDL1 expression in 48 eQTL datasets. We observed significant (P < 0.01 after the Bonferroni correction) colocalization between the UK Biobank GWAS results and eQTL associations in four tissues (Supplemental Fig. 4). Consistently significant colocalization of the CCSS-Rad-study GWAS results and eQTL associations were observed in two tissues (Supplemental Fig. 5).
Comparison of tinnitus GWAS results between Pt-study and UK Biobank
Alignment between Pt-study19, UK Biobank and the genetic reference panel resulted in 7,612,105 variants (Supplemental Fig. 2). LD clumping resulted in 339,616 variants from the Pt-study and 341,202 variants from the UK Biobank GWAS for tinnitus. The p-value cutoff of 10–5 was used for selection of variants from the base dataset, resulting in 16 variants from Pt-study and 85 variants from UK Biobank. Pairwise comparison of the lead variants across datasets shows that none of the lead variants are replicated across studies (Fig. 4, top panel). Despite a relative enrichment of the lead variants from Pt-study at the 50th percentile of the UK Biobank dataset (75%), the enrichment was not pronounced at the higher percentiles, with 1.18% of the lead variants from UK Biobank in the 99th percentile of the results of Pt-study. None of the lead variants from Pt-study were within the 99th percentile of GWAS p-values in the UK Biobank dataset (Fig. 4, middle panel).
Pairwise comparison of tinnitus GWAS results across UK Biobank and Pt-study using cutoff of 10–5. (A) UK Biobank as the base GWAS. (B) Pt-study as the base GWAS. Top panel shows − log10(p) of the lead variants from the base GWAS in the secondary GWAS. Solid lines show the 10–5 threshold and dashed lines show the Bonferroni corrected thresholds. Middle panel shows position of the lead variants in the base GWAS (red) in the QQ plot of the target GWAS results (grey). Dotted lines show the 50th, 99th and 99.9th percentiles of the target dataset and values next to them show percent of the lead variants of the base dataset above each line. Bottom panel shows colocalization results for loci consisting of variants within 250 kb of the lead variants in the base dataset with p-values below the 10–5 cutoff. Dashed lines show 0.5 colocalization cutoffs for the posterior probabilities. PP.H4 is the posterior probability that the GWAS signals are colocalized and PP.H3 is the posterior probability that there are two independent signals in the locus.
Follow up comparisons using p-value cutoffs of 10–4 and 10–3 confirm that the lead variants were not replicated across the two studies (Supplemental Fig. 6). Consistently, only 0.76% (cutoff 10–4) and 0.97% (cutoff 10–3) of the lead variants from UK Biobank as the base dataset were within the 99th percentile in the Pt-study GWAS results. Similarly, only 1.24% (cutoff 10–4) and 0.51% (cutoff 10–3) of the lead variants from Pt-study as the base dataset were positioned in the 99th percentile of the UK Biobank GWAS results. Colocalization test across studies using the association p-value threshold of 10–5 in the base dataset (Fig. 4, bottom panel) did not show a significantly colocalized locus.
Comparison with previously reported tinnitus-associated variants using UK Biobank
There are 55 variants representing loci significantly associated with tinnitus in the three studies using UK Biobank15,16,34 alongside a meta-analysis across UK Biobank and the Million Veteran Program31. None of the reported variants associated with tinnitus in UK Biobank reached the Bonferroni corrected threshold for significance in Pt-study or CCSS-Rad-study (Supplemental Table 6). One of the variants from the meta-analysis cohort (rs6825241) reached the Bonferroni corrected significance threshold in Pt-study. In cases that the reported variants were absent in the cancer GWAS, none of the variants within 250 kb of the lead variant reached the Bonferroni corrected threshold for significance, indicating a lack of significant proxy variants in these loci.
Discussion
We performed a GWAS for de novo tinnitus on 134,837 subjects in UK Biobank, resulting in 150 variants from nine loci reaching genome-wide significance (p < 5 × 10–8). To the best of our knowledge, significant genome wide association for four of these loci are unreported in the context of de novo tinnitus (represented by rs7336872, rs115125870, rs1532898 and rs2537), with rs7336872 showing borderline significance in the study by Wells et al.14 (p = 5.9 × 10–8). The remaining five loci have been previously reported as significantly associated with tinnitus14,15,31. We used the UK Biobank GWAS for paired comparisons of results between loci associated with de novo and radiation- or cisplatin-induced tinnitus. Interestingly, using cutoffs of 10–5 and 10–4 in comparisons of the CCSS-Rad-study and the UK Biobank results, two intronic variants from a locus within PD-L1 reached the replication thresholds of significance. However, we did not identify overlapping genetic risk factors between cisplatin-induced tinnitus and de novo tinnitus. Importantly, these results underscore the complexity of the underlying biological pathways, suggesting the need for genetic evaluation and management strategies for tinnitus, dependent on the etiology or pathophysiology.
