Abstract
Accurate diagnosis of oral lesions, early indicators of oral cancer, is a complex clinical challenge. Recent advances in deep learning have demonstrated potential in supporting clinical decisions. This paper introduces a deep learning model for classifying oral lesions, focusing on accuracy, interpretability, and reducing dataset bias. The model integrates three components: (i) a Classification Stream, utilizing a CNN to categorize images into 16 lesion types (baseline model), (ii) a Guidance Stream, which aligns class activation maps with clinically relevant areas using ground truth segmentation masks (GAIN model), and (iii) an Anatomical Site Prediction Stream, improving interpretability by predicting lesion location (GAIN+ASP model). The development dataset comprised 2765 intra-oral digital images of 16 lesion types from 1079 patients seen at an oral pathology clinic between 1999 and 2021. The GAIN model demonstrated a 7.2% relative improvement in accuracy over the baseline for 16-class classification, with superior class-specific balanced accuracy and AUC scores. Additionally, the GAIN model enhanced lesion localization and improved the alignment between attention maps and ground truth. The proposed models also exhibited greater robustness against dataset bias, as shown in ablation studies.
Similar content being viewed by others
Introduction
More than 300,000 oral cancers (OC) are diagnosed annually worldwide.1 These cancers have a much lower probability of survival (\(<50\%\)) than common cancers (e.g., breast, prostate)2, and one of the highest morbidity and suicide rates of all cancers. Importantly, more than half of all patients present with an advanced-stage disease; this proportion has not decreased in the past 40 years3,4. An advanced stage at diagnosis is associated with a lower probability of survival, higher health care cost, and increased risk of significant impairment and deformity5,6. Hence, early diagnosis plays a key role in reducing the burden of this disease.
An abnormal change in the soft tissue of the oral cavity (oral lesion) can be one of the earliest signs of OC. Hence, early diagnosis of OC can be achieved by correctly situating any oral soft tissue lesion in the continuum from normal mucosa to malignancy. For example, screening for abnormal oral lesions, even based on visual examination alone, has been shown to significantly reduce the morbidity and mortality associated with OC in high-risk populations7,8,9,10. However, many general dentists do not appear to adopt a systematic approach for evaluating oral lesions11,12 and also report major challenges in diagnosing oral lesions12,13,14. For example, half of the general dentists surveyed in a study felt that their knowledge and training were not up to date in diagnosing oral lesions14. Furthermore, there is a wide variety of oral lesions with similar clinical appearances, and many systemic conditions have oral manifestations, which make them more difficult to diagnose13,15,16,17. These difficulties, combined with limited clinical training in oral pathology, lead to high diagnostic uncertainty and delay16,18,19.
Deep learning algorithms have demonstrated remarkable success in various image recognition tasks20. In the context of oral lesion classification, there have been several attempts to develop CNN-based models to aid in clinical decision-making21,22,23,24,25,26,27 Almost all of these previous attempts have been focused on the classification task of cancer vs no-cancer. This approach may miss a large group of potentially malignant oral lesions, where an early and accurate diagnosis and excision could prevent a malignant transformation. Furthermore, two key challenges need to be addressed to enhance the practicality and reliability of these models in clinical practice: interpretability and dataset bias28,29,30.
Interpretability refers to the ability to understand and explain the model’s decision-making process, enabling clinicians to trust and interpret the predictions from the model effectively. While CNNs have demonstrated impressive performance in various domains, they are often considered black-box models31, making it difficult to ascertain the underlying features and reasoning behind their predictions32. This lack of interpretability hinders their adoption in clinical settings, where explanations and insights are crucial for informed decision-making and building trust between the model and healthcare professionals33,34.
Furthermore, dataset bias, including oral lesion classification, is common in medical image analysis35. Biases can arise due to variations in imaging conditions, equipment, patient demographics, and lesion characteristics, among other factors28,36. These biases can introduce unwanted confounding factors and affect the generalization ability of deep learning models37. Consequently, the models may exhibit poor performance when presented with data that significantly deviates from the training distribution, thereby limiting their clinical utility and potentially leading to misdiagnosis or inadequate treatment plans38,39.
To address these challenges, we propose a deep learning model for oral lesion classification that emphasizes interpretability and robustness against dataset bias. Our method draws inspiration from the existing GAIN (Guided Attention Inference Network) framework, a weakly supervised semantic segmentation approach40. While GAIN has been previously proposed and utilized in other domains41,42, our contribution lies in its adaptation and application specifically for the domain of oral lesions classification task. Moreover, our approach also aims to address the issue of dataset bias43 by leveraging segmented masks to guide the attention of a convolutional neural network (CNN) during training. These masks highlight the relevant regions within oral cavity images, enabling the model to focus on the informative areas while suppressing the influence of irrelevant or confounding factors. By incorporating this guidance mechanism, our approach reduces the impact of variations in imaging conditions, patient demographics, and lesion characteristics that contribute to dataset bias44. Furthermore, we augment the network with the task of predicting the anatomical location of the lesion within the oral cavity image, considering eight potential anatomical locations. This augmentation serves a dual purpose: It enriches the model’s interpretability and capitalizes on the standard set of CNN features shared between the anatomical site prediction and lesion classification tasks. Our underlying hypothesis is rooted in the idea that capturing the inherent relationship between lesion types and their corresponding anatomical sites can yield performance improvements for the overall model, thereby amplifying its clinical utility45.
