Abstract
Pompe disease, a severe lysosomal storage disorder, is marked by heart problems, muscle weakness, and respiratory difficulties. This study aimed to identify novel markers for infantile-onset Pompe disease by analyzing key genes and immune cells infiltrating affected skeletal muscles, building on existing research linking its progression to the immune cell infiltration. The datasets GSE38680 and GSE159062 were downloaded and differential expression genes (DEGs) were identified. Potential markers were screened using support vector machine recursive feature elimination (SVM-RFE) analysis and least absolute shrinkage and selection operator (LASSO) regression models. To analyze DEGs and immune processes, 22 immune cell types in affected tissues were examined using CIBERSORT. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed significant enrichment in the calcium signaling pathway, phosphatidylinositol signaling system, ubiquitin-mediated proteolysis and JAK-STAT signaling pathway. Machine learning identified GPNMB, CALML6 and TRIM7 as key genes. Immune cell infiltration analysis showed that the expression levels of the above three genes were closely related to immune cells in our target population under study, including T cells regulatory (Tregs), monocytes, macrophages M0, resting and activated dendritic cells, naive and memory B cells. Overall, our study results may provide new clues for exploring markers related to Pompe disease by highlighting key genes and their relationship with immune infiltration, offering insights into the disease’s development. However, previous studies have shown limited evidence of significant immune cell infiltration in muscle biopsies from Pompe patients, and this lack of direct evidence necessitates further investigation and experimental validation in laboratory settings to confirm these associations.
Similar content being viewed by others

Introduction
Pompe disease, also known as acid alpha-glucosidase deficiency or glycogen storage disease type II, is a rare genetic metabolic disorder belonging to the group of LSDs1,2. It is caused by a deficiency or absence of the enzyme acid alpha-glucosidase (GAA), leading to the accumulation of glycogen, particularly in muscle cells. This accumulation results in damage to the heart, skeletal muscles, and other tissues, manifesting in various clinical symptoms3. The disease is categorized into infantile-onset and late-onset forms, based on the onset and severity of symptoms. Infantile-onset Pompe disease is particularly severe, with symptoms such as muscle weakness, heart problems, and respiratory difficulties appearing within the first few months after birth. Without treatment, this form is fatal in infancy4,5. Diagnosis typically involves genetic testing and enzyme activity assays. The primary treatment is enzyme replacement therapy (ERT), which entails regular injections of a synthetic GAA enzyme to supplement the missing enzyme, thereby slowing disease progression and improving the patient’s quality of life6,7,8. Although Pompe disease is an autosomal recessive genetic disorder, the establishment of genotype-phenotype correlations is challenging due to the vast variety of pathogenic mutations in the GAA gene9,10,11. Furthermore, while the causative gene for Pompe disease is well-defined, the specific pathological processes are not fully understood, and the clinical severity is influenced by other factors. Current research has reported that modifier alleles, based on their haplotype background, can affect the severity of specific mutations12,13,14.
The occurrence and progression of Pompe disease are closely related to cellular stress mechanisms. In patients with Pompe disease, the deficiency or absence of GAA leads to the accumulation of glycogen within cells, particularly in skeletal and cardiac muscle cells. This accumulation results in lysosomal storage and significant disruption of muscle cell structure, yet it fails to robustly activate satellite cells, leading to impaired muscle regeneration and repair15. Some studies suggest that immune system abnormalities may occur in Pompe disease patients, such as macrophage infiltration into muscle tissue, but there is limited evidence of significant infiltration by other inflammatory cells, such as B or T lymphocytes. Additionally, changes in cytokine levels and potential dysfunction of immune cells have been observed. However, the immune responses seen in Pompe disease may not necessarily exacerbate the severity of the disease in the same way as in classic inflammatory myopathies. The muscle damage in Pompe disease is primarily driven by lysosomal glycogen accumulation, and while immune cells like macrophages may play a role in clearing damaged muscle fibers, the overall contribution of the immune system to disease progression remains less clear compared to other muscular disorders15. In addition, although ERT therapy is the primary treatment for Pompe disease, some patients may develop an immune response to the recombinant enzyme used in the treatment and form antibodies, which can reduce the effectiveness of the treatment16. Therefore, monitoring and managing the immune response in patients is crucial for optimizing treatment outcomes and improving prognosis. As a result, understanding the relationship between Pompe disease and the immune system is essential for the treatment and management of the disease.
