Abstract
This study aimed to address the limitations of conventional methods for measuring skeletal muscle mass for sarcopenia diagnosis by introducing an artificial intelligence (AI) system for direct computed tomography (CT) analysis. The primary focus was on enhancing simplicity, reproducibility, and convenience, and assessing the accuracy and speed of AI compared with conventional methods. A cohort of 3096 cases undergoing CT imaging up to the third lumbar (L3) level between 2011 and 2021 were included. Random division into preprocessing and sarcopenia cohorts was performed, with further random splits into training and validation cohorts for BMI_AI and Body_AI creation. Sarcopenia_AI utilizes the Skeletal Muscle Index (SMI), which is calculated as (total skeletal muscle area at L3)/(height)2. The SMI was conventionally measured twice, with the first as the AI label reference and the second for comparison. Agreement and diagnostic change rates were calculated. Three groups were randomly assigned and 10 images before and after L3 were collected for each case. AI models for body region detection (Deeplabv3) and sarcopenia diagnosis (EfficientNetV2-XL) were trained on a supercomputer, and their abilities and speed per image were evaluated. The conventional method showed a low agreement rate (κ coefficient) of 0.478 for the test cohort and 0.236 for the validation cohort, with diagnostic changes in 43% of cases. Conversely, the AI consistently produced identical results after two measurements. The AI demonstrated robust body region detection ability (intersection over Union (IoU) = 0.93), accurately detecting only the body region in all images. The AI for sarcopenia diagnosis exhibited high accuracy, with a sensitivity of 82.3%, specificity of 98.1%, and a positive predictive value of 89.5%. In conclusion, the reproducibility of the conventional method for sarcopenia diagnosis was low. The developed sarcopenia diagnostic AI, with its high positive predictive value and convenient diagnostic capabilities, is a promising alternative for addressing the shortcomings of conventional approaches.
Similar content being viewed by others
Introduction
Artificial intelligence (AI) represents the embodiment of human intelligence in computer programs and is widely used across various facets of our lives. In the medical field, diagnostic imaging is an area in which AI can offer substantial value1,2,3,4. AI exhibits the capability to diagnose pathological conditions, as evidenced by its effectiveness in identifying pancreatic lesions5,6. Moreover, AI’s potential extends to assessing body composition7,8, including muscle and fat mass, which is recognized as a factor influencing the clinical outcomes of various diseases.
Sarcopenia, which is characterized by age-related muscle loss, has become a focal point in recent years. Initially proposed by Irwin Rosenberg, the European Working Group on Sarcopenia in Older People (EWGSOP) defined it as a syndrome involving generalized and progressive loss of skeletal muscle mass and strength associated with adverse outcomes, such as decreased quality of life and mortality9. Sarcopenia is estimated to affect 5–10% of the elderly population over the age of 65 years10 and is projected to increase to over 200 million by 205011. Its management incurs significant costs, with the United States spending approximately $18.5 billion in 200012.
With the recognition of sarcopenia as an imaging biomarker with implications on clinical outcomes, there is an urgent need for accessible and straightforward diagnostic tools. The 2018 revision13 of the diagnostic algorithm for sarcopenia emphasizes early detection and intervention, focusing on muscle strength assessment. However, the accurate assessment of both muscle quantity and quality remains challenging.
Although Computed Tomography (CT) scans have become vital for diagnosing various diseases, including malignancies, the current method of measuring skeletal muscle mass relies on manual segmentation, which is hampered by its complex and resource-intensive nature14.
This study aimed to utilize deep learning, specifically deep convolutional neural networks (CNN), to develop an AI-based sarcopenia diagnostic tool to overcome the time and measurement errors associated with conventional methods, increase clinical applicability, and detect changes in body composition more efficiently and accurately. Overcoming these drawbacks would provide valuable information for the prevention of adverse events.