Associated variants with tinnitus using an expanded UK Biobank cohort
Although our GWAS of the UK Biobank data uses a tinnitus definition similar to the one applied in an earlier report14, we combine samples across four iterations of the UK Biobank data, which is expected to improve power in our analysis due to the larger number of cases (n = 19,907) and stricter definition of controls (n = 114,930). We identified nine loci below the association threshold (p < 5 × 10–8). We have evaluated these loci in the context of three previous publications of de novo tinnitus using UK Biobank (Table 1, Supplemental Table 7)14,15,16. We also included a recent meta-analysis of tinnitus using UK Biobank and the Million Veteran Program (MVP)31. However, comparisons with this meta-analysis should be interpreted with care due to expected differences in environmental exposures in the veteran population and, considering that tinnitus is the leading disability reported by veterans35. Therefore, this meta-analysis might not accurately represent de novo tinnitus in the general population. To the best of our knowledge, the significant associations for four of these loci with tinnitus are novel. The nine loci also include three of the six significant loci reported by Clifford et al.15 using UK Biobank, all of which are also present in their more recent meta-analysis that incorporates the MVP31. An additional significantly associated locus from this meta-analysis is also present among our associated loci. The significantly associated locus reported by Wells et al. alongside the locus showing borderline significance in their study (rs7336872) are also present in our loci associated with tinnitus14. In contrast, the significant locus reported by Bhatt et al. is not among our associated loci16, which could be due to the differences in the definition of tinnitus and analyses approaches.
The SNP heritability estimate (h2g) for de novo tinnitus from our GWAS is 0.08 which is comparable to earlier estimates of SNP heritability (h2g) reported by Wells et al. (h2g = 0.105)14 and h2g of approximately 0.06 reported by Clifford et al.15.
The nearest protein coding genes for the loci lead variants in the UK Biobank tinnitus GWAS
The nearest protein coding genes within 20 kb proximity of the lead variants are provided in Table 1. Among these, MSRA, ZNF318 and COL11A1 have been previously reported by Clifford et al. in the context of association with tinnitus15. In mice, elevated cochlear expression of MSRA after acoustic stimulation has been reported and knockout of MSRA leads to progressive hearing loss and increased vulnerability to acoustic trauma36. ZNF318 encodes a zinc finger protein37 and expression of ZNF318 in cochlear hair cells of adult mice has been observed38. Significant associations of variants at the vicinity of ZNF318 have also been reported in GWAS for hearing difficulty and age-related hearing loss15,34,39. COL11A1, encodes an alpha chain of type XI collagen40. Genetic variations of COL11A1 have been associated with hearing loss in the context of Stickler Syndrome and non-syndromic deafness41,42,43. Expression of COL11A1 in the otic vesicle, cochlea and inner ear hair cells in animal models has been reported44,45,46. Another gene near a significantly associated variant is TNRC6B and this locus has been recently reported in a meta-analysis of tinnitus using UK Biobank and a cohort of veterans31. TNRC6B encodes a scaffolding protein involved in gene silencing mediated by small RNA47. Elevated expression of TNRC6B by mitochondrial rich cells during development of the endolymphatic sac of the inner ear has been reported in a mouse model48.
RCOR1 is another gene from our results that has been previously reported as the nearest gene for the significantly associated variant in GWAS for tinnitus by Wells et al.14. Animal studies have shown that RCOR1 plays a critical role in promoting differentiation of neural and hematopoietic progenitor cells49,50,51. It is important to note that RCOR3 which encodes a paralog of RCOR1, is also the nearest gene for a suggestively associated variant (rs12743163; p = 5.0 × 10–7) in our results. RCOR1 and RCOR3 exhibit distinct expression patterns during differentiation of both neural progenitor cells and hematopoietic lineages52,53, indicating that they play varying roles. Presence of both paralogs in our results suggests a pronounced role of this protein family in the context of tinnitus. Both RCOR1 and RCOR3 can form transcription repressor complexes with histone demethylase LSD1 and transcription factor GFI153. GFI1 is expressed in the mouse otic vesicle and is essential for differentiation and survival of inner ear hair cells54. Likewise, LSD1 is expressed in mouse otic progenitors and regulates differentiation of sensory neurons in the inner ear55. Collectively these observations suggest that RCOR1 and RCOR3 might also play important roles during the sensorineural differentiation of the inner ear.