Here, we present a comprehensive evaluation of our proposed approach on a dataset of oral cavity images encompassing 16 different types of lesions. We compare the performance of our method with state-of-the-art approaches and demonstrate its effectiveness in achieving accurate and interpretable oral lesions classification. Additionally, we assess the robustness of our model to dataset bias by conducting experiments on diverse data subsets.
Results
Our approach of incorporating guided attention considerably improved the performance of the model compared to the standard fine-tuning of EfficientNet-B5 for the oral lesion classification task, with the added advantage of improved interpretability and robustness against dataset bias.
Oral lesion classification performance
The class-specific and overall performance metrics are presented in Table 1. The GAIN model showed 7.2% relative improvement in balanced accuracy, increasing from 73.4 to 78.7%. Additionally, the model showed an improved brier score and comparable AUC values. Although the GAIN+ASP model exhibited modest incremental performance gains compared to the GAIN model, it still surpassed the baseline performance by 2.8% relative improvement.
Class-specific metrics revealed that the GAIN model achieved clear improvements in balanced accuracy for all classes except two, compared to the baseline model. However, for those two classes the GAIN+ASP model showed comparable accuracy to the baseline model. Notably, adding anatomical site classification improved the discriminatory power of the model for the majority (11 out of 16) classes, with AUC values ranging from 83.7 to 99.7%. The GAIN model showed significant improvements in balanced accuracy for Squamous Cell Carcinoma (oral cancer) and Mucocele lesion classes, with 10% and 16% relative improvements over the baseline, respectively. Moreover, the GAIN and GAIN+ASP models improved balances accuracy and F1 scores when considering broader class labels based on prognosis of lesions (non-benign vs benign vs premalignant vs malignant), suggesting a improvement in potential clinical utility (Appendix Table A1).
Anatomical site classification performance
The GAIN+ASP model showed exceptional performance in the classification of anatomical sites, achieving an overall balanced accuracy of 91.6%. Site-specific accuracies ranged from 76.9 to 94.9%, as detailed in Table 1. Furthermore, the model showed strong discriminatory abilities between different anatomical sites, with AUC values spanning from 92.3 to 98.78%. The results underscore the model’s robustness and effectiveness in distinguishing between various anatomical locations.
Improvement in interpretability
The interpretability of our proposed methodology was evaluated by comparing the Intersection over Union (IoU), and Dice-Sørensen coefficient (DSC) between the baseline model and our methods, GAIN and GAIN+ASP. The IoU measures the overlap between the attention maps and ground truth annotations, indicating the alignment between the generated maps and the actual lesion regions. Our GAIN+ASP model showed a 6.5% relative improvement in IoU compared to the baseline model, suggesting improved agreement. Similarly, the DSC, which measures the similarity between the attention maps and annotations, showed a 5% relative enhancement with the GAIN+ASP method compared to the baseline. We note that these improvements were also observed in the GAIN model, which employs guided attention alone. Improving the overall agreement between the generated attention maps and ground truth segmentation maps may not fully capture the dimensions of performance gain. It is particularly important for the guidance approach to enhance the attention on those test images where the baseline model performs poorly. As shown in Tables 2 and 3 , our models, GAIN and GAIN+ASP, significantly improved the IoU for test images falling into the lowest quartile of attention scores in the baseline model, with relative improvements exceeding 100%. The gradient of relative improvement across quartiles at baseline may indicate the inherent performance limitations of the baseline model. However, given that our primary task is image-level classification rather than segmentation of oral lesions, the implications of these improvements in attention are only evaluated as a secondary outcome.
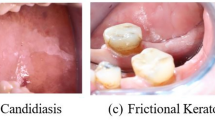
Furthermore, the visual comparison of attention maps between the baseline and GAIN, as shown in Fig. 1, reveals the improved alignment of attention with ground truth regions in our approach. The attention maps clearly indicate the regions of interest that the model focused on when making predictions. These regions aligned well with known characteristics and diagnostic indicators of the respective oral lesions, enabling clinicians to gain insights into the model’s decision-making process and identify key features contributing to oral lesion classification. This interpretability aspect is crucial for building trust and confidence in the model’s predictions, thereby making it more suitable for integration into clinical practice.
Mitigating dataset bias: prediction of implausible combinations (true zeros)
We quantified the number of implausible combinations of oral lesion class and anatomical site (based on ground truth labels) predicted by each model, as shown in Table 4. By definition, all implausible combination predictions are incorrect classifications. The GAIN model reduced the number of implausible predictions to 62, a significant improvement from the 109 seen in the baseline model, reflecting a 43% relative improvement. Whereas, the GAIN+ASP only reduced the number of implausible predictions marginally compared to the baseline. Albeit surprising, this increase in number of implausible predictions with the addition of anatomical site classification task to the GAIN model could be attributed to the shared parameters of the CNN feature extractor backbone and the approximately similar contributions of the loss functions \(L_cl\) and \(L_{as}\) as identified by the optimal hyper-parameter values of \(\alpha\) and \(\gamma\) in the combined loss function, \(L_{total}\) (see Appendix Tables A2 and A3).
