Abstract
The increasing occurrence of hospital-associated infections, particularly bacteremia, caused by extensively drug-resistant (XDR) carbapenemase-producing colistin-resistant Klebsiella pneumoniae highlights a critical requirement to discover new therapeutic alternatives. Bacteriophages having host-specific bacteriolytic effects are promising alternatives for combating these pathogens. Among 12 phages isolated from public wastewater in Thailand, two phages-vB_kpnM_05 (myovirus) and vB_kpnP_08 (podovirus) showed broad-host range, producing bacteriolytic activities against 81.3% (n = 26) and 78.1% (n = 25) of 32 XDR carbapenemase-producing colistin-resistant K. pneumoniae, with capsular types—K15, K17, K50, K51, K52/wzi-50 and K2/wzi-2. Both phages showed short replication times, large burst sizes with rapid adsorptions. They exhibited significant stability under various environmental conditions. Genomic analysis revealed that both phages are genetically distinct phages from Myoviridae and Podoviridae family, with the lack of toxin, virulence, lysogeny and antibiotic resistance genes. These characteristics highlighted their promising potential for utilizing in phage therapy for combating XDR K. pneumoniae. Although phage cocktail combining vB_kpnM_05 and vB_kpnP_08 provided significant bacteriolysis for longer duration (8 h) than its monophage (6 h), bacterial regrowth was observed which suggested an evitable development of phage resistance under phages’ selection pressures. Future study will be undertaken to elucidate the precise mechanisms by which these XDR K. pneumoniae developed phage resistance and their associated fitness cost. Remarkably, combining phage cocktail with amikacin at their sub-inhibitory concentrations produced potent synergy by completely suppressing bacterial regrowth in vitro. Our study demonstrated the significant therapeutic and prophylactic effectiveness of a phage cocktail-amikacin combination as a promising alternative strategy for overcoming bacteremia associated with XDR K. pneumoniae having carbapenemase and colistin resistance in vivo.
Similar content being viewed by others
Introduction
Klebsiella pneumoniae, a Gram-negative Enterobacteriaceae pathogen, is commonly implicated in hospital-associated infections, particularly bacteremia1. Currently, there is an increasing number of multidrug-resistant clones among K. pneumoniae, which have become endemic in several countries1,2,3. These high-risk clones, designated as highly-virulent hypermucoviscous K. pneumoniae (hvKp), were discovered to be frequently associated with bloodstream infections in clinical settings4. The extensive administration of carbapenem and/or colistin, combined with the rapid dissemination of plasmid-mediated carbapenemases, mobilized colistin resistance (mcr) genes, and mutations in resistance determinants driven by antibiotic selective pressures, have contributed to the global rise of carbapenem and colistin resistance among K. pneumoniae, resulting in the emergence of extensively drug-resistant (XDR) pathogens5. The scarcity of efficient treatment options and limitations in the advancement of new antibiotics, emphasize the urgent requirement to discover a viable therapeutic strategy for addressing XDR K. pneumoniae-associated bacteremia6,7,8.
Bacteriophages (phages) are bacterial-infecting viruses abundantly found in environments, including wastewater9,10,11,12. Phages, with their abilities to attach to specific surface receptors and self-replicate within hosts to initiate host-specific bacteriolysis without disturbing other resident microbiota, have been suggested as potential therapeutic options for treating infections caused by XDR K. pneumoniae13. Phages with a broad host range, which specifically target an extensive number of K. pneumoniae, are preferable for therapeutic applications compared to K. pneumoniae-specific phages with a narrow host range14. The combination of K. pneumoniae-specific phages into a synergistic cocktail offers a feasible approach for addressing the limited host range of these phages and the emergence of more virulent phage-resistant hypermucoviscous strains following monophage treatment15,16,17. Nevertheless, a previous study documented the development of bacterial resistance to the 5-phage cocktail after several days, although such an occurrence was exceptionally uncommon11,18. This represents an important challenge that needs to be addressed to facilitate the therapeutic applications of phages feasible19.
One further approach to tackle the emergence of phage resistance is the phage-antibiotic synergy (PAS) generated by the combination of phages with an antibiotic20,21,22. Complementing phage or phage cocktails with an antibiotic not only enhanced their combined antibacterial effects, but also restored the efficacy of antibiotic even against pathogens which are resistant to that antibiotic20,21,22. Earlier case reports have revealed that phage-antibiotic combination significantly decreased treatment failures, increased microbiological cure, and improved patients’ survival in clinical settings23,24. To the best of our knowledge, no study has investigated the potential effectiveness of a phage cocktail-antibiotic combination for tackling bacteremia associated with XDR K. pneumoniae having carbapenemase and colistin resistance. This pre-clinical study aimed to isolate and characterize potent K. pneumoniae-specific phages from public wastewater in Thailand, followed by investigating the synergistic efficacy of the phage cocktail-antibiotic combination for overcoming XDR K. pneumoniae-associated bacteremia both in vitro and in vivo.
Results
Phages—vB_kpnM_05 and vB_kpnP_08 showed broad-host range, producing bacteriolytic activities against XDR K. pneumoniae
Among 12 phages isolated from public wastewater, two phages-vB_kpnM_05 and vB_kpnP_08 showed broad-host range, producing bacteriolytic activities against 81.3% (n = 26) and 78.1% (n = 25) of 32 XDR carbapenemase-producing colistin-resistant K. pneumoniae from different clinical sources, particularly those with ERIC-A profile and capsular types of K15, K17, K50, K51, K52/wzi-50 and K2/wzi-2 (Fig. 1) (Table 1) (Supplementary Fig. 1) (Supplementary Table 1).
Heatmap illustrating the host range of isolated phages (n = 12) by determination of phages bacteriolytic activities against XDR carbapenemase-producing colistin-resistant K. pneumoniae clinical isolates (n = 32) collected between 2016 and 2021. Blue: Isolates with high sensitivity to phage’s bacteriolytic activities; white: Isolates with low sensitivity to phage’s bacteriolytic activities.
When their bacteriolytic efficiencies were quantified, bacteriolysis of high (EOP ≥ 0.5), medium (0.1 ≤ EOP < 0.5) and low (0.001 ≤ EOP < 0.1) efficiencies were observed in 21, 3 and 2 XDR K. pneumoniae isolates for vB_kpnM_05; and 20, 3 and 2 XDR K. pneumoniae isolates for vB_kpnP_08, respectively (Table 1). Both phages showed targeted host specificities, having no bacteriolytic activities against non-target bacteria (Supplementary Table 2).
On double-layer plates, vB_kpnM_05 produced small, clear plaques of 0.5–1 mm with halo and vB_kpnP_08 possessed large, clear plaques of 3-5 mm with halo (Fig. 2a,b). According to TEM, vB_kpnM_05 was a myovirus possessing icosahedral head (58.2 ± 0.7 nm), and long contractile tail (96.6 ± 0.8 nm), whereas vB_kpnP_08 was a podovirus having icosahedral head (56.7 ± 0.4 nm) and short noncontractile tail (14 ± 0.5 nm) (Fig. 2a,b).
Morphological, and biological characteristics of phage. Plaque morphologies and electron micrographs of (a) vB_kpnM_05 and (b) vB_kpnP_08; (c) optimal MOIs of vB_kpnM_05 and vB_kpnP_08; (d) adsorption of vB_kpnM_05 and vB_kpnP_08, (e) one-step growth curve of vB_kpnM_05, and (f) one-step growth curve of vB_kpnP_08.
Both phages exhibited rapid adsorptions, short latent periods, and large burst sizes with low optimal MOIs for propagation
The MOI of 0.01 and 0.1 were observed to produce maximum phage titers for vB_kpnM_05 and vB_kpnP_08, signifying their low optimal MOIs for propagation (Fig. 2c).