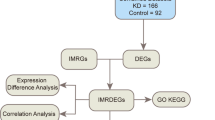
This study explores the hypothesis that pathogenic mutations in the GAA gene in Pompe disease patients not only lead to a decrease in GAA enzyme activity but also initiate a series of transcriptional responses, including dysregulation of the immune response, thereby affecting the disease process and clinical spectrum. To better clarify the focus of our study on the immune cells infiltrating skeletal muscle in infantile-onset Pompe disease, we analyzed two GEO datasets detailing the gene expression profiles specifically in skeletal muscle from these patients. By employing bioinformatics and machine learning, we aim to identify DEGs between Pompe disease patients and control samples, to uncover genes associated with Pompe disease, and to explore their roles in immunity. This will further deepen our understanding of the pathogenesis of Pompe disease and may serve as prognostic biomarkers and alternative therapeutic targets for adjunctive treatment.
Materials and methods
Online data obtaining and processing
By searching the GEO database (https://www.ncbi.nlm.nih.gov/geo/) and using “Pompe disease” as a keyword, we identified the mRNA expression profiling datasets GSE38680 and GSE159062 for inclusion in our analysis. GSE38680 comprised two cohorts: Study 1, with 11 Pompe disease patients and 7 controls, and Study 2, with 9 Pompe disease patients and 10 controls. In the final analysis, we excluded sample C.b.10 from Study 2, which was ultimately diagnosed with MELAS. GSE159062 consisted of 3 Pompe disease patients and 4 controls. It is important to note that GSE159062 was generated from 3D-muscle bundles derived from cells isolated from muscle biopsies of Pompe patients, not directly from muscle tissue, which could lead to gene expression changes during the culturing period. Baseline transcriptomic data from both Pompe patients and controls were used for subsequent analysis, ultimately involving 23 Pompe patients and 20 controls. The two GEO datasets originate from distinct platforms. GSE38680 utilizes the GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array, whereas GSE159062 employs the GPL24676 Illumina NovaSeq 6000 (Homo sapiens). The script processed genomic data by reading the GPL file to link gene symbols with probe IDs and loading the GSE file to merge these IDs with expression data. It averaged expression data per gene symbol and imputed missing values. High expression values were log-transformed, and batch normalization was applied across arrays. The script then merged two GEO databases based on the “ID” grouped by “ID” to compute averages, and created a model matrix for different sample types to run batch effect corrections using ComBat from the ‘sva’ package, setting up data for detailed statistical analysis and controlling for potential batch effects. The file containing gene expression levels adjusted for batch effects has been placed in the supplementary materials (Batch_gseExp.txt).
Analysis of DEGs in integrated data
DEGs between Pompe diseases patients and control samples were identified using the Limma package in R, with thresholds set at |Log2 Fold Change (FC)| > 0.585, p-value < 0.05, and False Discovery Rate (FDR) < 0.05.
Functional enrichment analyses
Functional enrichment analysis was performed for DEGs between Pompe diseases patients and controls using Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis17,18,19. GO and KEGG was conducted using the ‘clusterProfiler’ package in R20. After adjusting for multiple testing using the Benjamini-Hochberg method, GO terms and KEGG pathways with an adjusted p-value < 0.05 were considered significantly enriched.
Candidate signatures selection
LASSO and SVM were used for feature selection in patients with Pompe disease. LASSO minimizes squared residuals and coefficients’ absolute value, using cross-validation to optimize λ and ‘glmnet’ in R for analysis. SVM iteratively removes the least weighted feature until reaching the desired number, implemented with ‘e1071’ in R and a custom script. Both methods helped identify key features for our predictive model. LASSO’s advantage lies in feature selection and regularization to prevent overfitting, though it selects fewer features, while SVM-RFE identifies the most contributory features for prediction but requires more computational resources and is less stable with many features.
CIBERSORT analysis
The computational method of CIBERSORT (http://cibersort.stanford.edu/) is based on deconvolution analysis of gene expression profiles, and it is used to evaluate the differences in a specific gene set compared to the overall gene expression within a sample. Through the use of CIBERSORT, our team was able to determine the immune responses of 22 types of immune cells and examine the relationship between these immune cells and the expression of key genes in both control and Pompe diseases samples. CIBERSORT uses a linear support vector regression model to estimate cell proportions from gene expression data, assessing significance with permutation tests. It outputs relative proportions and statistical parameters for each immune cell type, ensuring accurate estimates. However, CIBERSORT may struggle to detect small amounts or rare cell types, potentially leading to inaccurate estimates for these cells21,22.