Mitigating dataset bias: induced data zeros
To evaluate our models for robustness against dataset bias, we removed all images with ground truth diagnosis of Mucocele occurring at the outer lip site from the training set, effectively introducing Data Zero combination as described in methods section. We then evaluated the models’ ability to mitigate this bias by examining the number of correctly predicted Mucocele lesions at the outer lip site in the test dataset (Table 5). Our GAIN+ASP model showed significant improvement, correctly predicting 9 out of 15 images, compared to the baseline model’s 3 out of 15 correct predictions. Similarly, the GAIN model, without anatomical site prediction, also showed similar ability to mitigate dataset bias by correctly predicting 8 out of 15 images.
In summary, our experimental results validate the effectiveness of our proposed methodology for oral lesion classification. Our approach outperforms the baseline model in terms of accuracy, discrimination ability, and calibration. Additionally, the generated attention maps exhibit better alignment with the ground truth annotations, enhancing interpretability. These findings highlight the benefits of integrating additional guidance loss and anatomical site prediction in our methodology, resulting in improved performance, interpretability and robustness against dataset-bias compared to the baseline model.
Related work
Rokshad et al. recently reviewed the literature on previous attempts on oral lesion classification from intra-oral images using deep learning approaches27. Majority of primary studies identified in that review focused on binary classification task (e.g., suspicious vs non-suspicious46) or broad multi-class classification (e.g, ulcer vs papule vs macule23, normal vs benign vs premalignant vs malignant47). Although, these approaches might have some clinical value, fine-grained multi-class classification as clinical diagnosis level has higher clinical utility. In our work we have focused on predicting 16 histopatholgically confirmed diagnosis classes. Moreover, a key limitation of all most all of the previous work in this area is the lack of investigation of explainability methods.
Comparable to the work presented here, Figueroa et al. implemented a guided attention network, with minimal adaptation to the original GAIN approach, to classify intra-oral images into suspicious and non-suspicious lesions48. In addition to the difference in fundamental task of binary classification and use of human labelled ground truth, in this work, we introduced anatomical site classification task in addition to the diagnostic classification task. Our adaptation was particularly motivated to mitigate dataset bias, which the previous work did not explore.
Welikala et al. on the other had used a soft attention approach with the attention weights estimated from specific layers of the architecture rather than a shared parameter approach of GAIN and did not investigate the impact of their approach on dataset bias46. Moreover, both of these previous approaches reported only marginal improvement in classification accuracy, whereas our approach shows significant improvement in classification accuracy and explainability with the added advantage of mitigation of dataset bias.
Conclusion
In this paper, we propose a deep learning-based approach for oral lesion classification that emphasizes interpretability and tackles dataset bias. Our methodology, utilizing the Guided Attention Inference Network (GAIN) framework, generates attention maps to highlight important regions in oral cavity images and promote trust between the model and healthcare professionals. By incorporating guidance loss and pixel-level annotations, our approach mitigates dataset bias and enhances generalization. Experimental results demonstrate the effectiveness of our approach, outperforming baselines in accuracy and interpretability, with robustness to dataset bias.
Our research has significant implications for clinical practice, aiding clinicians in making informed decisions, fostering trust in automated systems, and improving patient outcomes. Future research can explore advanced interpretability techniques and incorporate multi-modal information to further improve accuracy and clinical utility. Overall, our work contributes to accurate and interpretable oral lesion classification, advancing patient care in oral healthcare.
Methods
Our method integrates three key components: (i) classification stream, (ii) guidance stream, and (iii) anatomical site prediction stream.
Classification stream
The classification stream is analogous to a typical multiclass classification using CNNs. In the classification stream, the network is trained using the oral cavity images, where each image is labeled with one of the 16 types of different lesions. During training, the network learns to extract relevant features from the input images and optimize its parameters to minimize the classification loss; typically computed using a suitable loss function, such as cross-entropy loss.
Guidance stream
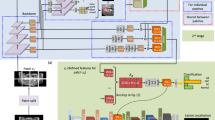
The guidance stream is introduced to provide additional guidance to the network about the classification task. This guidance stream, denoted as \(S_g\), utilizes external guidance, such as pixel-level segmentation masks, to guide the network’s attention more precisely. By leveraging pixel-level labels, we are able to provide detailed guidance to the network regarding the specific regions of interest in the oral cavity images. This stream operates in parallel with the Classification Stream and shares the same network parameters (Fig. 2). The first step in this stream is to generate the trainable attention maps to influence the network’s learning. The attention maps reflect the areas in the oral cavity images which contributes to the network’s final prediction. These attention maps can be utilized further during training in order to promote the network to focus it’s attention on region of interest in the image. By training the network to focus on the relevant regions, we aim to improve the overall classification performance and interpretability of the network’s decision-making process as well as mitigate the dataset bias for improved generalization.