Approximately 60% of vB_kpnM_05 and 40% of vB_kpnP_08 were adsorbed to their hosts within 5 min and their adsorption were increased with time (Fig. 2d). Eventually, the adsorption of vB_kpnM_05 and vB_kpnP_08 to their hosts reached more than 99% by 15 min and 20 min (Fig. 2d). The adsorption rate constant, k, of vB_kpnM_05 and vB_kpnP_08 were 3.5 × 10−9 and 2.1 × 10−9 ml/min, indicating the rapid and efficient adsorptions of these phages to their hosts (Fig. 2d).
According to one-step growth curves, the latent period—the interval between phage adsorption to its host and the beginning of the release of new phages from infected host cells, were observed to be short—approximately 35 min for vB_kpnM_05 (Fig. 2e) and 30 min for vB_kpnP_08 (Fig. 2f). The burst size – the number of new phages released from host lysis, exhibited to be large—104 ± 5 PFU/host for vB_kpnM_05, and 220 ± 7 PFU/host for vB_kpnP_08 (Fig. 2e,f).
Phages were stable under different environmental conditions
Both vB_kpnM_05 and vB_kpnP_08 were thermostable between − 20 and 60 °C, but complete inactivation of their bacteriolytic activities was observed at 80 °C (Fig. 3a). Both phages showed stable bacteriolytic activities between pH 4–10, however, their activities were completely diminished at pH outside of this range (Fig. 3b). UV irradiation completely inhibited bacteriolytic activities at 60 min postexposure (Fig. 3c). Both phages were long-term stable with no significant changes in their activities at 4 °C up until 6 months which was the endpoint (Fig. 3d). Chemically, both phages were stable in chloroform (10–40%) but their activities were adversely affected with ethanol (70, 99%) (Fig. 3e).
Stability of phages under different environmental conditions. (a) Thermostability of vB_kpnM_05 and vB_kpnP_08; (b) stability of vB_kpnM_05 and vB_kpnP_08 under different pH, (c) stability of vB_kpnM_05 and vB_kpnP_08 under UV exposure; (d) stability of vB_kpnM_05 and vB_kpnP_08 for long-term storage; (e) stability of vB_kpnM_05 and vB_kpnP_08 under chloroform and ethanol exposure.
Genomic characteristics of phages
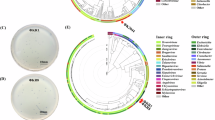
Both vB_kpnM_05 and vB_kpnP_08 were DNA phages having different banding patterns of RAPD-PCR and restriction digestions (Supplementary Fig. 2a–b). Whole-genome sequencing analysis revealed that both phages were double-stranded DNA phages, whereas vB_kpnM_05 was 42,756 bp in length with 52.76% GC content belonging to Myoviridae family, and vB_kpnP_08 possessed 40,238 bp genome with 52.84% GC content belonging to Podoviridae family, respectively (Fig. 4a,b) (Accession number: European Nucleotide Archive: PRJEB75204). The genome annotation analysis indicated that a total of 75 and 70 CDS were identified in vB_kpnM_05 and vB_kpnP_08. Among their CDS, 50 CDS of vB_kpnM_05 and 45 CDS of vB_kpnP_08 had annotated functions, while the remaining 25 CDS in both vB_kpnM_05 and vB_kpnP_08 were annotated as hypothetical proteins. The functional proteins of both vB_kpnM_05 and vB_kpnP_08 were classified into three categories: DNA replication/metabolism, structure/packaging, and host lysis. The genes related to toxin, virulence, lysogeny and antibiotic resistance were not detected in the genomes of both phages. The phylogenetic analysis of vB_kpnM_05 and vB_kpnP_08 revealed that these phages shared close evolutionary relationships with other Klebsiella phages (Fig. 4c,d), indicating a shared evolutionary history within the broader landscape of Klebsiella phages.
Bacterial regrowth was observed with monophage and phage cocktail therapies
When we investigated antibacterial efficacies by giving vB_kpnM_05 or vB_kpnP_08 as monophage at different MOIs, phage-induced bacteriolysis was observed in dose-dependent manners with most significant effects at MOI-10 and MOI-100 of each monophage (Fig. 5a,b; p < 0.01). Compared to its monophage (MOI-10), phage cocktail combining vB_kpnM_05 and vB_kpnP_08 (MOI-10 each) suppressed bacterial growth for longer duration-8 h (Fig. 5c; p < 0.01). Nevertheless, bacterial regrowth was observed after 6 and 8 h of monophage and cocktail therapies (Fig. 5a–c). Upon our preliminary evaluation, phage cocktails developed by combining vB_kpnM_05 or vB_kpnP_08 with other phages as 3–12 phage-containing cocktails did not exhibit a better inhibition of bacterial growth when compared to a 2 phage-containing cocktail. Therefore, a phage cocktail containing vB_kpnM_05 and vB_kpnP_08 was selected to further investigate its synergistic effects in conjunction with different antibiotics.
(a) Bacteriolytic activities of vB_kpnM_05 as monophage therapy at different MOIs (0.01–100); (b) bacteriolytic activities of vB_kpnP_08 as monophage therapy at different MOIs (0.01–100): (c) bacteriolytic activities of vB_kpnM_05 and vB_kpnP_08 as monophage and combined phage cocktail at MOI-10 in vitro.
Phage cocktail-amikacin combination exhibited potent synergistic activities, completely inhibiting bacterial regrowth in vitro
In all tested isolates, MIC for monophage was MOI of > 1000, and MIC for phage cocktail was MOI-100, which were observed at 8 h of the checkerboard assay (Supplementary Fig. 3–4). Combining vB_kpnM_05 or vB_kpnP_08 as either monophage with antibiotics (amikacin, colistin, imipenem, ciprofloxacin, ceftazidime) or phage cocktail with antibiotics (colistin, imipenem, ciprofloxacin, ceftazidime) did not exhibit any synergistic effects in our preliminary checkerboard screening (Supplementary Fig. 3–4). Meanwhile, combining subinhibitory concentrations of phage cocktail (MOI-10 each) and amikacin (8 μg/ml) produced the potent synergistic effects (FICI < 0.5) (Fig. 6a), (Supplementary Fig. 5). Bacterial regrowth was observed at 4–8 h after exposure to phage cocktail or amikacin monotherapies in all tested isolates with varying levels of amikacin susceptibility, including intermediate susceptibility and resistance (32–128 µg/ml). Meanwhile, a phage cocktail-amikacin combination significantly exhibited potent synergistic activities by substantially decreasing their growth (OD600) and viabilities (CFU/ml) within 2 h, and completely suppressing bacterial regrowth until 24 h (Fig. 6b,c; p < 0.01) (Supplementary Fig. 5).
(a) Checkerboard screening assay showing the synergistic activities of a phage cocktail-amikacin combination (FICI < 0.5); Time-kill confirmation assay showing the synergistic activity of a phage cocktail-amikacin combination for reducing the (b) bacterial growth (OD600nm), and (c) bacterial viability (CFU/ml) in vitro.
Phage cocktail-amikacin combination produced significant therapeutic efficacy in murine bacteraemia in vivo
Investigations on the safety of administering an IP phage cocktail to mice revealed that it was safe, with no symptoms of toxicity, discomfort, disease development, or death in the animals. Giving an IP phage cocktail displayed stability for up to 24 h post-administration (Supplementary Fig. 6a–b). We then evaluated an infective dose to induce bacteremia in mice by delivering different doses of the tested bacteria intraperitoneally (1 × 106, 1 × 107, 1 × 108 CFU/ml). All mice died within 48 h following a bacterial challenge with 1 × 108 CFU/ml, which was denoted as LD100 (100% lethal dose) (Supplementary Fig. 7a) and utilized to develop bacteremia in our subsequent experiment. Murine bacteremia developed at 2hpi with LD100, resulting in the progressive dissemination of bacteria into the bloodstream and internal organs (Supplementary Fig. 7b). The bacteria recovered from our animal studies was identical to the isolate we used to induce bacteremia, characterized by the presence of NDM and IS1-integrated mgrB with amikacin resistance.