Statistical analysis
Student’s t-tests were employed to compare the expression of GPNMB, CALML6, and TRIM7 between Pompe diseases patients and controls. The Mann-Whitney U test is used to compare the differences in immune cells between Pompe disease patients and controls due to non-normal distributions. Pearson correlation coefficients are used for correlation analysis. Differences were deemed statistically significant at *p < 0.05, **p < 0.01, ***p < 0.001, or ****p < 0.0001.
Results
Identification of DEGs in Pompe disease patients
For our retrospective analysis, we incorporated a total of 43 samples from two GEO datasets (GSE38680 and GSE159062), comprising 23 patients with Pompe disease and 20 control individuals. The DEGs in the aggregated dataset were examined using the Limma package after correcting for batch effects. We identified 38 DEGs, with 19 genes significantly upregulated and 19 genes markedly downregulated (Fig. 1(a) and 1(b)).
Volcano plot (a) and heat map (b) of DEGs between Pompe patients and control samples. (a) Dashed lines define significance thresholds for differential expression. The x-axis, Log2FC (Log2 Fold Change), shows gene expression changes, with positives indicating upregulation and negatives indicating downregulation. The y-axis, Log10FDR (False Discovery Rate), plots the False Discovery Rate’s logarithm, where lower values denote higher significance. (b) The x-axis represents each sample, combining two GEO datasets followed by cluster analysis. The gene expression levels are displayed as normalized values on a scale, where negative values indicate downregulation relative to a reference point, and positive values indicate upregulation.
Functional enrichment analyses
To investigate the biological functions of the 38 DEGs in Pompe disease, we performed GO and KEGG analyses using the ClusterProfiler R package. The analysis revealed that these DEGs were mainly involved in processes such as endoplasmic reticulum membrane organization, nuclear membrane structure, cleavage furrow formation, cell division site organization, GTPase activity, ubiquitin-like protein ligase activity, and transmembrane transporter binding (Fig. 2(a)). Additionally, KEGG pathway analysis identified significant enrichment in pathways including the calcium signaling pathway, phosphatidylinositol signaling system, ubiquitin-mediated proteolysis, JAK-STAT signaling pathway, tuberculosis, alcoholism, Kaposi sarcoma-associated herpesvirus infection, and human cytomegalovirus infection (Fig. 2(b)).
GO analysis (a) and KEGG analysis (b) of 38 DEGs via the ClusterProfile. Figure (a) displays the results of the Gene Ontology (GO) analysis for differentially expressed genes. The left side of the figure categorizes the results into Biological Process, Molecular Function, and Cellular Component. The size of each circle represents the number of genes enriched in each category, and the color gradient from blue to pink indicates progressively smaller p-values. Figure (b) displays the results of the KEGG analysis. The left side shows the names of the KEGG metabolic or signal transduction pathways, and the circle sizes have the same meaning as in the GO analysis.
Determination and verification of signatures of Pompe disease
Two distinct algorithms were utilized to pinpoint potential biomarkers for Pompe disease. The LASSO regression algorithm was applied to the DEGs, identifying 7 variables as potential disease signatures for Pompe disease (Fig. 3(a)). The SVM-RFE algorithm was employed to select the top 10 feature genes from the DEGs (Fig. 3(b)). The selection of 10 features was based on a balanced consideration of model performance and feature importance. The three common features (GPNMB, CALML6, TRIM7) shared by these two algorithms were ultimately selected (Fig. 3(c)). LASSO and SVM-RFE agreed on these 3 genes because they were identified as the most important features for model prediction by both methods. This suggests that these 3 genes may have significant biological relevance or play a key role in the prediction task.
The expression of selected signatures in Pompe disease
We analyzed the expression differences of three intersection genes between patients with Pompe disease and control individuals. The GPNMB gene exhibited a significant increase in expression in these patients (Fig. 4(a)), while the CALML6 and TRIM genes showed a significant decrease in expression in Pompe disease patients (Fig. 4(b, c)).