Result
Background factors of all patients
The study population consisted of 3096 patients diagnosed at the Aichi Cancer Center in Japan. The study population was comprised of 2152 males (69.5%) and 944 females (30.5%). The mean age was 66.3 ± 9.3 years (Table 1). The mean BMI was 21.4 ± 3.2 kg/m2. The mean visceral fat mass (VFM) was 90.5 ± 57.9 cm2, the mean subcutaneous fat mass (SFM) was 94.8 ± 58.6 cm2, the total SMA in the L3 region was 109.1 ± 32.2 cm2, and the mean SMI was 42.01 ± 8.88 (cm2/m2). The background diseases at the time of the CT scan were advanced esophageal cancer in 1291 patients (41.7%), advanced colorectal cancer in 326 patients (10.5%), and scheduled endoscopic submucosal dissection (ESD) for early-stage cancer without overlapping cancer in 1479 patients (47.8%), which includes 334 esophageal cancer cases, 822 gastric cancer cases, and 323 colorectal cancer cases.
Reproducibility of sarcopenia diagnosis by conventional methods
For conventional methods, the test cohort exhibited a concordance rate of 68.4% and a κ value of 0.47 (95% confidence interval (CI) 0.354–0.601). In contrast, the validation cohort showed a concordance rate of 47.5% and a κ value of 0.236 (95% CI 0.143–0.330) (Table 2).
Diagnostic performance of Body_AI
The median IoU for the Body_AI system in the validation cohort was 0.93, and the IoU of all images in the validation cohort was > 0.5, indicating that body regions were successfully extracted from all images of the validation cohort.
Diagnostic performance for the diagnosis of BMI_AI
The validation cohort was comprised of 140 patients (82 men and 58 women). The mean age was 62.8 ± 12.0 years, and the mean BMI was 22.2 ± 3.4 kg/m2 (Table 3). The Area Under the Receiver Operating Characteristic (AUROC) for each patient’s ability to diagnose BMI using AI was 0.959. The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and their 95% CIs were 0.960 (0.888–0.992), 0.862 (0.753–0.935), 0.889 (0.800–0.948), and 0.914 (0.855–0.955), respectively. These values were determined using output value of 0.50 from the ROC analysis as the cutoff points (Table 4).
Diagnostic performance for the diagnosis of Sarcopenia_AI
In the test cohort of 114 cases, there were 97 men and 17 women, with a mean age of 66.7 ± 8.0 years and a mean BMI of 21.4 ± 2.9 kg/m2. The mean VFM was 77.7 ± 62.1 cm2, mean SFM was 113.9 ± 34.6 cm2, mean total SMA in the L3 region was 88.2 ± 60.7 cm2, and mean SMI was 44.7 ± 9.8 (cm2/m2). The SMI low, middle, and high groups comprised 36, 16, and 62 individuals, respectively.
In the validation cohort of 118 patients, there were 98 men and 20 women, with a mean age of 64.9 ± 8.2 years and a mean BMI of 20.9 ± 2.8 kg/m2. The mean VFM was 79.3 ± 45.7 cm2, mean SFM was 117.3 ± 41.2 cm2, mean total SMA in the L3 region was 87.1 ± 50.7 cm2, and mean SMI was 44.9 ± 8.9 (cm2/m2). The SMI low, middle, and high groups was comprised of 30, 14, and 74 individuals, respectively (Table 5).
The AUROC for the ability of the AI to diagnose sarcopenia was 0.892. The sensitivity, specificity, PPV, NPV, and accuracy, along with their 95% CIs, were 0.803 (0.682–0.894), 0.981 (0.897–1.000), 0.980 (0.894–0.999), 0.812 (0.695–0.899), and 0.886 (0.813–0.938) (Table 4).
The created Sarcopenia AI was able to recognize sarcopenia quickly, in only approximately 0.18 s (supple).
Discussion
Our investigation of the performance of AI-based sarcopenia diagnostic tools revealed a substantial leap forward from traditional manual segmentation methods. The conventional approach, characterized by consistently low inter-rater reliability (κ values) across multiple measurements, posed significant challenges. By contrast, the AI models, particularly Body_AI, showed remarkable precision in sarcopenia detection. Notably, Sarcopenia_AI exhibited commendable sensitivity and specificity and a swift decision-making time of approximately 0.18 s. These results underscore the transformative potential of the AI tools developed in this study, which includes reduced measurement time, procedural simplification, and minimized measurement errors, effectively mitigating the shortcomings associated with conventional methods.