To generate the attention maps, we leverage the intermediate feature maps obtained from the Classification Stream. These attention maps are generated using the fundamental framework of Grad-CAM (Gradient-weighted Class Activation Mapping)49. Given an input image, we obtain the activations of the units in a specific convolutional layer, denoted as \(f_{l, k}\), where l represents the layer index and k represents the unit index. For each class c corresponding to the ground-truth label, we calculate the gradient of the class score \({\hat{y}}^c\) with respect to the activations \(f_{l, k}\). These gradients are then subjected to global average pooling to derive the neuron importance weights \(w_{l, k}^{c}\).
where GAP\((\cdot )\) represents the global average pooling operation. These importance weights reflect the contribution of each activation map to the prediction of class c. We then apply a 2D convolution operation on the activations \(f_l\) using \(w_c\) as kernel weights, which effectively generates map with a combination of activations. Finally, the ReLU operation is applied to obtain the attention map \(A^c\) as shown in the equation.
The resulting attention map \(A^c\) represents the regions in the image that contribute most to the network’s prediction of class c. Our hypothesis is that the end-to-end trainable attention map \(A^c\) influences the network’s learning by effectively focusing on relevant regions, thereby making predictions more plausible within the area of interest. These attention maps, combined with pixel-level annotations, provide supplementary information that guides the network in accurately identifying and understanding the characteristics of oral lesions. The discrepancy between the attention maps and the provided pixel-level annotations is calculated by the guidance loss, denoted as \(L_g\). This loss encourages the attention maps to align with the precise regions of interest defined by the external supervision (e.g. pixel-level segmentation masks), leading to more accurate and detailed attention maps. We define the guidance loss \(L_g\) as:
where \(A^c\) is the generated attention map and the \(H^c\) represents the pixel-level mask highlighting the region of interest.
Anatomical site prediction stream
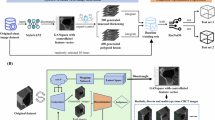
Expanding upon our approach, we introduce an auxiliary anatomical site prediction task in the pipeline. Illustrated in Fig. 3, this extension involves the integration of the anatomical site classification task alongside the existing lesion classification stream and the guidance stream with attention loss. Notably, all three streams utilize a common set of CNN features. However, a new MLP head is introduced specifically for the anatomical site prediction task. The neural network is tasked with predicting the precise anatomical location of the lesion, selecting from a set of eight potential locations. To achieve this, we employ a cross-entropy loss function. It is important to highlight that this multifaceted approach aims to maximize the benefits derived from shared features, thus enhancing the model’s performance that comprehensively addresses both lesion classification and anatomical site prediction tasks.
Our hypothesis posits a fundamental correlation between anatomical sites and lesion types (see the Dataset bias section below), asserting that the ability to learn these correlation structures may enhance the performance of the model in the lesion prediction task. This premise builds on the notion that recognizing the likelihood of certain lesions appearing in specific anatomical areas can improve the model’s ability to identify those lesions.
The model, by recognizing the anatomical context, gains a richer set of features that are inherently valuable for distinguishing between different types of lesions. This is not a unidirectional benefit; the insight gained from predicting the anatomical site feeds back into the lesion prediction task. Consequently, understanding where a lesion is likely to occur can provide additional learning signal to the network during training. This dual-task learning framework not only improves the model’s predictive accuracy but also its interpretability. When the model predicts a lesion, it also provides the anatomical context, offering a more comprehensive understanding of its prediction.
Combining the streams
We bring together the three streams by optimizing a finely-tuned loss function, denoted as \(L_{total}\). This function harmonizes the insights drawn from each stream by combining the classification loss (\(L_{cl}\)) with the guidance loss (\(L_g\)), and the anatomical site classification loss (\(L_{as}\)). This combined loss function is given as:
Here, \(\alpha\), \(\beta\), and \(\gamma\) serve as weighting parameters that adjust the contribution of each loss component to the total loss. By optimizing the combined loss function, the network learns to enhance its focus on input images, incorporating both the discriminative features captured by the classification stream and the refined attention maps from the guidance stream. This joint optimization process not only sharpens the model’s focus on relevant image regions but also equips it with an understanding of the intricacies involved in oral lesion identification, thereby reducing dataset bias and enhancing overall predictive performance. The detailed algorithm of our method is presented in the appendix and implementation in pytorch is provided https://gist.github.com/MadathilSA/48ca84378e393b7737311d18d1c0b5aa.
Dataset characteristics. (a) Illustrates the distribution of oral lesion classes, with geographic tongue being the most prevalent. (b) Presents the data zero and true zero combinations. The grey-shaded cells (data zeros) indicate plausible combinations for which no data is available in our dataset, the black-shaded cells (true zeros) represent implausible combinations, and the white cells represents plausible combinations where data is available.
Dataset and pre-processing
Data source
We established a retrospective cohort of patients who consulted with the oral pathologist in our team (PC) from 1999 to 2021. During this period, 2,765 images across 16 oral lesion classes were compiled from 1079 patients referred to the oral pathologist. Dental experts in the team, meticulously cleaned the dataset for quality by removing images with invisible lesions or multiple lesions in the same image, resulting in 1,888 typical images of 16 oral lesion types (Fig. 4a).