Selected signatures are related to immunocyte infiltration levels
Immune factors play a crucial role in the occurrence and progression of Pompe disease. The characteristics of immune cells were analyzed using the CIBERSORT method. The primary objective of this part was to analyze the immune cell composition in the muscle tissues of patients with Pompe disease compared to a control group, utilizing the CIBERSORT tool to characterize the differences in immune cell types based on their gene expression profiles. This analysis will provide insights into the specific immune cell populations present in both groups and explore how these populations differ, potentially contributing to the pathophysiology of Pompe disease. Notably, it must be acknowledged that this algorithm can only provide the relative proportions of immune cells. The Fig. 5(a) illustrates that in most samples, T cells, particularly CD4 memory T cells, constitute a larger proportion, with B cells, NK cells, and monocytes/macrophages also prominently represented. A comparative analysis shows that neutrophils are more abundant in the Pompe group compared to the control group. In Fig. 5(b), the interactions among immune cells within the study samples are displayed, revealing potential synergistic or inhibitory relationships between different types of immune cells. This allows for an understanding of how these cells might coordinate or counteract each other in the immune response. However, due to the low number of certain cell types, this may lead to insufficient statistical power and limit the robustness of the results. Moreover, we observed significant dysregulation in the levels of B cells memory, plasma cells, T cells regulatory (Tregs), activated NK cells, monocytes, macrophages M0, resting dendritic cells, eosinophils, and neutrophils in Pompe disease patients compared to controls (Fig. 5(c)). We further explored the relationship between the expression of GPNMB, CALML6, and TRIM7 and the level of immune infiltration. Since immune factors play an indispensable role in Pompe disease, we hypothesize that these genes not only play a significant role in muscle cells but may also modulate immune responses within these tissues. Our findings revealed that the expression of these three genes was associated with Tregs, monocytes, macrophages M0, resting and activated dendritic cells, naive and memory B cells. Additionally, as depicted in Fig. 6(a), GPNMB was associated with T cells gamma delta, plasma cells, activated NK cells, and eosinophils. Concurrently, we found that CALML6 was also linked to T cells gamma delta, plasma cells, resting and activated NK cells, neutrophils, and eosinophils (Fig. 6(b)). Furthermore, the TRIM7 gene was found to be related to resting NK cells, neutrophils, and activated mast cells (Fig. 6(c)). Our results suggest that these three genes may play a crucial role in the pathogenesis of Pompe disease by regulating various immune cells.
(a) The percentage of the 22 immunocyte populations identified via the CIBERSORT arithmetic. The X-axis features pink and blue triangles representing controls and Pompe disease patients, respectively. (b) Displays correlations between different infiltrative immune cell populations in skeletal muscle samples from Pompe disease patients and controls. The strongest positive correlation is between monocytes and macrophages M0, and the strongest negative correlation is between T cells CD4 memory resting and T cells follicular helper. (c) The diversities in the architecture of immunocytes between controls and Pompe patients.
Correlation between GPNMB (a), CALML6 (b), TRIM7 (c) expression levels and infiltrating immune cells in muscle tissue of infantile-onset Pompe patients. The left side of the figure displays the names of immune cells. The larger the circle in the figure, the greater the correlation with the immune cells. The color transition from red to green in the circles indicates increasingly smaller p-values. Specific p-values are displayed on the right side of the figures.
Discussion
Pompe disease, although a rare genetic disorder, places a considerable burden on society and families due to its severe symptoms and limited treatment options23. Consequently, a thorough investigation of its pathogenesis is essential to advance the development of more precise and early diagnostic techniques, enhancing both the rate of diagnosis and early detection, and facilitating the creation of targeted therapies. In this study, we analyzed the GEO dataset and identified 38 DEGs between Pompe disease samples and control samples. The GO and KEGG pathway analyses underscore several biological processes and pathways, such as endoplasmic reticulum and nuclear membrane organization, GTPase and ubiquitin-like protein ligase activities, as well as calcium and phosphatidylinositol signaling systems, that may be pivotal in the pathogenesis of Pompe disease. Particularly, the involvement of ubiquitin-mediated proteolysis and the JAK-STAT signaling pathways could suggest novel therapeutic targets or biomarkers. This discussion integrates these results into our current understanding of Pompe disease, proposing how these specific pathways might contribute to its development and highlighting potential areas for further research.