Afterwards, we assessed the therapeutic efficacy of different treatments using our established bacteremia model. Compared to PBS and their monotherapies, IP phage cocktail-amikacin combination therapy significantly reduced bacterial burden in internal organs (Fig. 7a; p < 0.01) and improved murine survival to 100% by Day 7 (Fig. 7b; p < 0.05). The survival of animals was significantly improved by giving a therapeutic phage cocktail-amikacin combination at 2hpi, in contrast to administration at 8hpi or 24hpi (Supplementary Fig. 8; p < 0.01). Significant and comparable concentrations of both phages were detected only in mice which received phage cocktail monotherapy or phage cocktail-amikacin combination (Fig. 7c), (Supplementary Fig. 9a–b).
Therapeutic effectiveness of phage cocktail and amikacin as monotherapy and combination therapy in bacteremia by determination of (a) bacterial burden in internal organs (blood, kidney, liver, and spleen), (b) survival of treated mice, and (c) viable titre of phage in internal organs (blood, kidney, liver, and spleen) of mice receiving different treatments in vivo.
Phage cocktail-amikacin combination showed remarkable prophylactic efficacy in murine bacteraemia in vivo
Prophylactic administration of IP phage cocktail-amikacin combination therapy significantly reduced bacterial burden in internal organs (Fig. 8a; p < 0.01) and protected all treated mice from fatalities, as compared to their monotherapies (Fig. 8b; p < 0.01). Significant and comparable concentrations of both phages were observed only in animals exposed to the administration of phage cocktail monotherapy or phage cocktail-amikacin combination (Fig. 8c), (Supplementary Fig. 8c–d).
Prophylactic effectiveness of phage cocktail and amikacin as monotherapy and combination therapy in bacteremia by determination of (a) bacterial burden in internal organs (blood, kidney, liver, and spleen), (b) survival of treated mice, and (c) viable titre of phage in organs (blood, kidney, liver, and spleen) of mice receiving different treatments in vivo.
Discussion
Nowadays, XDR K. pneumoniae resistant to carbapenem and colistin, are frequently reported in fatal bacteremia due to the lack of potent therapeutic alternatives, and limitations in development of new antibiotics. This highlights the critical need for discovering viable therapeutic approach to overcome bacteremia associated with this pathogen in clinical settings5. Phages have recently been proposed as potential therapeutic alternatives for addressing these difficult-to-treat pathogens9. Among 12 phages isolated from public wastewater in Thailand, vB_kpnM_05 (myovirus) and vB_kpnP_08 (podovirus) demonstrated broad-host range, producing bacteriolytic activities against 81.3% and 78.1% of XDR K. pneumoniae clinical isolates, particularly those with ERIC-A profile and capsular types of K15, K17, K50, K51, K52 and K2. Furthermore, vB_kpnM_05 and vB_kpnP_08 demonstrated host specificities and high bacteriolytic efficiencies in their target XDR K. pneumoniae isolates. Differences in these phages’ host range therefore suggested the possible differences in their receptors for recognition, adsorption and lysis while targeting its pathogens25. In accordance with prior studies26,27,28, we discovered short latent periods, large burst sizes with rapid absorption rates, and low optimal MOIs for propagation of vB_kpnM_05 and vB_kpnP_08, revealing that both phages required short replication times to generate a significant number of progenies at low MOIs for triggering highly-efficient, targeted antibacterial effects against XDR K. pneumoniae. These findings highlighted their appealing potential for utilizing in phage therapy in XDR K. pneumoniae-associated infections9,28.
Comparable to prior studies26,27,28, both phages were thermostable between − 20 and 60 °C, but their activities were completely inactivated at 80 °C, highlighting the detrimental impact of extremely high temperature on their lytic activities. In agreement with earlier studies26,27,28, both phages demonstrated stable bacteriolytic activities between pH 4–10. Their bacteriolytic activities were dramatically decreased at pH 2, 12, and 14, suggesting that phages were denaturized in these environmental conditions. Consistent with previous studies26,27,28, both phages displayed stabilities for long-term storage and chloroform (10–40%), while exposure to UV and 70–90% ethanol exhibited negative impacts on their viabilities. Both phages were DNA phages belonging to Myoviridae and Podoviridae families, with the absence of genes related to toxin, virulence, lysogeny and antibiotic resistance, which further highlighted their safety for therapeutic applications. The phylogenetic analysis of both phages revealed a shared evolutionary history within the broader landscape of Klebsiella phages. As the stability and safety of genetically-different phages under various environmental conditions is an important selection criteria for their usage as ideal therapeutic phages in clinical settings, these phages presented a promising role for utilizing in phage therapy for combating XDR K. pneumoniae-associated infections9,28.
When we tested bacteriolytic efficacies by giving vB_kpnM_05 or vB_kpnP_08 as monophage, we found dose-dependent bacteriolysis, with most potent effects at MOI-10 and MOI-100. In agreement with prior studies13,29, phage cocktail (MOI-10 each) provided better bacteriolysis that lasted longer duration than their monophage (MOI-10). It will be necessary to conduct additional research on the target attachment site of each phage because an enhanced bacteriolysis produced by phage cocktail suggested that each phage could target different receptors and use distinct antibacterial tactics to effectively limit their target pathogens, rather than competing with one another for the same antibacterial mechanisms or receptors25. Consistent with earlier studies, bacterial regrowth was observed after 6 and 8 h of monophage and phage cocktail treatments, revealing that neither treatment was effective at preventing phage-resistant bacterial regrowth18,30,31,32. It also suggested that evolution of phage-resistant bacteria under phages selection pressures is unavoidable challenge that might contribute to limited therapeutic effectiveness of phages31,33. Bacteria develop phage resistance by different mechanisms including altered phage adsorption through surface structures modifications, phage DNA interference by inhibiting its entry or cleaving phage DNA, and abortive phage infections34. Future investigations will therefore be undertaken to elucidate the precise mechanisms by which these XDR K. pneumoniae developed phage resistance and their associated fitness cost.
Upon screening monophage or phage cocktail in combination with different antibiotics, combining sub-inhibitory concentrations of phage cocktail (MOI-10 each) with amikacin (8 μg/ml) produced potent synergistic effects. Interestingly, any potential interference that could jeopardize the therapeutic benefits of this phage cocktail-amikacin combination triggered by regrowth bacteria was not observed because bacterial growth was completely suppressed when subjected to a phage cocktail-amikacin combination therapy. These results were consistent with prior studies which revealed the superior efficacy of phage-antibiotic combinations for suppressing bacterial regrowth and overcoming the emergence of resistance in different pathogens22,35,36. Phage produces antibacterial activity by binding to its specific receptors on target pathogen, self-proliferating inside and triggering bacteriolysis9. Amikacin exhibits bactericidal effects by interfering with bacterial protein synthesis and enhancing cellular respiration37,38. By exposing XDR K. pneumoniae to these distinct selective pressures, phage-induced bacterial membrane disintegration could mitigate permeability-associated barriers and increase amikacin entry into its target, whereas they could interact synergistically to produce potent bactericidal effects against these pathogens. Furthermore, because of an association between phage exposure and restoration of bacterial antibiotic susceptibilities from lower virulence as evolutionary trade-offs39,40,41, it is suggested that exposure to phage cocktail in this study could resensitize XDR K. pneumoniae to amikacin, allowing it to produce potent PAS at sub-inhibitory doses. Future studies will therefore be conducted to determine how phage cocktail and amikacin interact inside XDR K. pneumoniae to produce potent PAS observed in this study.