The images were collected by oral pathologists as part of the routine clinical examination of patients with oral lesions and were included in their electronic health records. The images were captured using digital cameras of varying quality under dental clinic lighting conditions. Ground truth labels for the oral lesion types were obtained from histopathology reports (the gold-standard diagnosis) for lesions requiring a biopsy (e.g., pre-malignant lesions) and from expert clinician diagnoses for lesions not requiring a biopsy. Two dentists annotated the oral lesions, generating the ground truth labels for the segmentation masks.
All steps of this research were conducted in accordance with the Declaration of Helsinki. No identifiable information or images are presented in this manuscripts. An informed consent was obtained from all participants during the data collection process. The project received approval from the Research Ethics Review Board at McGill University [Protocol no: A07-M40-21B].
Class imbalance
Class imbalance is a prevalent challenge in oral lesion classification tasks, where certain lesion types occur infrequently50. Figure 4a illustrates the distribution of oral lesion classes in our dataset. To address the impact of class imbalance, we employed class weighting during the training process51. Class weights were calculated as the inverse of the class frequencies in the training dataset. Lesion types with lower occurrences were assigned higher weights, while lesion types with higher occurrences received lower weights. This approach aimed to balance the influence of each lesion type during the training process, giving more emphasis to the underrepresented classes. By assigning appropriate class weights, we aimed to alleviate the adverse effects of class imbalance and improve the overall performance of our oral lesion classification model.
Dataset bias
The classification of oral lesions from intra-oral images presents unique challenges, notably due to the variability of lesion incidence based on anatomical sites within the oral cavity. Certain lesions are exclusive to specific sites, while others may never occur in those areas. For instance, Hairy Tongue is condition that only appears on the tongue’s surface and is implausible on other sites such as the buccal mucosa. We define such implausible combinations as True Zero combinations. The oral pathology expert in the team was consulted in creating the True Zero combination based on the expert evidence synthesis. On the other hand, oral squamous cell carcinoma (OSCC), a rare lesion, can theoretically occur at any anatomical site within the oral cavity. However, despite being plausible, there may be no training images available for this lesion type at certain locations. We refer to these plausible but absent combinations in the training data as Data Zero combinations. Figure 4b illustrates the True Zero and Data Zero combinations of lesions and anatomical sites pertinent to this study.
Additionally, the limited accessibility to certain anatomical sites (e.g., the palate) means that images of lesions in these areas can only be captured in specific ways. These factors contribute to a high correlation between anatomical sites and lesion types, leading to potential bias in CNN models. While datasets that are heterogeneous, exhaustive, and contain a sufficiently large number of images for each unique combination of anatomical site and oral lesion type may help mitigate this bias, real-world datasets often make it challenging for models to distinguish between Data Zero and True Zero combinations. Our group has previously developed a method to incorporate external knowledge of implausible combinations by augmenting the loss function of CNN models45. In contrast, this study explores an alternative approach, aiding the model in learning the correlation structure through guided attention to the lesion’s location and supervised learning of the lesion’s anatomical site.
Data leakage
A common challenge in intra-oral image classification tasks is data leakage, which occurs when the algorithm learns using information from the data that should not be utilized for the task at hand, such as the shape of teeth52. This issue is further exacerbated by the use of multiple angles of the same lesion, leading to an illusion of high performance. To avoid data leakage, we first created clusters of images representing the same oral lesion using the EXIF data, patient IDs, lesion types, and anatomical sites. These clusters were then randomly assigned to either training or test sets, resulting in a training set of 1,200 images and a test set of 688 images (Fig. 4a). This approach helps ensure that the model does not gain an unfair advantage by seeing the same lesion in both training and testing phases.
Data augmentation
To improve the diversity of our oral lesions dataset, we employed a range of data augmentation techniques. These included random rotations within a certain range, horizontal and vertical flips, random translations, and adjustments in brightness and contrast, all while maintaining the aspect ratio of the images. These transformations were meticulously designed to preserve the intrinsic characteristics of oral cavity images, ensuring that the shape and size of the lesions remained unaltered. This careful approach allowed us to create a more robust dataset, suitable for training our models.
Experimental and training setup
We conducted a comprehensive series of experiments to evaluate the effectiveness of our approach in three key areas: (i) accurately classifying oral lesions, (ii) improving interpretability through generated attention maps, and (iii) mitigating dataset bias.
Models
We employed a transfer learning approach with EfficientNet-B5 (EB5) model as the backbone architecture53. Initially, a baseline model was established by fine-tuning the EB5 model pre-trained on ImageNet-1K54, incorporating only the Classification stream. Building upon this, we introduced the guidance loss through the Guidance stream resulting in (GAIN) model. Subsequently, we added the anatomical site classification loss via the Anatomical site prediction stream, yielding the (GAIN+ASP) model. Except for the hyper-parameter tuning procedure detailed below, all other aspects of fine-tuning were kept consistent across the three model variants. By comparing the performance of our approach with the baseline model, we aim to demonstrate the additional benefits and improvements achieved by the integration of guidance stream, which trains the network’s attention to enhance interpretability and mitigate dataset bias. The inclusion of anatomical site prediction further enhances interpretability and reduces dataset bias.