To identify biomarkers for Pompe disease, we applied two machine learning algorithms to 38 previously mentioned DEGs, identifying three genes: GPNMB, CALML6, and TRIM7. These genes show different expression levels between Pompe disease samples and controls, suggesting their potential as diagnostic biomarkers. Glycoprotein Nonmetastatic Melanoma Protein B (GPNMB) is a type 1 transmembrane glycoprotein involved in various physiological processes, including immune response modulation, particularly influencing macrophages24,25. It mediates mesenchymal stem cell survival, proliferation, and migration through a CD44-dependent mechanism26. Although there’s no direct evidence linking GPNMB to Pompe disease, its role in neuroinflammatory processes could be relevant in the context of lysosomal storage disorders. In our study, GPNMB gene expression was linked with various immune cells, including gamma delta T cells, plasma cells, activated NK cells, and eosinophils. These cells are integral to both innate and adaptive immune responses, indicating that GPNMB may play a crucial role in regulating key immune pathways, which could either exacerbate or mitigate muscle cell damage in Pompe disease. Calmodulin Like 6 (CALML6) is part of the calmodulin family, involved in calcium signaling and regulation of cellular processes27. CALML6 has been identified as a negative regulator of the NF-κB signaling pathway, which is crucial for the innate immune response. It interacts with TAK1, a key kinase in this pathway, to inhibit its activity and thereby modulates the inflammation process and immune response28. In Pompe disease, calcium homeostasis may be affected due to glycogen accumulation and cellular damage, which could relate to the pathophysiological processes of the disease29. Additionally, our findings show that CALML6 is associated with a range of immune cells, such as gamma delta T cells, plasma cells, as well as both resting and activated NK cells, neutrophils, and eosinophils. This widespread association underscores CALML6’s potential influence on modulating both innate and adaptive immune responses, highlighting its significance in the inflammatory processes and immune surveillance within Pompe disease. The protein encoded by TRIM7 belongs to the Tripartite Motif (TRIM) family, involved in cell proliferation, differentiation, development, and immune response30. While the specific role of TRIM7 in Pompe disease is unclear, its general role in antiviral defense and regulation of inflammatory signaling pathways suggests a potential involvement in immune regulation. Furthermore, the association of TRIM7 with resting NK cells, neutrophils, and activated mast cells suggests its significant role in regulating the inflammatory response. This could potentially alter the immune environment surrounding the impaired muscle tissue in Pompe disease, impacting disease progression and symptom manifestation.
The accumulation of glycogen in muscle cells of Pompe disease patients leads to lysosomal storage and significant disruption of muscle cell structure, triggering a cellular stress response and activation of satellite cells involved in muscle repair31. In treating Pompe disease, the development of anti-drug antibodies (ADA) in response to recombinant human alpha-glucosidase (rhGAA), especially in CRIM-negative patients, reduces enzyme replacement therapy (ERT) effectiveness; immune tolerance induction (ITI) strategies are proposed to counter ADA’s impact32,33,34. In this study, we discovered that GPNMB, CALML6, and TRIM7are all associated with Tregs. Regulatory T cells (Tregs) not only control immune responses but also accumulate in skeletal muscles post-injury, moderating the immune environment to enhance muscle repair by fostering an anti-inflammatory state and interacting with essential cells like stem cells35,36,37. In Pompe disease, glycogen accumulation causes endoplasmic reticulum stress, triggering NF-κB signaling pathways and leading to exacerbated inflammation in muscles and other tissues, which can be mitigated by factors like CALML628,38,39. GPNMB, a protein expressed in immune cells, is known for its role in mediating inflammation and fibrosis, particularly in liver repair contexts. It influences macrophage behavior, promoting an environment that can either support tissue repair or contribute to fibrosis, depending on the context26. Additionally, a study on Pompe disease by Nina Raben and others demonstrates that targeting autophagy by deactivating the Atg7 gene in muscle significantly reduces glycogen accumulation when combined with enzyme replacement therapy. This method, which improves skeletal muscle response, could provide a new therapeutic strategy for diseases involving disrupted autophagy40.