To our knowledge, the remaining four genes (UBAC2, NUDT9, TGM4 and MPP2) have not been previously reported as the nearest genes to a significantly associated variant for tinnitus including in the three GWAS for de novo tinnitus using UK Biobank14,15,16 and the recent meta-analysis of tinnitus incorporating the veteran dataset31. Although the study by Wells et al. reported borderline significance for the variant near UBAC2 (rs7336872)14. UBAC2 encodes a component of an endoplasmic reticulum-associated degradation system56. Association of polymorphism in the UBAC2 with increased susceptibility to noise-induced hearing loss was recently reported57. NUDT9 encodes a mitochondrial member of NUDIX hydrolysis family and has a high specificity for breakdown of ADP-ribose58. Oxidative stress is a potential causal factor for damage to the inner ear and tinnitus59,60,61. It has been proposed that during oxidative stress, ADP-ribose from mitochondria is released into the cytosol, activating TRPM2 calcium channels62. Consistently, knock down of NUDT9 in cultured cells has been shown to result in elevated calcium influx through TRPM2 channels and, increased accumulation of master transcription factor HIF-1a during hypoxia63. TGM4 encodes a prostatic secretory protein and is an antigen in autoimmune polyendocrine syndrome type 164. Currently there is a limited understanding of the potential role of TGM4 in the context of tinnitus. It is important to note that rs1532898 has known QTL roles for an array of other genes at its vicinity (Supplemental Table 2) which could drive or contribute to the observed association of its loci with tinnitus. MMP2 encodes a member of the membrane-associated guanylate kinases (MAGUKs) protein family that act as scaffolding proteins that organize protein complexes at cell junctions65. MMP2 is shown to control the activity of c-Src, an important regulator of both cellular proliferation and migration66. MMP2 has also been reported as a component of post synaptic receptor complexes in the rat brain67. A recent study links a mutation in MMP2 with increased risk for Vogt-Koyanagi-Harada (VKH) disease68 with tinnitus and hearing loss as its two common manifestations69.
Comparison of tinnitus GWAS results between the UK Biobank and the cancer survivor cohorts
Our results from comparison of associated variants across de novo tinnitus and two cohorts of cancer survivors show a limited reciprocal replication. Only variants rs7023227 and rs3780395 from the same locus reached replication significance when the UK Biobank and the CCSS-Rad-study results were compared using 10–5 and 10–4 thresholds. Colocalization analysis across the two GWAS results provides further evidence for the presence of a shared genetic risk factor at this locus. Both variants are located within introns of the PD-L1 coding sequence and are eQTL variants for its expression. PD-L1 is a transmembrane protein and a key player in the PD1/PDL pathway which suppresses the adaptive immune response and is important in an array of autoimmune diseases70. Importantly, tinnitus is one of the main auditory complications associated with autoimmune disorders71. Beside the direct role of PD-L1 in autoimmune conditions, several cases or case series studies have reported the onset of ototoxic symptoms (including tinnitus) after administration of PD-1 blockers in cancer treatment72,73,74,75,76. Conversely, in a mouse model, administration of a PD-1 blocker appears to show a protective effect, preventing inner ear hair cell loss77. The expression of PD-L1 impacts the development and healing of the nervous system78,79,80. PD-L1 is also expressed by sensory neurons. Neurons that express PD-L1 exhibit reduced nocifensive activity in the presence of PD-181. There is a paucity of data on the role of the PD-1/PD-L1 pathway in auditory neurons. We also observed significant colocalization of local variant statistics from the tinnitus GWAS results and eQTL associations for PD-L1 expression in multiple tissues. Significant colocalizations in “whole blood cells”, “brain cerebellum” and “brain cerebellar hemisphere” tissues are consistent with the potential role of PD-L1 in regulation of the immune and nervous systems. However, colocalization with eQTL associations could be masked due to limiting factors including sample size or low level of expression. Additionally, the whole tissue transcriptomes used in tissue eQTL analyses would not necessarily mirror the transcriptome of individual contributing cellular lineages82. This could prevent the detection of consequential yet lineage specific expression patterns. While our colocalization results support that change in PD-L1 expression is related with tinnitus, care is needed in extrapolating these results to determine the impacted tissues. Considering the wide-ranging impact of the PD-1/PD-L1 pathway across the immune and nervous systems, the role of PD-L1 in the context of tinnitus would be an important avenue for further investigation.