Hyper-parameter optimization
Our method requires three additional hyper-parameters to be optimized for the combined loss function. We utilized Bayesian Optimization, implemented via BoTorch55 on the Adaptive Experimentation Platform56, to tune these parameters. Specifically, we kept the hyper-parameters for EfficientNet-B5 and AdamW Optimizer57 at their recommended settings and focused the optimization process on our method’s hyper-parameters. Details of the optimization procedure and the optimal hyper-parameters identified are provided in Appendix. We fine-tuned all model variants on our dataset, adjusting weights of all the layers to optimize performance using appropriate loss functions and the optimal parameters identified.
Metrics
Our choice of evaluation metrics was guided by the recent ‘Metrics Reloaded’ framework from58. This framework recommends a problem driven approach that enhances the user-centric application of the model. Based on to this framework, we have a combination of metrics for image-level classification (classifying the image into one of 16 classes) and object localization (network’s attention on the lesion area). We chose the following metrics for image-level classification: (i) per-class balanced accuracy (BA), (ii) per-class brier score (BS), and (iii) ROC-One-vs-Rest AUC (AUC). To assess the interpretability of the generated attention maps, we utilized recommended segmentation metrics: (i) Intersection over Union (IoU), and (ii) Dice-Sørensen coefficient (DSC). These metrics collectively ensure a comprehensive evaluation of our model’s performance and interpretability.
Mitigating dataset bias
To evaluate our method’s ability to mitigate dataset bias, we conducted two ablation studies focusing on True Zero and Data Zero combinations. First, we quantified the implausible lesion predictions made by each model. A prediction is considered implausible if the predicted lesion type, given the ground truth anatomical site, contradicts established anatomical norms (Fig. 4b).
Second, we introduced a category of Data Zero combination by removing a specific combination of anatomical site and oral lesion type from the training dataset (specifically, Mucocele lesions occurring on outer-lip). We then retrained all model variants from scratch on this modified dataset. The subsequent evaluation focused exclusively on the models’ performance on this particular lesion type. The rationale behind this approach is that if a model is influenced by dataset bias, its performance would deteriorate when encountering certain lesion types in anatomical locations it hasn’t been trained on. By conducting these studies, we aimed to assess the robustness of our models and their ability to generalize beyond the biases inherent in the training data.
Data availability
Due to the limited access to the oral cavity, images of specific anatomical locations (e.g. palate) could only be taken in a limited number of ways. Further, some images may contain identifiable patient information (e.g., parts of eyes, skin color, dentition), preventing us from providing dataset publicly. However, dataset can be made available for reproducing results or collaborative research upon request to the corresponding author.
References
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74, 229–263 (2024).
Marcu, L. & Yeoh, E. A review of risk factors and genetic alterations in head and neck carcinogenesis and implications for current and future approaches to treatment. J. Cancer Res. Clin. Oncol. 135, 1303–14 (2009).
Yu, T., Wood, R. & Tenenbaum, H. Delays in diagnosis of head and neck cancers. J. Can. Dent. Assoc. 74 ( 2008).
Brinkerhoff, B. T. & Choong, N. W. Diagnosis to treatment interval and outcome in patients with locally-advanced squamous cell carcinoma of the head and neck in a veterans affairs medical center. J. Cancer Sci. Ther.[SPACE]https://doi.org/10.4172/1948-5956.1000122 (2012).
Hammerlid, E., Silander, E., Harnestam, L. & Sullivan, M. Health-related quality of life three years after diagnosis of head and neck cancer: A longitudinal study. Head Neck 23, 113–25 (2001).
Jacobson, J. J. et al. The cost burden of oral, oral pharyngeal, and salivary gland cancers in three groups: Commercial insurance, medicare, and medicaid. Head Neck Oncol.[SPACE]https://doi.org/10.1186/1758-3284-4-15 (2012).
Sankaranarayanan, R. et al. Effect of screening on oral cancer mortality in Kerala, India: A cluster-randomised controlled trial. The Lancet 365, 1927–33 (2005).
Subramanian, S. et al. Cost-effectiveness of oral cancer screening: Results from a cluster randomized controlled trial in india. Bull. World Health Organ. 87, 200–6 (2009).
Sankaranarayanan, R. et al. Long term effect of visual screening on oral cancer incidence and mortality in a randomized trial in Kerala, India. Oral Oncol. 49, 314–21 (2013).
Olson, C., Burda, B., Beil, T. & Whitlock, E. Screening for oral cancer: A targeted evidence update for the us preventive services task force. U.S. Preventive Services Task Force Evidence Syntheses, formerly Systematic Evidence Reviews ( 2013).
Brocklehurst, P., Baker, S. & SR, P. M. A qualitative study examining the experience of primary care dentists in the detection and management of potentially malignant lesions. 1. factors influencing detection and the decision to refer.. Br. Dent. J. 208, 72–3 (2010).
Choudhary, A. et al. Dentist’s knowledge regarding oral mucosal lesions: Revealing the diagnostic dilemma. Int. J. Health Allied Sci. 8(1), 68 (2019).