In accordance with a prior study42, all mice died within 48 h from bacteremia after IP inoculation with 1 × 108 CFU/ml of XDR K. pneumoniae (LD100) which developed 2 h after bacterial challenge. Our findings demonstrated that giving mice an IP phage cocktail was stable and safe in vivo, with no occurrence of toxicity, illnesses, or death. Comparable to earlier studies43,44, therapeutic administration of phage cocktail-amikacin combination significantly reduced bacterial burden in internal organs and increased murine survival to 100%, demonstrating its significant in vivo therapeutic effectiveness for overcoming XDR K. pneumoniae-associated bacteremia. Consistent with other studies45,46, giving a phage cocktail-amikacin combination at 2 hpi produced better survival outcomes than administering at 8 hpi or 24 hpi. This could be attributable to the fact that early phage cocktail-amikacin administration may encourage the significant reduction of bacterial burden and reduce the progression of bacteremia when compared to delayed administration47,48. Administering prophylactic phage cocktail-amikacin combination significantly reduced bacterial burdens and prevented fatalities in all mice, indicating its synergistic prophylactic efficacy against XDR K. pneumoniae-associated bacteremia. The observation that animals receiving either a therapeutic and prophylactic phage cocktail or a phage cocktail-amikacin combination showed comparable levels of phages in their internal organs, while mice given PBS or amikacin exhibited no phages, suggested the lack of endogenous XDR K. pneumoniae-specific phages in the treated animals45. Despite having viable phages in animals treated with therapeutic and prophylactic phage cocktail monotherapy, the lack of bacterial load reduction or mortality protection in these animals further highlighted the synergistic significance of a phage cocktail-amikacin combination in murine bacteremia in vivo. Recent human studies have reported the clinical applicability of phage-antibiotic combination for eliciting favorable therapeutic outcomes in K. pneumoniae-infected patients where conventional antibiotics have consistently failed23,49,50. However, using high dosages of phages were reported to have unfavorable effects due to accelerated phage-induced bacteriolysis51. In our study, phage cocktail-amikacin combination at sub-inhibitory doses demonstrated significant therapeutic and prophylactic efficacies in treated mice without triggering undesirable effects, suggesting that phage cocktail-amikacin combination at their subinhibitory concentrations could also prevent the occurrence of adverse effects and evolution of resistance under its dual selection pressures.
Limitation of this study
Owing to discrepancies in animal and human biology, the findings of our pre-clinical study may not be directly transferable to humans, as they were derived exclusively from in vitro and in vivo mouse studies. Additional human clinical studies and investigations into the impacts of phages on host immune responses are necessary to validate the therapeutic applicability of phage cocktail-amikacin combination in XDR K. pneumoniae-associated bacteremia. Further research is required to assess its efficacy in treating XDR K. pneumoniae-associated infections, along with its pharmacokinetic and safety characteristics.
Conclusion
Our study demonstrated the remarkable therapeutic and prophylactic effectiveness of a phage cocktail-amikacin combination as a potential alternative therapy for overcoming bacteremia associated with XDR K. pneumoniae having carbapenemase and colistin resistance.
Materials and methods
Bacterial strains
The previously characterized XDR K. pneumoniae clinical isolates having carbapenemase and different mechanisms of colistin resistance (n = 32) (Supplementary Table 3)7,52 and Escherichia coli, Acinetobacter baumannii, Proteus mirabilis, Enterobacter cloacae, Pseudomonas aeruginosa, and methicillin-resistant Staphylococcus aureus (n = 6) isolated from routine clinical specimens were used. Bacterial isolates were stored at − 80 °C. For bacterial capsule typing, PCR amplification and sequencing of wzi gene was conducted using wzi-F (GTG CCG CGA GCG CTT TCT ATC TTG GTA TTC C) and wzi-R (GAG AGC CAC TGG TTC CAG AA [C or T] TT [C or G] ACC GC), as published previously53.
Phage screening, isolation, and purification
As previously described27,54, public wastewater from 7 different locations in Thailand were firstly collected. Then the phages in these wastewaters were screened and isolated by using kp924 and kp291 as hosts. These hosts were XDR K. pneumoniae clinical isolates which exhibited different carbapenemase and distinct mechanisms of colistin resistance (Supplementary Table 3). Briefly, the wastewater samples were centrifuged at 12,000×g for 10 min and filtered through 0.22 μm syringe filters. Then the filtrates (10 ml) were mixed with TSB (10 ml) and exponential cultures of host bacteria (200 μl) for phage enrichment by incubating overnight at 37 °C with shaking (150 rpm). After enrichment, bacteria were harvested by centrifuging at 8000×g for 30 min, and the supernatant was filtered using a sterile 0.22 μm filter. The filtered supernatant (200 μl) was mixed with exponential-phase bacterial culture (200 μl). This mixture was then mixed with top agar (3 ml, 0.7% agar in TSB) and it was quickly overlaid onto TSA to form a double-layer plate. After overnight incubation of these plates at 37 °C, phage activity in wastewater was screened by the presence of lytic plaque on the double-layer agar plates. For isolation and purification, a single plaque was then obtained from these double-layer agar plates and mixed in SM buffer, followed by centrifugation at 6000×g for 10 min at 4 °C, and filtration. The filtrates were then serially diluted in SM buffer and mixed with exponential-phase host bacterial culture to conduct double-layer agar method as above and the phage morphology was observed. This procedure of single-plaque purification was repeated five times until the purified phages were isolated27,54.
To determine phage titres (PFU/ml), the isolated phages were serially diluted (100 to 10−12) in SM buffer and mixed with exponential-phase bacterial culture to conduct double-layer agar method. After overnight incubation at 37 °C, the number of plaques formed by phages in each plate was counted and the phage titers were calculated by C = \(\frac{n}{d * v (ml)}\), whereas C was the phage titre, n was the number of plaques, d and v represented the dilution factor and the volume of dilution added.
Host range, specificity, and efficiency of plating (EOP)
To determine phage host range and host specificity, bacteriolytic activities of these newly isolated phages (n = 12) were tested against XDR K. pneumoniae clinical isolates having carbapenemase and different mechanisms of colistin resistance (n = 32) and other pathogenic bacteria (n = 6), by using standard spot assay as previously published27,55. Briefly, 200 μl of exponential culture of tested bacteria (108 CFU/ml) was mixed with 3 ml of top agar. This mixture was then overlaid onto TSA plates. After cooling at room temperature for 5 min, 10 μl of purified phages (109 PFU/ml) were spotted onto bacterial lawn of the double-layer plates, each carrying a different test bacterial strain. The plates were incubated at 37 °C overnight to observe for the presence of clear lytic plaques which indicated the susceptibility of this bacterial strain to the tested phage. This experiment was repeated triplicates. Among 12 phages, the most potent phages -vB_kpnM_05 and vB_kpnP_08 exhibiting broad-host range were selected for further analysis.
To further quantify their bacteriolytic efficiencies, the efficiency of plating (EOP) was determined as previously described27,55,56, exponential cultures of host bacteria-kp924 and kp291 and test isolates (108 CFU/ml) were mixed with each tested phage (109–105 PFU/ml) and the plaque-forming abilities (PFU/ml) of vB_kpnM_05 and vB_kpnP_08 in both test isolates and its hosts were evaluated by double-layer agar method. The EOP was calculated by the ratio of average PFU/ml of phage on the test isolate to average PFU/ml of the same phage on its host, and interpreted as high (EOP ≥ 0.5), medium (0.1 ≤ EOP < 0.5), and low (0.001 ≤ EOP < 0.1)56.