Comparison with previously reported variants associated with tinnitus in UK Biobank shows that none of the overlapping variants reach the significance threshold in the cancer cohorts (Supplemental Table 6). Only rs6825241 from a meta-analysis that incorporated both the UK Biobank and the MVP cohorts reached the statistical significance in the Pt-study results. Due to the mixed background of the population used in the meta-analysis (which includes the veteran population with expectedly distinct profile including higher noise exposure and noise-induced tinnitus), it is difficult to attribute the observed associations in the meta-analysis to de novo tinnitus in the general population. Overall, these comparisons suggest that a minority of genetic risk factors are shared across different tinnitus etiologies tested, consistent with our observation from comparisons with our GWAS in UK Biobank.
Considering the heterogeneity of tinnitus, several strategies for the subtyping, categorization or multidimensional analysis of tinnitus have been proposed83,84. The limited existence of shared genetic risk factors for tinnitus across cohorts in our study is noteworthy and is in line with the observed heterogeneity in the biological mechanisms and pathophysiology of tinnitus. Importantly, our results suggest that the majority of genetic risk factors for tinnitus play distinctive roles in the context of (and likely through interaction with) primary environmental risk factors, such as exposure to loud noise and cancer treatments. Therefore, in the absence of a sizeable shared genetic architecture for tinnitus, studies aimed at understanding its genetic underpinnings are expected to benefit from separate analyses of subtypes when primary risk factors can be identified, for instance treatments linked to high incidents of tinnitus in cancer survivor cohorts.
Limitations
Despite the strengths of our study, there are several limitations, including the lack of a replication cohort. However, two of the three significantly associated loci that were replicated by Clifford et al.15 (represented by rs553448379 and rs143424888) are also among the four most significantly associated loci here. This suggests that our results are consistent with the previously reported replicated tinnitus findings. Another potential limitation is the relatively limited sample size of the cancer survivor cohorts compared to the large UK Biobank. Nevertheless, to our knowledge, the two cancer cohorts used here are the only cohorts reported so far that would allow comparison of GWAS results for tinnitus after treatment for cisplatin or radiation. Future studies using extended or novel cohorts of tinnitus in the presence of specific primary environmental risk factors would be required to expand on our findings. Another limitation in our study is difference in age across cohorts. The median age of the UK Biobank patients (62 years) is greater than in Pt-study and in CCSS-Rad-study (37 and 43 years at time of evaluation, respectively). This difference might preclude efforts to identify shared genetic risk factors that might play distinctive roles in different age groups. However, the presence and abundance of these age-specific genetic risk factors are currently unknown. To our knowledge, there currently are no age-matched tinnitus GWAS results available from cancer survivor cohorts that would permit direct comparisons with UK Biobank.
Conclusion
We report a new GWAS for de novo tinnitus using UK Biobank with nine loci showing variants with significant associations. To our knowledge, these include novel significant associations in four of these loci. Comparing results from our UK Biobank GWAS with those of the two cancer survivor cohorts showed limited shared genetic risk factors across studies. These observations cast doubt on the existence of sizable shared genetic risk factors across different etiologies of tinnitus. This observation does not negate the potential for identification of additional shared genetic risk factors in future studies with larger cohorts or the potential of polygenic risk score models incorporating them. However, in the absence of a sizable pool of shared genetic risk factors, tinnitus models are expected to be limited in predictability and scope across tinnitus subtypes. Therefore, separate analyses of tinnitus based on the primary environmental risk factor are expected to improve future efforts to understand the genetic aspects of tinnitus and to pave the way for improved prevention and management strategies.
Data availability
The GWAS summary statistics for de novo tinnitus in UK Biobank can be obtained from GWAS Catalog (https://www.ebi.ac.uk/gwas/, GCP ID: GCP001026). The UK Biobank database is available to researchers upon application and approval (https://www.ukbiobank.ac.uk/enable-your-research/apply-for-access). The Childhood Cancer Survivor Study is a US National Cancer Institute funded resource (U24 CA55727) to promote and facilitate research among long-term survivors of cancer diagnosed during childhood and adolescence. CCSS data are publicly available on dbGaP at https://www.ncbi.nlm.nih.gov/gap/ through its accession number phs001327.v2.p1. and on the St Jude Survivorship Portal within the St. Jude Cloud at https://survivorship.stjude.cloud/. In addition, utilization of the CCSS data that leverages the expertise of CCSS Statistical and Survivorship research and resources will be considered on a case-by case basis. For this utilization, a research Application Of Intent followed by an Analysis Concept Proposal must be submitted for evaluation by the CCSS Publications Committee. Users interested in utilizing this resource are encouraged to visit http://ccss.stjude.org. Full analytical data sets associated with CCSS publications since January of 2023 are also available on the St. Jude Survivorship Portal at https://viz.stjude.cloud/community/cancer-survivorship-community~4/publications.