Ali, M., Joseph, B. & Sundaram, D. Dental students’ ability to detect and diagnose oral mucosal lesions. J. Dent. Educ. 79, 140–5 (2015).
Lopez-Jornet, P., Camacho-Alonso, F. & Molina-Minano, F. Knowledge and attitudes about oral cancer among dentists in spain. J. Eval. Clin. Pract. 16, 129–33 (2010).
Bacci, C., Donolato, L., Stellini, E., Berengo, M. & Valente, M. A comparison between histologic and clinical diagnoses of oral lesions. Quintessence Int. 45, 789–94 (2014).
Morgan, R., Tsang, J., Harrington, N. & Fook, L. Survey of hospital doctors’ attitudes and knowledge of oral conditions in older patients. Postgrad. Med. J. 77, 392–4 (2001).
Bataineh, A. B., Hammad, H. M. & Darweesh, I. A. Attitude toward oral biopsy among general dental practitioners: Awareness and practice. J. Orofac. Sci. 7(1), 19 (2015).
Williams, P., Poh, C., Hovan, A., Ng, S. & Rosin, M. Evaluation of a suspicious oral mucosal lesion. J. Can. Dent. Assoc. 74, 275–80 (2008).
W, I. Potentially malignant disorders of the oral and oropharyngeal mucosa; present concepts of management.. Oral Oncol. 46, 423–5 (2010).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inform. Process. Syst. 25 ( 2012).
Jubair, F. et al. A novel lightweight deep convolutional neural network for early detection of oral cancer. Oral Dis. 28, 1123–1130 (2022).
Warin, K., Limprasert, W., Suebnukarn, S., Jinaporntham, S. & Jantana, P. Performance of deep convolutional neural network for classification and detection of oral potentially malignant disorders in photographic images. Int. J. Oral Maxillofac. Surg. 51, 699–704 (2022).
Gomes, R. F. T. et al. Use of artificial intelligence in the classification of elementary oral lesions from clinical images. Int. J. Environ. Res. Public Health 20, 3894 (2023).
Welikala, R. A. et al. Automated detection and classification of oral lesions using deep learning for early detection of oral cancer. IEEE Access 8, 132677–132693. https://doi.org/10.1109/ACCESS.2020.3010180 (2020).
Tanriver, G., Soluk Tekkesin, M. & Ergen, O. Automated detection and classification of oral lesions using deep learning to detect oral potentially malignant disorders. Cancers 13, 2766 (2021).
Shamim, M. Z. M. et al. Automated detection of oral pre-cancerous tongue lesions using deep learning for early diagnosis of oral cavity cancer. Comput. J. 65, 91–104 (2022).
Rokhshad, R. et al. Artificial intelligence for classification and detection of oral mucosa lesions on photographs: A systematic review and meta-analysis. Clin. Oral Invest.[SPACE]https://doi.org/10.1007/s00784-023-05475-4 (2024).
Alabi, R. O. et al. Machine learning in oral squamous cell carcinoma: Current status, clinical concerns and prospects for future-a systematic review. Artif. Intell. Med. 115, 102060 (2021).
Salahuddin, Z., Woodruff, H. C., Chatterjee, A. & Lambin, P. Transparency of deep neural networks for medical image analysis: A review of interpretability methods. Comput. Biol. Med. 140, 105111 (2022).
Alabi, R. O., Almangush, A., Elmusrati, M. & Mäkitie, A. A. Deep machine learning for oral cancer: From precise diagnosis to precision medicine. Front. Oral Health 2, 794248 (2022).
Castelvecchi, D. Can we open the black box of AI?. Nat. News 538, 20 (2016).
Zhang, Q.-S. & Zhu, S.-C. Visual interpretability for deep learning: A survey. Front. Inform. Technol. Electron. Eng. 19, 27–39 (2018).
Vellido, A. The importance of interpretability and visualization in machine learning for applications in medicine and health care. Neural Comput. Appl. 32, 18069–18083 (2020).
ElShawi, R., Sherif, Y., Al-Mallah, M. & Sakr, S. Interpretability in healthcare: A comparative study of local machine learning interpretability techniques. Comput. Intell. 37, 1633–1650 (2021).
Corbella, S., Srinivas, S. & Cabitza, F. Applications of deep learning in dentistry. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 132, 225–238 (2021).
Kelly, C. J., Karthikesalingam, A., Suleyman, M., Corrado, G. & King, D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 17, 1–9 (2019).
Wang, F., Casalino, L. P. & Khullar, D. Deep learning in medicine-promise, progress, and challenges. JAMA Intern. Med. 179, 293–294 (2019).
Kleppe, A. et al. Designing deep learning studies in cancer diagnostics. Nat. Rev. Cancer 21, 199–211 (2021).
Gerber, M., Pillay, N. & Khammissa, R. A comparative study of supervised and unsupervised neural networks for oral lesion detection. In 2021 IEEE Symposium Series on Computational Intelligence (SSCI), 01–07 (IEEE, 2021).
Li, K., Wu, Z., Peng, K.-C., Ernst, J. & Fu, Y. Tell me where to look: Guided attention inference network. In Proceedings of the IEEE conference on computer vision and pattern recognition, 9215–9223 ( 2018).