The limitations of BMI as a comprehensive indicator of body composition are well-established. BMI, which is primarily based on weight and height, lacks the granularity required to accurately reflect nuances in body composition accurately11,15. In contrast to BMI, sarcopenia, often considered a representative marker of body composition abnormalities, consistently exhibits a correlation with mortality, particularly in individuals aged 79 years and above12. The impact of sarcopenia on prognosis is similar in patients diagnosed with cancer16,17,18. Therefore, recognizing the effect of sarcopenia on clinical outcomes and therapeutic interventions targeting these conditions may lead to improvements in complications, quality of life (QOL), and overall prognosis.
However, the diagnosis of sarcopenia has not yet reached a point where it can be applied in clinical practice. A major obstacle is the lack of readily available diagnostic tools for sarcopenia. Although tools that can measure skeletal muscle mass are currently available, the conventional method is manual segmentation analysis, which has several drawbacks and has not yet been applied clinically.
First, segmentation is complicated by various CT-specific factors19. Therefore, manual segmentation has drawbacks such as inter-image and inter-observer variations20,21 and overestimation of muscle mass owing to intramuscular fat21. In this study as well as in other studies, manual segmentation is used as an evaluation criterion in AI development. However, the manual segmentation exhibited strong variations. Therefore, to compensate for the drawback of manual segmentation, which is the variation between measurements, manual segmentation was performed three times and used as a reference, making it possible to develop an AI with accurate values.
Furthermore, the tools used for conventional sarcopenia diagnosis have the disadvantage of being time-consuming due to manual segmentation22. Although recent efforts have been made to reduce the measurement time and improvements of 1–3 min per slice have been reported, this process remains time-consuming23. Furthermore, sarcopenia diagnosis requires calculation of skeletal muscle index after measurement and obtaining clinical information necessary for sarcopenia diagnosis; considering this additional time, the entire process is considerably time-consuming. Our AI simplified the diagnosis of sarcopenia by enabling the immediate recognition of the presence of sarcopenia using routine CT images without the need for specialized protocols. In addition, since the diagnosis can be made with a plain CT scan without contrast, even patients with poor renal function can be diagnosed. This diagnostic tool has sufficient utility in patient populations for whom CT9, the gold standard in the diagnosis of sarcopenia, is available.
In addition to our study, there have been other reports on the measurement of skeletal muscle mass using AI23,24,25,26,27. Although these data have a publication bias, both Dice similarity coefficients (DSC) and Jaccard similarity coefficients (JSC) have very high accuracies of 0.941 and 0.967, respectively28. However, most of these studies use only 30–60 scans. This can lead to overlearning in deep learning, which is useful for the segmentation of similar datasets. However, the performance is degraded on newer datasets. Therefore, these findings are limited to research publications and have not yet reached broad clinical practice. In this study, we developed a model by analyzing 3096 CT images, which is a much larger number of images than those used in previous studies. Additionally, these are tested using an external cohort model to prevent performance deterioration.
Although our study has made significant strides in the realm of AI-based CT body composition measurements, certain limitations must be acknowledged. The retrospective design and relatively small sample size introduced potential biases in the evaluated imaging cases. We consider this limitation not only due to the sample size, but also because this study was biased by the large number of patients with cancer. Moreover, this study was not explicitly designed to ascertain the treatability of sarcopenia, and future research should delve into this aspect.
To strengthen future studies, a prospective study design and inclusion of diverse patient populations, especially elderly people, can improve the generalizability and applicability of findings to the aging society. It is crucial to integrate muscle strength assessment, which is one of the predictors of functional outcomes and quality of life in elderly people. Longitudinal tracking of muscle mass over time will provide insight into age-related changes and their impact on disease prognosis and management. Furthermore, incorporating external validation in multiple centers will improve the robustness and applicability of findings. Future studies considering these points may provide a more comprehensive and practical assessment framework that is better suited to the needs of the aging society.
In conclusion, we developed an AI-based diagnostic tool that overcomes the time and measurement errors associated with conventional methods, efficiently and accurately recognizes body composition, and diagnoses sarcopenia. Using the AI that we researched and developed, it is possible to diagnose sarcopenia quickly and accurately. Furthermore, in the future, we hope to develop an AI that can not only recognize sarcopenia from CT images but also evaluate muscle strength, which is necessary for the diagnosis of sarcopenia and individual physical functions, and predict complications, side effects, and prognosis of various diseases. We believe that this method can contribute not only to sarcopenia research, but also to geriatrics in the future.