Transmission electron microscopy (TEM)
Morphological characterization of vB_kpnM_05 and vB_kpnP_08 was conducted by spotting 5 µl of phages (109 PFU/ml) onto a copper grid and negatively stained with 2.5% (v/v) uranyl acid for 30 s, followed by visualization under TEM (JEM-1400, JEOL, Japan) at 200 kV57. The sizes of vB_kpnM_05 and vB_kpnP_08 were measured and analyzed using ImageJ57.
Optimal multiplicity of infection (MOI)
MOI is the ratio of infecting phages to bacterial hosts, and the MOI producing the highest phage titre was defined as the optimal MOI27,55. To determine the optimal MOI, the exponentially grown hosts and phages (kp924 for vB_kpnM_05, and kp291 for vB_kpnP_08) were incubated at different MOIs (0.01–100) at 37 °C with shaking (150 rpm) for 4 h, followed by measuring phage titre (PFU/ml) using double-layer agar method to determine the optimal MOI of each phage27,55. The experiments were conducted three times.
Phage adsorption assay
To determine phage adsorption, exponentially grown hosts and phages were mixed at their optimal MOIs (kp924 and vB_kpnM_05 at MOI-0.01, kp291 and vB_kpnP_08 at MOI-0.1) and incubated at 37 °C without shaking for a total of 25 min. The samples (100 μl) were collected every 5 min and diluted with TSB (0.9 ml), followed by centrifugation at 13,000×g for 30 s to remove the bacteria and adsorbed phages. The supernatants were collected and the titre of unabsorbed free phages in filtered supernatant were determined by double-layer agar method. The experiments were conducted three times independently27,55. The adsorption rate constant, k (ml/min), was then calculated by, k= \(\frac{2.3}{ {B}_{t}}\) log \(\frac{\begin{array}{c}{P}_{0}\\ \end{array}}{P}\), whereas P was the number of unabsorbed phages, P0 was the initial number of phages, B was the bacterial titer, and t was the time after infection58.
One-step growth curve
As previously published27,55, exponentially grown hosts (kp924, kp291) firstly centrifuged at 6000×g for 20 min at 4 °C. The resulting bacterial pellet was then suspended in TSB and then infected with phages at their optimal MOIs (MOI-0.01 for vB_kpnM_05, MOI-0.1 for vB_kpnP_08). Both phages were allowed to adsorb their hosts by incubation at 37 °C with shaking (150 rpm) for 15 min. These mixtures were then centrifuged at 13,000×g for 10 min to remove the unabsorbed phages. After the supernatants were discarded, and the resulting pellets containing the mixture of host and phage were resuspended in TSB and incubated at 37 °C with shaking (150 rpm). Triplicate samples from each phage were collected every 5 min until 90 min, and double-layer agar method was used to determine phage titre for generation of one-step growth curve and determination of latent period and burst size of each phage. The experiments were conducted three times independently. Latent period was the time between phage adsorption to host and the release of phage progeny from the host cell lysis. Burst size is the number of phage progeny released from its host, and it was calculated as the ratio of phages released at plateau level to their initial number during latent period27,55.
Stability of phages under different environmental conditions
To determine thermostability and pH stability as previously published27,55, phages (vB_kpnM_05, vB_kpnP_08) were exposed to different temperatures (− 20, 4, 25, 37, 40, 60 and 80 °C) and pH (2, 4, 6, 7, 8, 10, 12 and 14) for 180 min, and their abilities to form lytic plaques were assessed by double-layer agar method. Stabilities under UV radiation was assessed by exposing phage suspensions to UV lamp at an intensity of 1.722 ± 0.112 w/cm2, followed by taking samples every 10 min for 80 min to determine the presence of lytic plaque by double-layer agar method27,55. For assessing stabilities for long-term storage, phages stored at 4 °C were tested for their lytic activities monthly for 6 months by double-layer agar method27,55. To evaluate phage stability during chemical exposure, both phages treated with chloroform (10%, 20%, 40%), and ethanol (70%, 99%) for 180 min were assessed for the presence of lytic plaques using double-layer agar method27,55. All experiments were conducted triplicates independently.
Molecular analysis of phages
After treating purified phage lysates (500 µl, 1012 PFU/ml) with 1 µL DNase I (1 U/µl), and 1 µl RNase A (10 mg/ml) (Thermo Scientific) at 37 °C for 1.5 h, phage genomic DNAs were extracted using the QIAGEN® Blood & Tissue Kit (QIAGEN, Germany)59. Phage DNAs (10 ng) were subjected to RAPD-PCR for genetic diversity assessment using primers—RAPD, RAPD-P1 and RAPD-P2 as previously published16,60. Phages’ DNAs (0.5 μg) were also subjected to restriction digestions using enzymes (NcoI, HindIII, EcoRV, EcoRI) as per manufacturer’s protocol (Thermo Scientific). The digested products were separated with 1% agarose gel electrophoresis at 80 V for 45 min and visualized under gel documentation system (BioRad). The phage genomes were sequenced using an Illumina HiSeq2500 and assembled into circular contigs using Unicycler v0.5.061,62,63. Both phage genomes were annotated and visualized using Pharokka v1.7.164. Open reading frames (ORFs) were predicted using GeneMark (http://topaz.gatech.edu/GeneMark/), and the functions of the proteins encoded by the ORFs were predicted using BLASTp (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins). Glimmer 3.02 was used for gene prediction. The virulence factor database (VFDB), the Comprehensive Antibiotic Research Database (CARD) and NCBI were used to search and analyze virulence, antibiotic resistance and lysogeny genes. The nucleotide sequences of both phages were deposited in European Nucleotide Archive (https://www.ebi.ac.uk/) (Accession number: PRJEB75204). A phylogenetic tree for each phage was constructed based on the comparison of whole genome sequences using the Molecular Evolutionary Genetic Analysis software (version 7.0).
Bacteriolytic efficacies of phages as monophage or cocktail
In vitro bacteriolytic efficacy of phages as monophage versus phage cocktail were tested as previously described27,55. Briefly, the phage cocktail was prepared by combining equal concentrations and volumes of phages. The exponential culture of test isolates (108 CFU/ml) was mixed with monophage (1010–107 PFU/ml, MOI-100-0.01) or phage cocktail (1010 PFU/ml each, MOI-10) at 37 °C for 24 h, followed by monitoring bacterial growth by measuring OD600 every hour using a spectrophotometer.
Evaluating synergistic efficacy of vB_kpnM_05 and vB_kpnP_08 as monophage or phage cocktail in combination with different antibiotics in vitro
By checkerboard screening assay, the synergistic activities of vB_kpnM_05 and vB_kpnP_08 as either monophage or phage cocktail in combination with antibiotics having different antibacterial mechanisms (colistin, imipenem, amikacin, ciprofloxacin, ceftazidime), were firstly screened as previously published52,65. Briefly, 50 µl of antibiotics were serially diluted (2–2048 µg/ml) in checkerboard test plates. Subsequently, 50 µl of vB_kpnM_05 and vB_kpnP_08, at different MOIs (0.001–1000), were added to their respective plates, as either monophage or phage cocktail. The plates were inoculated with 100 µl of test isolates (105 CFU/ml) and incubated at 37 °C. The antibacterial activities of antibiotics alone, monophage or phage cocktail alone, and untreated bacterial culture as control were assessed in parallel. Five representative clinical isolates of XDR K. pneumoniae, each displaying different carbapenemase and distinct mechanisms colistin resistance, were subjected to checkerboard screening assay in vitro. The synergistic activities were then interpreted by fractional inhibitory concentration index (FICI)—Synergy:FICI ≤ 0.5 and No interaction:0.5 < FICI ≤ 466. The checkerboard screening assay revealed that phage cocktail (MOI-10) in combination with amikacin (8 µg/ml) exhibited significant synergy (FICI < 0.5) across all 5 representative clinical isolates of XDR K. pneumoniae with varying levels of amikacin susceptibility, including intermediate susceptibility and resistance (32–128 µg/ml) (Supplementary Fig. 3–5). To further prevent the occurrence of unfavorable effects and development of resistance due to the selection pressure superimposed by these antimicrobials, we selected the subinhibitory concentrations of both phage cocktail (MOI-10) and amikacin (8 µg/ml), which demonstrated potent synergy in checkerboard assay, for subsequent experiments.