While our research has highlighted the potential role of immune factors in Pompe disease, it is important to recognize several limitations in our study. First, our analysis was restricted to immune cells in skeletal muscle; therefore, immune responses in secondary lymphoid organs were not considered. This narrow focus could potentially exclude significant immunological dynamics pertinent to the overall disease mechanism. Additionally, bulk RNA sequencing, the technique we used to analyze gene expression, can provide an overall profile of gene expression in the sample, but it cannot directly quantify specific cell types, such as inflammatory cells. Although tools like CIBERSORT can deconvolute RNA expression data to infer the relative proportions of different immune cells, they only provide relative ratios and not absolute numbers. Therefore, the limited number of certain cell types analyzed might result in findings that are disproportionately influenced by random variation. We look forward to future research focusing on the immunological aspects of Pompe disease and the publication of corresponding datasets, so that we can increase our sample sizes and thereby obtain more robust results. Additionally, although the sample size is relatively small, expanding it along with the study cohort is necessary to better understand the specificity and sensitivity of the genes under investigation. This would help in differentiating more effectively between Pompe disease patients and healthy controls. Finally, the data presented only supports correlation analyses and does not establish causality between Pompe disease and changes in immune cell profiles. Future studies incorporating experimental validation, such as flow cytometry, will be crucial in confirming these associations.
Conclusion
Through bioinformatics and machine learning, it has been demonstrated that GPNMB, CALML6, and TRIM7 are key DEGs in Pompe disease compared to control specimens. The findings of this study highlight the significant role of immune factors in the pathogenesis and progression of Pompe disease, underscoring the importance of monitoring and managing the immune response in patients to optimize treatment outcomes and improve prognosis. Furthermore, these results may offer potential targets for the treatment of Pompe disease.
Data availability
All data generated or analysed during this study are included in this published article. All data are available from the corresponding author on reasonable request.
References
Farah, B. L., Yen, P. M. & Koeberl, D. D. Links between autophagy and disorders of glycogen metabolism - perspectives on pathogenesis and possible treatments. Mol. Genet. Metab. 129 (1), 3–12 (2020).
Peruzzo, P., Pavan, E. & Dardis, A. Molecular genetics of pompe disease: Acomprehensive overview. Ann. Transl Med. 7 (13), 278 (2019).
Kishnani, P. S. et al. Pompe disease diagnosis and management guideline. Genet. Med. 8 (5), 267–288 (2006).
Kishnani, P. S. et al. A retrospective, multinational, multicenter study on the natural history of infantile-onset pompe disease. J. Pediatr. 148 (5), 671–676 (2006).
van den Hout, H. M. et al. The natural course of infantile pompe’s disease: 20 original cases compared with 133 cases from the literature. Pediatrics. 112 (2), 332–340 (2003).
Kishnani, P. S. et al. Recombinant human acid [alpha]-glucosidase: Major clinical benefits in infantile-onset pompe disease. Neurology. 68 (2), 99–109 (2007).
van der Ploeg, A. T. et al. A randomized study of alglucosidase alfa in late-onset pompe’s disease. N Engl. J. Med. 362 (15), 1396–1406 (2010).
Amalfitano, A. et al. Recombinant human acid alpha-glucosidase enzyme therapy for infantile glycogen storage disease type II: Results of a phase I/II clinical trial. Genet. Med. 3 (2), 132–138 (2001).
Kroos, M. A. et al. Broad spectrum of pompe disease in patients with the same c.-32-13T->G haplotype. Neurology. 68 (2), 110–115 (2007).
Laforet, P. et al. Juvenile and adult-onset acid maltase deficiency in France: Genotype-phenotype correlation. Neurology. 55 (8), 1122–1128 (2000).
Wokke, J. H. et al. Genotype-phenotype correlation in adult-onset acid maltase deficiency. Ann. Neurol. 38 (3), 450–454 (1995).
Kroos, M. A. et al. p.[G576S; E689K]: pathogenic combination or polymorphism in pompe disease? Eur. J. Hum. Genet. 16 (8), 875–879 (2008).
Labrousse, P. et al. Genetic heterozygosity and pseudodeficiency in the Pompe disease newborn screening pilot program. Mol. Genet. Metab. 99 (4), 379–383 (2010).
Tajima, Y. et al. Structural and biochemical studies on Pompe disease and a pseudodeficiency of acid alpha-glucosidase. J. Hum. Genet. 52 (11), 898–906 (2007).
Schaaf, G. J. et al. Lack of robust satellite cell activation and muscle regeneration during the progression of Pompe disease. Acta Neuropathol. Commun. 3, 65 (2015).