Kattenborn, T., Leitloff, J., Schiefer, F. & Hinz, S. Review on convolutional neural networks (cnn) in vegetation remote sensing. ISPRS J. Photogr. Remote Sens. 173, 24–49 (2021).
Venkataramanan, S., Peng, K.-C., Singh, R. V. & Mahalanobis, A. Attention guided anomaly localization in images. In European Conference on Computer Vision, 485–503 (Springer, 2020).
Tommasi, T., Patricia, N., Caputo, B. & Tuytelaars, T. A deeper look at dataset bias. Domain adaptation in computer vision applications 37–55 ( 2017).
Mehrabi, N., Morstatter, F., Saxena, N., Lerman, K. & Galstyan, A. A survey on bias and fairness in machine learning. ACM Comput. Surv. (CSUR) 54, 1–35 (2021).
Besombes, C., Patel, A. & Madathil, S. A. Incorporating expert prior knowledge for oral lesion recognition. In Maughan, K., Liu, R. & Burns, T. F. (eds.) The First Tiny Papers Track at ICLR 2023, Tiny Papers @ ICLR 2023, Kigali, Rwanda, May 5, 2023 ( OpenReview.net, 2023).
Welikala, R. A. et al. Clinically guided trainable soft attention for early detection of oral cancer. In Tsapatsoulis, N. et al. (eds.) Computer Analysis of Images and Patterns, 226–236, ( Springer International Publishing, Cham, 2021). https://doi.org/10.1007/978-3-030-89128-2_22
Song, B. et al. Mobile-based oral cancer classification for point-of-care screening. JBO 26, 065003. https://doi.org/10.1117/1.JBO.26.6.065003 (2021).
Figueroa, K. C. et al. Interpretable deep learning approach for oral cancer classification using guided attention inference network. J. Biomed. Opt. 27, 015001. https://doi.org/10.1117/1.JBO.27.1.015001 (2022).
Selvaraju, R. R. et al. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE international conference on computer vision, 618–626 ( 2017).
Song, B. et al. Classification of imbalanced oral cancer image data from high-risk population. J. Biomed. Opt. 26, 105001–105001 (2021).
Barandela, R., Sánchez, J. S., Garcıa, V. & Rangel, E. Strategies for learning in class imbalance problems. Pattern Recogn. 36, 849–851 (2003).
Mohammad-Rahimi, H., Rokhshad, R., Bencharit, S., Krois, J. & Schwendicke, F. Deep learning: A primer for dentists and dental researchers. J. Dent. 130, 104430 (2023).
Tan, M. & Le, Q. V. Efficientnet: Rethinking model scaling for convolutional neural networks. In Chaudhuri, K. & Salakhutdinov, R. (eds.) Proceedings of the 36th International Conference on Machine Learning, ICML 2019, 9-15 June 2019, Long Beach, California, USA, vol. 97 of Proceedings of Machine Learning Research, 6105–6114 ( PMLR, 2019).
Russakovsky, O. et al. ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. (IJCV) 115, 211–252. https://doi.org/10.1007/s11263-015-0816-y (2015).
Balandat, M. et al. BoTorch: Programmable Bayesian optimization in PyTorch. arxiv e-prints ( 2019).
Bakshy, E. et al. Ae: A domain-agnostic platform for adaptive experimentation. In NeurIPS Systems for ML Workshop ( 2018).
Loshchilov, I. & Hutter, F. Decoupled weight decay regularization. In 7th International Conference on Learning Representations, ICLR 2019, New Orleans, LA, USA, May 6-9, 2019 ( OpenReview.net, 2019).
Maier-Hein, L. et al. Metrics reloaded: Recommendations for image analysis validation. Nat. Methods 21, 195–212. https://doi.org/10.1038/s41592-023-02151-z (2024).
Acknowledgements
We would like to acknowledge the contributions of Mehak Khanna, and Mohammed Al-Shehri for their contribution toward data cleaning and pre-processing. This project was funded by the Canadian Institue of Health Research Project Grant [PJT-438778]. S Madathil is a recipient of a Career Award from the Fonds de Recherche du Québec-Santé. This research was partly enabled by support provided by Calcul Quebec (calculquebec.ca) and the Digital Research Alliance of Cancer (alliancecan.ca).
Author information
Authors and Affiliations
Contributions
Conceptualization: A.P., F.T., M.D., P.C., S.M.; Data curation: A.P., C.B., T.D., M.S., S.M.; Formal analysis: C.B., S.M.; Funding acquisition: F.T., P.C., S.M.; Investigation: A.P.; Methodology: A.P., C.B., S.M.; Project administration: A.P., S.M.; Resources: S.M.; Software: A.P., C.B., S.M.; Supervision: S.M.; Validation: A.P.; Visualization: A.P.; Writing—original draft: A.P., S.M.; Writing—review & editing: A.P., C.B., T.D., M.S., F.T., M.D., P.C., S.M.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Patel, A., Besombes, C., Dillibabu, T. et al. Attention-guided convolutional network for bias-mitigated and interpretable oral lesion classification. Sci Rep 14, 31700 (2024). https://doi.org/10.1038/s41598-024-81724-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-81724-0