Using a time-kill assay, we then confirmed the synergistic activity of a phage cocktail-amikacin combination in these 5 representative K. pneumoniae clinical isolates with varying levels of amikacin susceptibility, including intermediate susceptibility and resistance (32–128 µg/ml), as previously described52,65. Briefly, after adding amikacin (8 µg/ml) and phage cocktail (MOI-10) as monotherapies and combination therapy, test isolates (105 CFU/ml) were inoculated and incubated at 37 °C. Bacterial growth (OD600) and viabilities (CFU/ml) were measured at different timepoints (2, 4, 6, 8, 12, 24 h). The significant synergistic activities were interpreted with ≥ 2log10-fold decrease in combination as compared to monotherapies52,65.
Animal study
Animal studies were performed using 6–8-week-old C57BL/6 mice purchased from Nomura Siam International (Pathumwan, Bangkok, Thailand) and carried out in compliance with ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines10,67,68,69.
Safety and stability of phage cocktail
Prior to assessing the effect of phage cocktail in vivo, its safety was firstly evaluated by administering phage cocktail (100 μl, 109 PFU/ml per mouse) intraperitoneally to healthy mice (n = 5), and animals were observed for a month for toxicities, disease development or death, as previously published16. After intraperitoneal (IP) administration of phage cocktail, its titre in murine blood (PFU/ml) at different timepoints (0, 2, 4, 6, 8, 12, 24 h) was investigated by double-layer agar method to assess its stability in vivo16.
Lethal dose100 (LD100)
Following a previously established murine bacteremia model via IP inoculation52,70, we induced bacteremia in vivo using a clinical isolate of XDR K. pneumoniae obtained from the blood of a bacteremia patient. This clinical strain exhibited carbapenemase-NDM, IS1-integrated mgrB, and resistance to amikacin. Mice were inoculated intraperitoneally with bacterial suspensions at different doses (1 × 106, 1 × 107, 1 × 108 CFU/ml), (n = 5 for each dose). The survival of the animals was monitored to determine LD100, the infective dose that leads to 100% mortality from bacteremia. The LD100 obtained was subsequently utilized to induce murine bacteremia in our experiments. The bacteria recovered from our animal studies was assessed for the presence of carbapenemase-NDM, IS1-integrated mgrB, and amikacin resistance through PCR and broth microdilution. To evaluate the progression of bacteremia at different hour-post-infection (hpi) following bacterial challenge with LD100, bacterial burden (CFU/ml) in murine blood and internal organs (kidney, liver, spleen) was quantified by serial plating at different timepoints (2, 8, 24hpi), as previously published52,70.
Therapeutic efficacy of phage cocktail-amikacin combination in murine bacteraemia
The therapeutic efficacy of phage cocktail-amikacin combination in vivo was evaluated as previously published52,70. Briefly, mice were challenged with LD100 intraperitoneally, and bacteraemia was allowed to establish for 2 h. A single dose of either PBS (control), phage cocktail (MOI-10, 109 PFU/ml), amikacin (15 mg/kg/day), or phage cocktail-amikacin combination (MOI-10, 109 PFU/ml + 15 mg/kg/day) was administered intraperitoneally across four distinct treatment groups, each comprising five animals. All mice were euthanized at 14hpi to evaluate bacterial burden (CFU/ml) and phage titre (PFU/ml) in their blood and internal organs (kidney, liver, spleen) through serial plating and the double-layer agar method52,70. The same treatments were given once daily at various time points following bacterial challenge (2, 8, 24 hpi), and murine survival was monitored until either a clinical endpoint or experimental endpoint was reached, as published previously52,70. The clinical endpoint was determined using a five-point body condition score including weight loss, decreased body temperature, respiratory distress, hampered mobility, and hunched posture. The experimental endpoint was defined as 10 days post-infection for mice the did not reached the clinical endpoint45,52,70.
Prophylactic efficacy of phage cocktail-amikacin combination in murine bacteraemia
The prophylactic efficacies of IP phage cocktail and amikacin, as monotherapy or combination therapy in murine bacteraemia, were further evaluated by giving the same treatments as above to mice (n = 5 in each treatment group) at 24 h prior to bacteria challenge with LD100. All mice were euthanized at 14hpi to determine bacterial burden (CFU/ml) and phage titre (PFU/ml) in their blood and internal organs (kidney, liver, spleen) using serial plating and the double-layer agar method. Using the same treatments, murine survival was evaluated until clinical endpoint or experimental endpoint was reached45,52,70.
Statistical analysis
All statistical analysis was conducted using GraphPad Prism (9.3.1). Data were compared by two-tailed Student’s t-test, Mann–Whitney’s U test or Kaplan–Meier test. Statistical significance was accepted at p < 0.05.
Ethics statement
The study protocol was approved by Institutional Review Board (IRB) of the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand (IRB No.2865/65), and experiments were conducted in compliance with national and international ethical guidelines for human research as specified in the Declaration of Helsinki (1964) and its contemporary (2013) amendments, and comparable ethical standards including The Belmont Report, Council for International Organizations of Medical Sciences (CIOMS) Guidelines, and International Conference on Harmonization in Good Clinical Practice (ICH-GCP). For this retrospective study of anonymized clinical isolates, the requirement for informed consent from patients was waived by the IRB.
All protocols involving animals (C57BL/6 mice) conformed to the revised guidelines of the U.S. Institute of Laboratory Animal Resources, Commission on Life Sciences, National Research Council Guide for the Care and Use of Laboratory Animals, Washington, DC: National Academy Press; 1996, and the Animals for Scientific Purposes Act, 2015 (BE 2558), and experiments were performed following the ethical standards as laid down in the Basel Declaration on the use of animals in research. The Institutional Animal Care and Use Committee (IACUC) of the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand, approved the protocol (Certificate No.018/2566, Research Project No.001/2566) and protocols performed by operators licensed by the Thai Institute for Animals for Scientific Purpose Development and National Research Council of Thailand.
Inclusion and diversity
We, the authors of this paper, embrace inclusive, diverse, and equitable conduct of research. Our team comprises individuals who self-identify as underrepresented ethnic minorities, gender minorities, members of the LGBTQIA + community, and individuals living with disabilities. We actively promote gender balance in our reference list while maintaining scientific relevance.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its additional information. Genomic data were deposited to European Nucleotide Archive: PRJEB75204 (https://www.ebi.ac.uk/ena/browser/view/PRJEB75204).
References
Li, D. et al. Klebsiella pneumoniae bacteremia mortality: A systematic review and meta-analysis. Front. Cell. Infect. Microbiol. 13, 469 (2023).
Chirabhundhu, N. et al. Occurrence and mechanisms of tigecycline resistance in carbapenem- and colistin-resistant Klebsiella pneumoniae in Thailand. Sci. Rep. 14, 5215. https://doi.org/10.1038/s41598-024-55705-2 (2024).
Devanga Ragupathi, N. K. et al. Evaluation of mrkD, pgaC and wcaJ as biomarkers for rapid identification of K. pneumoniae biofilm infections from endotracheal aspirates and bronchoalveolar lavage. Sci. Rep. 14, 23572. https://doi.org/10.1038/s41598-024-69232-7 (2024).
Arena, F. et al. Resistance and virulence features of hypermucoviscous Klebsiella pneumoniae from bloodstream infections: Results of a nationwide Italian surveillance study. Front. Microbiol. 13, 983294 (2022).