Methods
Ethical approval
This study was approved by the Institutional Review Board of Aichi Cancer Center (No. 2019-1-549, March 26, 2019) and was performed in accordance with the Declaration of Helsinki29. The study was based on medical record review. Informed consent for all participants was obtained from the opt-out form, and those who did not provide consent were excluded. The need for obtaining informed consent was waived due to the nature of the study. All authors had access to the study data and reviewed and approved the final manuscript.
Patients
We included patients who underwent an abdominal CT scan at our hospital between January 2011 and December 2021 and whose images were stored in their medical records. These data were stored in the Endoscopy Department database. A total of 3096 participants were included, after excluding those with poor image quality or missing basic information at the time of image capture. These prospectively and consecutively registered databases were retrospectively analyzed. Height and weight were also measured in all patients, and the body mass index (BMI) was calculated using the formula: BMI = (weight in kilograms)/(height in meters)2.
Data sets and preprocessing
All included cases were randomly assigned to the preprocess and sarcopenia cohorts without any specific criteria or bias. Random assignment was conducted to ensure unbiased representation of the cases across the two cohorts. Subsequently, the preprocessing cohort was randomly split into training and validation sets to create BMI_AI and body region extraction AI (Body_AI). To develop and validate the sarcopenia model, the entire dataset containing CT images of the third lumbar (L3) region nearest a mid-vertebral slice was randomly divided into the development and test cohorts. This division was performed through random sampling based on the cases rather than on individual images. Within the development cohort, additional random division into training and validation cohorts was performed (Fig. 1). Images of the L3 region were extracted from these cohorts and stored as digital still images (JPEG format). The L3 level was identified for all patients in each cohort and the total skeletal muscle area (SMA) was extracted. The CT value of skeletal muscle was measured using a range of -30 to 150 Hounsfield Units. For the validation and test cohorts, 10 images were extracted consecutively from each patient. It is important to note that random extraction of images and assignment of patients to cohorts were conducted without any predetermined criteria to ensure a random distribution of the data. First, we divided the cohort into preprocess and sarcopenia cohorts; then, the preprocess cohort was divided into training and validation cohorts, and the sarcopenia cohort was divided into training, validation, and test cohorts.
Flowchart of this study. First, we divided the cohort into preprocess and sarcopenia cohorts; then, the preprocess cohort was divided into training and validation cohorts, and the sarcopenia cohort was divided into training, validation, and test cohorts. Body_AI and BMI_AI were created from the BMI cohort and Sarcopenia_AI from the sarcopenia cohort.
Body_AI and BMI_AI were created from the BMI cohort and Sarcopenia_AI from the sarcopenia cohort.
SMA measurement and sarcopenia identification measurement
The total SMA at the L3 level in the abdominal CT images was measured using SYNAPSE VINCENT (Fujifilm Medical Co., Ltd., Tokyo, Japan). In addition, visceral fat and subcutaneous fat masses were measured at the umbilicus level. If there was a measurement error of 5% or more, a third measurement was taken, and the final label was determined based on the most frequently occurring value among the three measurements. Three measurements were taken to compensate for the measurement error of the conventional method. Thus, a determination was considered a match when the results from two consistent measurements agreed, and a third measurement was only introduced in cases where a measurement error of 5% or more was identified. The final Skeletal Muscle Index (SMI) was calculated using re-measurements (SMI = SMA/(height in meters)2). The percentage of concordance and κ (kappa) value were measured in the validation and test cohort. The κ value provides a measure of inter-rater agreement, considering the agreement occurring by chance.
Development of AI systems
In this study, we developed three AI systems: Body_AI, which can only extract body images from CT images; BMI_AI, which can evaluate BMI; and Sarcopenia_AI, which can evaluate sarcopenia. Supercomputing resources (Shirokane, Human Genome Center, Tokyo, Japan) and local computing resources (GeForce GTX 2080 Ti and RTX A6000, Nvidia Corporation, California, USA) were used. The CNN layers in Body_AI and BMI_AI were fine-tuned from the pre-trained models using ImageNet-21 K. To avoid overfitting, we employed early stopping30, data augmentation techniques (vertical flip, image rotation (-10 to 10), random erasing, and RandAugment)31, and label smoothing. The AdamW optimizer32 was used as the optimization algorithm to train the network weights. The models were trained using the training data until early stopping, and the model performance was assessed using the validation data. After training, the final trained model with the lowest loss was selected, and its performance was evaluated using the validation data. The test data were used to evaluate the performance of the Sarcopenia_AI.