Abbreviations
- LD:
-
Linkage disequilibrium
- eQTLs:
-
Expression quantitative trait loci
- GTEx:
-
The Genotype-Tissue Expression Project
- h2g:
-
SNP heritability estimate
- Pt-study:
-
The Platinum Study
- CCSS-Rad-study:
-
The radiation-associated ototoxicity study in the Childhood Cancer Survivor Study
- RAP:
-
The UK Biobank Research Analysis Platform
- sQTLs:
-
Splicing quantitative trait loci
References
Jarach, C. M. et al. Global prevalence and incidence of tinnitus: A systematic review and meta-analysis. JAMA Neurol. 79, 888–900 (2022).
Shargorodsky, J., Curhan, G. C. & Farwell, W. R. Prevalence and characteristics of tinnitus among US adults. Am. J. Med. 123, 711–718 (2010).
Weidt, S. et al. Which tinnitus-related characteristics affect current health-related quality of life and depression? A cross-sectional cohort study. Psychiatry Res. 237, 114–121 (2016).
Lugo, A. et al. Sex-specific association of tinnitus with suicide attempts. JAMA Otolaryngol. Head Neck Surg. 145, 685–687 (2019).
Cederroth, C. R. et al. Editorial: Towards an understanding of tinnitus heterogeneity. Front. Aging Neurosci. 11, 53 (2019).
Simoes, J. P. et al. Multidisciplinary tinnitus research: Challenges and future directions from the perspective of early stage researchers. Front. Aging Neurosci. 13, 647285 (2021).
Baguley, D., McFerran, D. & Hall, D. Tinnitus. Lancet 382, 1600–1607 (2013).
Bokemeyer, C. et al. Analysis of risk factors for cisplatin-induced ototoxicity in patients with testicular cancer. Br. J. Cancer 77, 1355–1362 (1998).
Lau, S. K., Wei, W. I., Sham, J. S., Choy, D. T. & Hui, Y. Early changes of auditory brain stem evoked response after radiotherapy for nasopharyngeal carcinoma–A prospective study. J. Laryngol. Otol. 106, 887–892 (1992).
Whelan, K. et al. Auditory complications in childhood cancer survivors: A report from the childhood cancer survivor study. Pediatr. Blood Cancer 57, 126–134 (2011).
Vanneste, S. & De Ridder, D. Deafferentation-based pathophysiological differences in phantom sound: Tinnitus with and without hearing loss. Neuroimage 129, 80–94 (2016).
Schmidt, S. A., Carpenter-Thompson, J. & Husain, F. T. Connectivity of precuneus to the default mode and dorsal attention networks: A possible invariant marker of long-term tinnitus. Neuroimage Clin. 16, 196–204 (2017).
Gilles, A., Van Camp, G., Van de Heyning, P. & Fransen, E. A pilot genome-wide association study identifies potential metabolic pathways involved in tinnitus. Front. Neurosci. 11, 71 (2017).
Wells, H. R. R., Abidin, F. N. Z., Freidin, M. B., Williams, F. M. K. & Dawson, S. J. Genome-wide association study suggests that variation at the RCOR1 locus is associated with tinnitus in UK Biobank. Sci. Rep. 11, 6470 (2021).
Clifford, R. E., Maihofer, A. X., Stein, M. B., Ryan, A. F. & Nievergelt, C. M. Novel risk loci in tinnitus and causal inference with neuropsychiatric disorders among adults of European Ancestry. JAMA Otolaryngol. Head Neck Surg. 146, 1015–1025 (2020).
Bhatt, I. S., Wilson, N., Dias, R. & Torkamani, A. A genome-wide association study of tinnitus reveals shared genetic links to neuropsychiatric disorders. Sci. Rep. 12, 22511 (2022).
Smit, D. J. A. et al. A genome-wide association study of a rage-related misophonia symptom and the genetic link with audiological traits, psychiatric disorders, and personality. Front. Neurosci. 16, 971752 (2022).
El Charif, O. et al. Clinical and genome-wide analysis of cisplatin-induced tinnitus implicates novel ototoxic mechanisms. Clin. Cancer Res. 25, 4104–4116 (2019).