Rocha, V. F. D. et al. Prolonged outbreak of carbapenem and colistin-resistant Klebsiella pneumoniae at a large tertiary hospital in Brazil. Front. Microbiol. 13, 831770 (2022).
Petrosillo, N., Taglietti, F. & Granata, G. Treatment options for colistin resistant Klebsiella pneumoniae: Present and future. J. Clin. Med. 8, 934 (2019).
Hongsing, P., Kongart, C., Nuiden, N., Wannigama, D. L. & Phairoh, K. Quantitative analysis of Swertiamarin content from Fagraea fragrans leaf extract using HPLC technique and its correlation to antibacterial activity. J. Curr. Sci. Technol. (2024). https://doi.org/10.59796/jcst.V14N2.2024.34
Na, S., Wannigama, D. L. & Saethang, T. Antimicrobial peptides recognition using weighted physicochemical property encoding. J. Bioinform. Comput. Biol. 21, 2350006. https://doi.org/10.1142/S0219720023500063 (2023).
Lin, D. M., Koskella, B. & Lin, H. C. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Therap. 8, 162 (2017).
Sutnu, N. et al. Bacteriophages isolated from mouse feces attenuates pneumonia mice caused by Pseudomonas aeruginosa. PLoS ONE 19, e0307079. https://doi.org/10.1371/journal.pone.0307079 (2024).
Sithu Shein, A. M. et al. Phage therapy could be key to conquering persistent bacterial lung infections in children. npj Antimicrob. Resist. 2, 31. https://doi.org/10.1038/s44259-024-00045-4 (2024).
Cui, L. et al. A comprehensive review on phage therapy and phage-based drug development. Antibiotics 13 (2024).
Gan, L. et al. Bacteriophage effectively rescues pneumonia caused by prevalent multidrug-resistant Klebsiella pneumoniae in the early stage. Microbiol. Spectrum 10, e02358-e12322 (2022).
Concha-Eloko, R., Barberán-Martínez, P., Sanjuán, R. & Domingo-Calap, P. Broad-range capsule-dependent lytic Sugarlandvirus against Klebsiella sp. Microbiol. Spectrum 11, e04298-e14222 (2023).
Chan, B. K., Abedon, S. T. & Loc-Carrillo, C. Phage cocktails and the future of phage therapy. Future Microbiol. 8, 769–783 (2013).
Singh, A. et al. Evaluation of bacteriophage cocktail on Septicemia caused by colistin-resistant Klebsiella pneumoniae in mice model. Front. Pharmacol. 13, 778676 (2022).
Ichikawa, M. et al. Bacteriophage therapy against pathological Klebsiella pneumoniae ameliorates the course of primary sclerosing cholangitis. Nat. Commun. 14, 3261 (2023).
Yang, Y. et al. Development of a bacteriophage cocktail to constrain the emergence of phage-resistant Pseudomonas aeruginosa. Front. Microbiol. 11, 327 (2020).
Leptihn, S. & Loh, B. Vol. 17 643–646 (Future Medicine, 2022).
Li, X. et al. A combination therapy of phages and antibiotics: Two is better than one. Int. J. Biol. Sci. 17, 3573 (2021).
Duc, H. M. et al. The use of phage cocktail and various antibacterial agents in combination to prevent the emergence of phage resistance. Antibiotics 12, 1077 (2023).
Malik, S., Nehra, K. & Rana, J. Bacteriophage cocktail and phage antibiotic synergism as promising alternatives to conventional antibiotics for the control of multi-drug-resistant uropathogenic Escherichia coli. Virus Res. 302, 198496 (2021).
Nir-Paz, R. et al. Successful treatment of antibiotic-resistant, poly-microbial bone infection with bacteriophages and antibiotics combination. Clin. Infect. Dis. 69, 2015–2018 (2019).
Law, N. et al. Successful adjunctive use of bacteriophage therapy for treatment of multidrug-resistant Pseudomonas aeruginosa infection in a cystic fibrosis patient. Infection 47, 665–668 (2019).
Rakhuba, D., Kolomiets, E., Dey, E. S. & Novik, G. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol. J. Microbiol. 59, 145 (2010).
Kim, Y. et al. Characterization of Klebsiella pneumoniae bacteriophages, KP1 and KP12, with deep learning-based structure prediction. Front. Microbiol. 13, 5289 (2023).
Chen, C. et al. Isolation and characterization of novel bacteriophage vB_KpP_HS106 for Klebsiella pneumonia K2 and applications in foods. Front. Microbiol. 14 (2023).
Herridge, W. P., Shibu, P., O’Shea, J., Brook, T. C. & Hoyles, L. Bacteriophages of Klebsiella spp., their diversity and potential therapeutic uses. J. Med. Microbiol. 69, 176 (2020).
Jokar, J. et al. Antibacterial effects of single phage and phage cocktail against multidrug-resistant Klebsiella pneumoniae isolated from diabetic foot ulcer. Virus Genes 59, 1–8 (2023).
Fang, Q., Feng, Y., McNally, A. & Zong, Z. Characterization of phage resistance and phages capable of intestinal decolonization of carbapenem-resistant Klebsiella pneumoniae in mice. Commun. Biol. 5, 48 (2022).
Oechslin, F. Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses 10, 351 (2018).
Hesse, S. et al. Phage resistance in multidrug-resistant Klebsiella pneumoniae ST258 evolves via diverse mutations that culminate in impaired adsorption. MBio 11, 02530–02519 (2020). https://doi.org/10.1128/mbio.
McCallin, S. & Oechslin, F. Bacterial resistance to phage and its impact on clinical therapy. Phage therapy: A practical approach, 59–88 (2019).
Labrie, S. J., Samson, J. E. & Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327 (2010).
Coulter, L. B., McLean, R. J., Rohde, R. E. & Aron, G. M. Effect of bacteriophage infection in combination with tobramycin on the emergence of resistance in Escherichia coli and Pseudomonas aeruginosa biofilms. Viruses 6, 3778–3786 (2014).
Kalapala, Y. C., Sharma, P. R. & Agarwal, R. Antimycobacterial potential of mycobacteriophage under disease-mimicking conditions. Front. Microbiol. 11, 583661 (2020).
Kato, H. et al. Evaluation of amikacin pharmacokinetics and pharmacodynamics for optimal initial dosing regimen. Drugs R&D 17, 177–187 (2017).
Kohanski, M. A., Dwyer, D. J., Hayete, B., Lawrence, C. A. & Collins, J. J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130, 797–810 (2007).
Mangalea, M. R. & Duerkop, B. A. Fitness trade-offs resulting from bacteriophage resistance potentiate synergistic antibacterial strategies. Infect. Immunity 88, 00926–00919 (2020). https://doi.org/10.1128/iai.
Gao, D. et al. Fitness trade-offs in phage cocktail-resistant Salmonella enterica Serovar enteritidis results in increased antibiotic susceptibility and reduced virulence. Microbiol. Spectrum 10, e02914-02922 (2022).
Chan, B. K. et al. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 6, 26717 (2016).
Lee, W.-H. et al. Vaccination with Klebsiella pneumoniae-derived extracellular vesicles protects against bacteria-induced lethality via both humoral and cellular immunity. Exp. Mol. Med. 47, e183–e183 (2015).
Altamirano, F. L. G. et al. Phage-antibiotic combination is a superior treatment against Acinetobacter baumannii in a preclinical study. EBioMedicine 80 (2022).
Wang, Z. et al. Combination therapy of phage vB_KpnM_P-KP2 and gentamicin combats acute pneumonia caused by K47 serotype Klebsiella pneumoniae. Front. Microbiol. 12, 674068 (2021).
Hesse, S. et al. Bacteriophage treatment rescues mice infected with multidrug-resistant Klebsiella pneumoniae ST258. MBio 12, 00034–00021 (2021). https://doi.org/10.1128/mbio.