Kazi, Z. B. et al. Durable and sustained immune tolerance to ERT in Pompe disease with entrenched immune responses. JCI Insight ;1(11). (2016).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28 (11), 1947–1951 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51 (D1), D587–D92 (2023).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28 (1), 27–30 (2000).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 16 (5), 284–287 (2012).
Becht, E. et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 17 (1), 218 (2016).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 12 (5), 453–457 (2015).
Schoser, B. et al. The humanistic burden of pompe disease: Are there still unmet needs? A systematic review. BMC Neurol. 17 (1), 202 (2017).
Gillett, D. A., Wallings, R. L., Uriarte Huarte, O. & Tansey, M. G. Progranulin and GPNMB: Interactions in endo-lysosome function and inflammation in neurodegenerative disease. J. Neuroinflammation. 20 (1), 286 (2023).
Saade, M., Araujo de Souza, G., Scavone, C. & Kinoshita, P. F. The role of GPNMB in inflammation. Front. Immunol. 12, 674739 (2021).
Lazaratos, A. M., Annis, M. G. & Siegel, P. M. GPNMB: A potent inducer of immunosuppression in cancer. Oncogene. 41 (41), 4573–4590 (2022).
Wang, Z. et al. The EF-Hand protein CALML6 suppresses antiviral innate immunity by impairing IRF3 dimerization. Cell. Rep. 26 (5), 1273–85e5 (2019).
Sheng, C. et al. CALML6 controls TAK1 ubiquitination and confers protection against acute inflammation. J. Immunol. 204 (11), 3008–3018 (2020).
Lim, J. A., Li, L., Kakhlon, O., Myerowitz, R. & Raben, N. Defects in calcium homeostasis and mitochondria can be reversed in pompe disease. Autophagy. 11 (2), 385–402 (2015).
Hatakeyama, S. TRIM family proteins: roles in Autophagy, immunity, and Carcinogenesis. Trends Biochem. Sci. 42 (4), 297–311 (2017).
Thurberg, B. L. et al. Characterization of pre- and post-treatment pathology after enzyme replacement therapy for pompe disease. Lab. Invest. 86 (12), 1208–1220 (2006).
Desai, A. K., Li, C., Rosenberg, A. S. & Kishnani, P. S. Immunological challenges and approaches to immunomodulation in pompe disease: a literature review. Ann. Transl Med. 7 (13), 285 (2019).
Nayak, S. et al. Immune responses and hypercoagulation in ERT for Pompe disease are mutation and rhGAA dose dependent. PLoS One. 9 (6), e98336 (2014).
Chen, H. A. et al. Optimizing treatment outcomes: Immune tolerance induction in Pompe disease patients undergoing enzyme replacement therapy. Front. Immunol. 15, 1336599 (2024).
Burzyn, D. et al. A special population of regulatory T cells potentiates muscle repair. Cell. 155 (6), 1282–1295 (2013).
Chen, Z. et al. Regulatory T cells-centered regulatory networks of skeletal muscle inflammation and regeneration. Cell. Biosci. 12 (1), 112 (2022).
Coulis, G. & Villalta, S. A. Muscle immune cells protect mitochondrial organelles during exercise. Nature. 625 (7993), 35–36 (2024).
Kohler, L., Puertollano, R. & Raben, N. Pompe disease: From basic science to therapy. Neurotherapeutics. 15 (4), 928–942 (2018).
Ma, K., Zhang, Y., Zhao, J., Zhou, L. & Li, M. Endoplasmic reticulum stress: Bridging inflammation and obesity-associated adipose tissue. Front. Immunol. 15, 1381227 (2024).
Raben, N. et al. Suppression of autophagy permits successful enzyme replacement therapy in a lysosomal storage disorder–murine pompe disease. Autophagy. 6 (8), 1078–1089 (2010).
Acknowledgements
We thank all participants for their interest and participation to the study.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Number: 31970698) and the Sichuan Science and Technology Program (Grant Number: 2023YFS0109).
Author information
Authors and Affiliations
Contributions
MYX and LYL designed the study; ZJF and LXY wrote the manuscript; YL and SY completed the data retrieval and analysis; CXL, ZYC, JWH assisted in the data analysis and drawing. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, J., Lin, X., Yin, L. et al. Analyzing immune cell infiltrates in skeletal muscle of infantile-onset Pompe disease using bioinformatics and machine learning. Sci Rep 14, 27485 (2024). https://doi.org/10.1038/s41598-024-78634-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-78634-6