Zhang, X. et al. Pharmacogenomics of cisplatin-induced neurotoxicities: Hearing loss, tinnitus, and peripheral sensory neuropathy. Cancer Med. 11, 2801–2816 (2022).
Trendowski, M. R. et al. Clinical and genetic risk factors for radiation-associated ototoxicity: A report from the Childhood Cancer Survivor Study and the St. Jude Lifetime Cohort. Cancer 127, 4091–4102 (2021).
Sudlow, C. et al. UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. https://doi.org/10.1371/journal.pmed.1001779 (2015).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature https://doi.org/10.1038/s41586-018-0579-z (2018).
Oldenburg, J., Fosså, S. D. & Dahl, A. A. Scale for chemotherapy-induced long-term neurotoxicity (SCIN): Psychometrics, validation, and findings in a large sample of testicular cancer survivors. Qual. Life Res. 15, 791–800 (2006).
Chang, C. C. et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
McLaren, W. et al. The ensembl variant effect predictor. Genome Biol. 17, 122 (2016).
Cunningham, F. et al. Ensembl 2022. Nucleic Acids Res. 50, D988–D995 (2022).
Sollis, E. et al. The NHGRI-EBI GWAS catalog: Knowledgebase and deposition resource. Nucleic Acids Res. 51, D977–D985 (2023).
Purcell, S. et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. https://doi.org/10.1371/journal.pgen.1004383 (2014).
Panjwani, N. et al. LocusFocus: Web-based colocalization for the annotation and functional follow-up of GWAS. PLoS Comput. Biol. https://doi.org/10.1371/journal.pcbi.1008336 (2020).
Clifford, R. E. et al. Genetic architecture distinguishes tinnitus from hearing loss. Nat. Commun. https://doi.org/10.1038/s41467-024-44842-x (2024).
Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Wickham, H. Ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, 2016).
Wells, H. R. R. et al. GWAS identifies 44 independent associated genomic loci for self-reported adult hearing difficulty in UK biobank. Am. J. Hum. Genet. 105, 788–802 (2019).
Piccirillo, J. F., Rodebaugh, T. L. & Lenze, E. J. Tinnitus. JAMA 323, 1497–1498 (2020).
Alqudah, S. et al. Methionine sulfoxide reductase A knockout mice show progressive hearing loss and sensitivity to acoustic trauma. Audiol. Neurootol. 23, 20–31 (2018).
Inoue, A. et al. The transcript for a novel protein with a zinc finger motif is expressed at specific stages of mouse spermatogenesis. Biochem. Biophys. Res. Commun. 273, 398–403 (2000).
Pisciottano, F. et al. Inner ear genes underwent positive selection and adaptation in the mammalian lineage. Mol. Biol. Evol. 36, 1653–1670 (2019).
Trpchevska, N. et al. Genome-wide association meta-analysis identifies 48 risk variants and highlights the role of the stria vascularis in hearing loss. Am. J. Hum. Genet. 109, 1077–1091 (2022).
Li, Y. et al. A fibrillar collagen gene, Col11a1, is essential for skeletal morphogenesis. Cell 80, 423–430 (1995).
Richards, A. J. et al. Alternative splicing modifies the effect of mutations in COL11A1 and results in recessive type 2 Stickler syndrome with profound hearing loss. J. Med. Genet. 50, 765–771 (2013).
Booth, K. T. et al. Splice-altering variant in COL11A1 as a cause of nonsyndromic hearing loss DFNA37. Genet. Med. 21, 948–954 (2019).
Rad, A. et al. Aberrant COL11A1 splicing causes prelingual autosomal dominant nonsyndromic hearing loss in the DFNA37 locus. Hum. Mutat. 42, 25–30 (2021).
Yoshioka, H. et al. Developmental pattern of expression of the mouse alpha 1 (XI) collagen gene (Col11a1). Dev. Dyn. 204, 41–47 (1995).
Shi, M. et al. Acute noise causes down-regulation of ECM protein expression in guinea pig cochlea. Mol. Biotechnol. 65, 774–785 (2023).
Liu, L.-M. et al. Characterization of the transcriptomes of Atoh1-induced hair cells in the mouse cochlea. Am. J. Stem Cells 9, 1–15 (2020).
Johnson, S. T., Chu, Y., Liu, J. & Corey, D. R. Impact of scaffolding protein TNRC6 paralogs on gene expression and splicing. RNA 27, 1004–1016 (2021).
Honda, K. et al. Molecular architecture underlying fluid absorption by the developing inner ear. Elife https://doi.org/10.7554/eLife.26851 (2017).