Rhodes, A. et al. Surviving sepsis campaign: International guidelines for management of sepsis and septic shock: 2016. Intensive Care Med. 43, 304–377 (2017).
Kumar, A. Early antimicrobial therapy in severe sepsis and septic shock. Curr. Infect. Dis. Rep. 12, 336–344 (2010).
Morquin, D. et al. Time is of the essence: Achieving prompt and effective antimicrobial therapy of bloodstream infection with advanced hospital information systems. Clin. Infect. Dis. 78, 1434–1442 (2024).
Eskenazi, A. et al. Combination of pre-adapted bacteriophage therapy and antibiotics for treatment of fracture-related infection due to pandrug-resistant Klebsiella pneumoniae. Nat. Commun. 13, 302 (2022).
Cano, E. J. et al. Phage therapy for limb-threatening prosthetic knee Klebsiella pneumoniae infection: Case report and in vitro characterization of anti-biofilm activity. Clin. Infect. Dis. 73, e144–e151 (2021).
Liu, D. et al. The safety and toxicity of phage therapy: A review of animal and clinical studies. Viruses 13, 1268 (2021).
Shein, A. M. S. et al. High prevalence of mgrB-mediated colistin resistance among carbapenem-resistant Klebsiella pneumoniae is associated with biofilm formation, and can be overcome by colistin-EDTA combination therapy. Sci. Rep. 12, 12939 (2022).
Brisse, S. et al. wzi Gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J. Clin. Microbiol. 51, 4073–4078 (2013).
Peng, Q. et al. Characterization of bacteriophage vB_KleM_KB2 possessing high control ability to pathogenic Klebsiella pneumoniae. Sci. Rep. 13, 9815 (2023).
Cao, Y., Zhang, Y., Lan, W. & Sun, X. Characterization of vB_VpaP_MGD2, a newly isolated bacteriophage with biocontrol potential against multidrug-resistant Vibrio parahaemolyticus. Arch. Virol. 166, 413–426 (2021).
Khan Mirzaei, M. & Nilsson, A. S. Isolation of phages for phage therapy: A comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE 10, e0118557 (2015).
Cao, B., Xu, H. & Mao, C. Transmission electron microscopy as a tool to image bioinorganic nanohybrids: The case of phage-gold nanocomposites. Microscopy Res. Tech. 74, 627–635 (2011).
Vandersteegen, K. et al. Romulus and Remus, two phage isolates representing a distinct clade within the Twortlikevirus genus, display suitable properties for phage therapy applications. J. Virol. 87, 3237–3247 (2013).
Jakočiūnė, D. & Moodley, A. A rapid bacteriophage DNA extraction method. Methods Protocols 1, 27 (2018).
Gutiérrez, D. et al. Typing of bacteriophages by randomly amplified polymorphic DNA (RAPD)-PCR to assess genetic diversity. FEMS Microbiol. Lett. 322, 90–97 (2011).
Bankevich, A. et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595. https://doi.org/10.1371/journal.pcbi.1005595 (2017).
Keeratikunakorn, K. et al. First detection of multidrug-resistant and toxigenic Pasteurella aerogenes in sow vaginal discharge: a novel threat to swine health in Thailand. Sci. Rep. 14, 25510. https://doi.org/10.1038/s41598-024-76428-4 (2024).
Bouras, G. et al. Pharokka: a fast scalable bacteriophage annotation tool. Bioinformatics https://doi.org/10.1093/bioinformatics/btac776 (2023).
Gu Liu, C. et al. Phage-antibiotic synergy is driven by a unique combination of antibacterial mechanism of action and stoichiometry. MBio 11, 01462–01420 (2020). https://doi.org/10.1128/mbio.
Odds, F. C. Vol. 52 1–1 (Oxford University Press, 2003).
Percie du Sert, N. et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. J. Cereb. Blood Flow Metab. 40, 1769–1777 (2020).
Pinitchun, C. et al. Aging-induced dysbiosis worsens sepsis severity but is attenuated by probiotics in D-galactose-administered mice with cecal ligation and puncture model. PLOS ONE 19, e0311774. https://doi.org/10.1371/journal.pone.0311774 (2024).
Hongsing, P. et al. Cannabidiol demonstrates remarkable efficacy in treating multidrug-resistant enterococcus Faecalis infections in vitro and in vivo. Trends Sci. 21, Accepted Manuscript (2024). https://doi.org/10.48048/tis.2024.8150
MacNair, C. R. et al. Overcoming mcr-1 mediated colistin resistance with colistin in combination with other antibiotics. Nat. Commun. 9, 458 (2018).
Acknowledgements
We thank the staffs of the bacteriology division, Department of Microbiology at King Chulalongkorn Memorial Hospital, for providing the clinical K. pneumoniae isolates. The computing facilities was supported by Mahidol University and the Office of the Ministry of Higher Education, Science, Research, and Innovation under the Reinventing University project: the Center of Excellence in AI-Based Medical Diagnosis (AI-MD) sub-project, NSTDA Supercomputer center (ThaiSC), and the National e-Science Infrastructure Consortium.
Funding
This Research is funded by Thailand Science Research and Innovation Fund Chulalongkorn University (HEA663000020). Aye Mya Sithu Shein was supported by Chulalongkorn University (Second Century Fund-C2F Fellowship). Dhammika Leshan Wannigama was supported by Chulalongkorn University (Second Century Fund-C2F Fellowship), the University of Western Australia (Overseas Research Experience Fellowship), and Yamagata Prefectural Central Hospital, Yamagata, Japan (Clinical Residency Fellowship). T.W. and P.J. were supported by National Research Council of Thailand [Grant No. N42A660897]. The sponsor(s) had no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
Author information
Authors and Affiliations
Contributions
A.M.S.S., D.L.W., T.C., and S.A. conceived the idea, supervised the investigation, acquired funding, and contributed to data curation, formal analysis, and writing the original draft. C.H., T.W., P.J., P.G.H., and P.H. contributed equally to this work, with formal analysis, supervision, methodology, validation, and manuscript editing. T.W., P.J., T.S., and V.N.B. conducted bioinformatics and biochemicals analysis, methodology, and validation. N.C., S.L., S.N., U.R., S.S., N.K., and M.L. contributed to data acquisition, curation, microscopy, and bacteria identification. P.N.M., M.A., W.G.F.D., P.O., S.M.A.H.R., N.N., A.T., H.I., R.J.S., A.L., N.K.D.R., A.Y.C., T.K., D.P., K.M., and L.C. provided critical review and manuscript editing. S.W. and A.R. provided technical support. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Informed consent
For this retrospective study of anonymous clinical isolates, the requirement for informed consent from patients was waived by the Institutional Review Board (IRB) of the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand (IRB No.2865/65).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shein, A.M.S., Wannigama, D.L., Hurst, C. et al. Phage cocktail amikacin combination as a potential therapy for bacteremia associated with carbapenemase producing colistin resistant Klebsiella pneumoniae. Sci Rep 14, 28992 (2024). https://doi.org/10.1038/s41598-024-79924-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-79924-9
Keywords
This article is cited by
-
Therapeutic application of a jumbo bacteriophage against metallo-β-lactamase producing Pseudomonas aeruginosa clinical isolates
Journal of Biomedical Science (2025)
-
Augmenting phage therapy using green nanotechnology for promising infection control, wound healing and devoiding phage resistance in MDR Pseudomonas aeruginosa
Scientific Reports (2025)
-
Healthcare-Associated Infections: The Role of Microbial and Environmental Factors in Infection Control—A Narrative Review
Infectious Diseases and Therapy (2025)
-
tesG expression as a potential clinical biomarker for chronic Pseudomonas aeruginosa pulmonary biofilm infections
BMC Medicine (2025)