Body_AI system
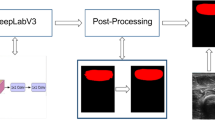
In this study, a semantic segmentation model comprising an encoder (efficientnet-b4) and a decoder (DeepLabV3 +) was utilized to develop Body_AI (Fig. 2) for body region extraction from CT images. The CT images from all cohorts were extracted and segmented to isolate the body regions and were then resized to 256 × 256 pixels. Body_AI, based on a CNN architecture, was trained using these images and could extract body regions from the CT images at the pixel level. The optimized model was then employed to extract body regions from CT images.
Creating Body_AI. (A) The extracted images may contain personal information or other artifacts that could be present in the image. (B) AI was created to automatically segment only the body region from the image. (C) The AI recognizes and extracts only the body region, removing any additional information from the original image.
BMI_AI system
BMI_AI was developed using the EfficientNetV2-L, a CNN architecture, designed to classify high/low BMI (low: BMI < 22, high ≥ 22) after extracting body images with Body_AI. The image input size was set to 512 × 512 pixels, and the output provided a predictive value ranging from zero to one. When the predictive value approached 1, the model predicted high BMI on the CT images. The model was trained and validated using images extracted by Body_AI.
Sarcopenia_AI system
Finally, Sarcopenia_AI was developed using an image classification model, specifically EfficientNetV2-L, to classify SMI into three levels: low (SMI < 38), middle (38 ≤ SMI < 42), and high (SMI ≥ 42). Considering these SMI levels, along with sex, Sarcopenia_AI assesses whether an individual has sarcopenia. The images used for the training and validation had an input size of 512 × 512 pixels. All CNN layers in the Sarcopenia_AI model were fine-tuned using weights derived from the BMI_AI model (Fig. 3). Similar to BMI_AI, Sarcopenia_AI employs images extracted using Body_AI.
We created a BMI_AI model from the labels we generated, and then fine-tuned the BMI_AI model to create a Sarcopenia_AI model. Fine-tuning is used to optimize the performance of a model for a new or different task. A pre-trained model (BMI_AI) trained on a large amount of labeled data is fine-tuned for a specific task. Once training is complete, the fine-tuned BMI_AI can be used to apply the model to a similar new task or dataset with limited labeled data of Sarcopenia_AI. openia_AI.
Outcome definition
For segmentation evaluation, intersection over union (IoU) was used. When IoU > 0.5, the prediction is a true positive (TP); when IoU < 0.5, it is a false positive (FP); and when the model detection area is 0, it is a false negative (FN).
For the classification evaluation, the predictive value for each case was calculated as the median of the predictive values for all images in that case.
The primary endpoint is the accuracy of assessment of sarcopenia for each case in the test cohort. The secondary endpoints were 1) the sensitivity and specificity of the assessment of sarcopenia for each case in the test cohort, 2) the accuracy, sensitivity, and specificity of assessment of sarcopenia for each case in the validation cohort, and those of assessment of BMI for each case in the validation/test cohort, and 3) the accuracy of detection of the body area and misalignment of sarcopenia evaluation (κ value) for the validation/test cohort.
Statistical methods
JMP version 13.2.0 (SAS Institute Inc.,Cary, NC, USA) was used for all statistical analyses. All tests were two-tailed, and statistical significance was set at P < 0.05. Continuous variables are presented as medians and ranges. Fisher’s exact test was used for categorical variables, and the Mann–Whitney U test was used for continuous variables. The McNemar test was used to compare differences in accuracy, sensitivity, and specificity. Inter-observer agreement was assessed using κ statistics: κ > 0.8, almost perfect agreement; 0.8 to 0.6, substantial agreement; 0.6 to 0.4, moderate agreement; 0.4 to 0.2, fair agreement; and < 0.2, slight agreement. A κ value of 0 indicated agreement equal to chance, and < 0 indicated disagreement.
Data availability
The datasets used and/or analyzed in the current study are available from the corresponding author upon reasonable request.
References
Cha, K. H. et al. Bladder cancer segmentation in CT for treatment response assessment: application of deep-learning convolution neural network-A pilot study. Tomography 2, 421–429. https://doi.org/10.18383/j.tom.2016.00184 (2016).