Fuentes, P., Cánovas, J., Berndt, F. A., Noctor, S. C. & Kukuljan, M. CoREST/LSD1 control the development of pyramidal cortical neurons. Cereb. Cortex 22, 1431–1441 (2012).
Monaghan, C. E. et al. REST corepressors RCOR1 and RCOR2 and the repressor INSM1 regulate the proliferation-differentiation balance in the developing brain. Proc. Natl. Acad. Sci. U. S. A. 114, E406–E415 (2017).
Yao, H. et al. Corepressor Rcor1 is essential for murine erythropoiesis. Blood 123, 3175–3184 (2014).
Sáez, J. E. et al. Decreased expression of CoREST1 and CoREST2 together with LSD1 and HDAC1/2 during neuronal differentiation. PLoS One 10, e0131760 (2015).
Upadhyay, G., Chowdhury, A. H., Vaidyanathan, B., Kim, D. & Saleque, S. Antagonistic actions of Rcor proteins regulate LSD1 activity and cellular differentiation. Proc. Natl. Acad. Sci. U. S. A. 111, 8071–8076 (2014).
Wallis, D. et al. The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development 130, 221–232 (2003).
Patel, D., Shimomura, A., Majumdar, S., Holley, M. C. & Hashino, E. The histone demethylase LSD1 regulates inner ear progenitor differentiation through interactions with Pax2 and the NuRD repressor complex. PLoS One 13, e0191689 (2018).
Christianson, J. C. et al. Defining human ERAD networks through an integrative mapping strategy. Nat. Cell Biol. 14, 93–105 (2011).
Wan, L. et al. Association between UBAC2 gene polymorphism and the risk of noise-induced hearing loss: A cross-sectional study. Environ. Sci. Pollut. Res. Int. 29, 32947–32958 (2022).
Perraud, A.-L. et al. NUDT9, a member of the Nudix hydrolase family, is an evolutionarily conserved mitochondrial ADP-ribose pyrophosphatase. J. Biol. Chem. 278, 1794–1801 (2003).
Atila, N. E. et al. The role of manganese, cadmium, chromium and selenium on subjective tinnitus. Biol. Trace Elem Res. 199, 2844–2850 (2021).
Neri, S. et al. Oxidative stress, nitric oxide, endothelial dysfunction and tinnitus. Free Radic. Res. 40, 615–618 (2006).
Neri, S. et al. Tinnitus and oxidative stress in a selected series of elderly patients. Arch. Gerontol. Geriatr. Suppl. 8, 219–223 (2002).
Perraud, A.-L. et al. Accumulation of free ADP-ribose from mitochondria mediates oxidative stress-induced gating of TRPM2 cation channels. J. Biol. Chem. 280, 6138–6148 (2005).
Yoon, B., Yang, E. G. & Kim, S. Y. The ADP-ribose reactive NUDIX hydrolase isoforms can modulate HIF-1α in cancer cells. Biochem. Biophys. Res. Commun. 504, 321–327 (2018).
Landegren, N. et al. Transglutaminase 4 as a prostate autoantigen in male subfertility. Sci. Transl. Med. 7, 292ra101 (2015).
Funke, L., Dakoji, S. & Bredt, D. S. Membrane-associated guanylate kinases regulate adhesion and plasticity at cell junctions. Annu. Rev. Biochem. 74, 219–245 (2005).
Baumgartner, M., Weiss, A., Fritzius, T., Heinrich, J. & Moelling, K. The PDZ protein MPP2 interacts with c-Src in epithelial cells. Exp. Cell Res. 315, 2888–2898 (2009).
Rademacher, N., Schmerl, B., Lardong, J. A., Wahl, M. C. & Shoichet, S. A. MPP2 is a postsynaptic MAGUK scaffold protein that links SynCAM1 cell adhesion molecules to core components of the postsynaptic density. Sci. Rep. 6, 35283 (2016).
Liu, X. et al. A de novo missense mutation in MPP2 confers an increased risk of Vogt-Koyanagi-Harada disease as shown by trio-based whole-exome sequencing. Cell Mol. Immunol. 20, 1379–1392 (2023).
Yang, P. et al. Development and evaluation of diagnostic criteria for Vogt-Koyanagi-Harada disease. JAMA Ophthalmol. 136, 1025–1031 (2018).
Zamani, M. R., Aslani, S., Salmaninejad, A., Javan, M. R. & Rezaei, N. PD-1/PD-L and autoimmunity: A growing relationship. Cell Immunol. 310, 27–41 (2016).
Ralli, M. et al. Audiovestibular symptoms in systemic autoimmune diseases. J. Immunol. Res. 2018, 5798103 (2018).
Rosner, S. et al. Immune-mediated ototoxicity associated with immune checkpoint inhibitors in patients with melanoma. J. Immunother. Cancer https://doi.org/10.1136/jitc-2020-001675 (2020).
Zibelman, M., Pollak, N. & Olszanski, A. J. Autoimmune inner ear disease in a melanoma patient treated with pembrolizumab. J. Immunother. Cancer 4, 8 (2016).
Tampio, A. J. F., Dhanireddy, S., Sivapiragasam, A. & Nicholas, B. D. Bilateral sensorineural hearing loss associated with nivolumab therapy for stage IV malignant melanoma. Ear Nose Throat J. 100, 286S-291S (2021).
Rajapakse, A., O’Leary, C., Gundelach, R., Deva, R. & O’Byrne, K. Unilateral autoimmune inner ear disease in a patient with lung cancer treated with nivolumab. Oxf. Med. Case Rep. 2020, omaa077 (2020).
Hobelmann, K. & Fitzgerald, D. A case of pembrolizumab induced autoimmune sensorineural hearing loss. J. Otol. Rhinol. 8, (2019).
Szepesy, J. et al. Anti-PD-1 therapy does not influence hearing ability in the most sensitive frequency range, but mitigates outer hair cell loss in the Basal Cochlear region. Int. J. Mol. Sci. https://doi.org/10.3390/ijms21186701 (2020).
Sham, C. W. et al. Neuronal programmed cell death-1 ligand expression regulates retinal ganglion cell number in neonatal and adult mice. J. Neuroophthalmol. 32, 227–237 (2012).
Kong, F. et al. PD-L1 improves motor function and alleviates neuropathic pain in male mice after spinal cord injury by inhibiting MAPK pathway. Front. Immunol. 12, 670646 (2021).
Han, R., Luo, J., Shi, Y., Yao, Y. & Hao, J. PD-L1 (Programmed Death Ligand 1) protects against experimental intracerebral hemorrhage-induced brain injury. Stroke 48, 2255–2262 (2017).
Meerschaert, K. A. et al. Neuronally expressed PDL1, not PD1, suppresses acute nociception. Brain Behav. Immun. 106, 233–246 (2022).
Aguet, F. et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science (1979) 369, (2020).
Genitsaridi, E., Hoare, D. J., Kypraios, T. & Hall, D. A. A review and a framework of variables for defining and characterizing tinnitus subphenotypes. Brain Sci. https://doi.org/10.3390/brainsci10120938 (2020).
Mohan, A., Leong, S. L., De Ridder, D. & Vanneste, S. Symptom dimensions to address heterogeneity in tinnitus. Neurosci. Biobehav. Rev. 134, 104542 (2022).
Acknowledgements
This research has been conducted using the UK Biobank Resource under Application Number 73962. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health (NIH), and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal (11/07/2023 for the UK Biobank GWAS variants; 11/28/2023 for rs7023227 and rs3780395) and dbGaP accession number phs000424.v8.p2 (11/07/2023 for the UK Biobank GWAS variants; 11/28/2023 for rs7023227 and rs3780395). The work was supported by the NIH Genetic Susceptibility and Biomarkers of Platinum-related Toxicities grant (R01 CA157823, L.B. Travis, Principal Investigator). The GWAS work for CCSS-Rad-study was supported by the National Cancer Institute (CA55727, G.T. Armstrong, Principal Investigator). Support to St. Jude Children’s Research Hospital also provided by the Cancer Center Support (CORE) grant (CA21765, C. Roberts, Principal Investigator) and the American Lebanese-Syrian Associated Charities (ALSAC).
Author information
Authors and Affiliations
Contributions
M.S.: conceptualization, methodology, formal analysis, writing-original draft preparation, visualization. H.E.W.: methodology, writing-reviewing and editing. G.T.A.: data curation, resources, writing-reviewing and editing. R.D.F.: methodology, writing-reviewing and editing. L.B.T.: resources, writing-reviewing and editing. M.E.D.: project administration, conceptualization, resources, writing-reviewing and editing, supervision. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shahbazi, M., Wheeler, H.E., Armstrong, G.T. et al. Comparison of GWAS results between de novo tinnitus and cancer treatment-related tinnitus suggests distinctive roles for genetic risk factors. Sci Rep 14, 27952 (2024). https://doi.org/10.1038/s41598-024-78274-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-78274-w