Trebeschi, S. et al. Deep learning for fully-automated localization and segmentation of rectal cancer on multiparametric MR. Sci. Rep. 7, 5301. https://doi.org/10.1038/s41598-017-05728-9 (2017).
Goh, K. H. et al. Artificial intelligence in sepsis early prediction and diagnosis using unstructured data in healthcare. Nat. Commun. 12, 711. https://doi.org/10.1038/s41467-021-20910-4 (2021).
Jin, C. et al. Development and evaluation of an artificial intelligence system for COVID-19 diagnosis. Nat. Commun. 11, 5088. https://doi.org/10.1038/s41467-020-18685-1 (2020).
Kuwahara, T. et al. Current status of artificial intelligence analysis for endoscopic ultrasonography. Dig. Endosc. 33, 298–305. https://doi.org/10.1111/den.13880 (2021).
Kuwahara, T. et al. Artificial intelligence using deep learning analysis of endoscopic ultrasonography images for the differential diagnosis of pancreatic masses. Endoscopy 55, 140–149. https://doi.org/10.1055/a-1873-7920 (2023).
Pamoukdjian, F. et al. Prevalence and predictive value of pre-therapeutic sarcopenia in cancer patients: A systematic review. Clin. Nutr. 37, 1101–1113. https://doi.org/10.1016/j.clnu.2017.07.010 (2018).
Haiducu, C., Buzea, A., Mirea, L. E. & Dan, G. A. The prevalence and the impact of sarcopenia in digestive cancers. A systematic review. Rom. J. Intern. Med. 59, 328–344. https://doi.org/10.2478/rjim-2021-0026 (2021).
Cruz-Jentoft, A. J. et al. Sarcopenia: European consensus on definition and diagnosis: Report of the European Working Group on Sarcopenia in older people. Age Ageing 39, 412–423. https://doi.org/10.1093/ageing/afq034 (2010).
Morley, J. E., Anker, S. D. & von Haehling, S. Prevalence, incidence, and clinical impact of sarcopenia: Facts, numbers, and epidemiology-update 2014. J. Cachexia Sarcopenia Muscle 5, 253–259. https://doi.org/10.1007/s13539-014-0161-y (2014).
Prado, C. M., Cushen, S. J., Orsso, C. E. & Ryan, A. M. Sarcopenia and cachexia in the era of obesity: Clinical and nutritional impact. Proc. Nutr. Soc. 75, 188–198. https://doi.org/10.1017/s0029665115004279 (2016).
Beaudart, C., Zaaria, M., Pasleau, F., Reginster, J. Y. & Bruyère, O. Health outcomes of sarcopenia: A systematic review and meta-analysis. PLoS One 12, e0169548. https://doi.org/10.1371/journal.pone.0169548 (2017).
Cruz-Jentoft, A. J. et al. Sarcopenia: Revised European consensus on definition and diagnosis. Age Ageing 48, 601. https://doi.org/10.1093/ageing/afz046 (2019).
Shi, B. et al. Sarcopenia is associated with perioperative outcomes in gastric cancer patients undergoing gastrectomy. Ann. Nutr. Metab. 75, 213–222. https://doi.org/10.1159/000504283 (2019).
Prado, C. M., Birdsell, L. A. & Baracos, V. E. The emerging role of computerized tomography in assessing cancer cachexia. Curr. Opin. Support Palliat. Care 3, 269–275. https://doi.org/10.1097/SPC.0b013e328331124a (2009).
Shachar, S. S., Williams, G. R., Muss, H. B. & Nishijima, T. F. Prognostic value of sarcopenia in adults with solid tumours: A meta-analysis and systematic review. Eur. J. Cancer 57, 58–67. https://doi.org/10.1016/j.ejca.2015.12.030 (2016).
Onishi, S. et al. Prognostic significance of sarcopenia in patients with unresectable advanced esophageal cancer. J. Clin. Med. https://doi.org/10.3390/jcm8101647 (2019).
Onishi, S. et al. Prognostic impact of sarcopenic obesity after neoadjuvant chemotherapy followed by surgery in elderly patients with esophageal squamous cell carcinoma. J. Clin. Med. https://doi.org/10.3390/jcm9092974 (2020).
Polan, D. F., Brady, S. L. & Kaufman, R. A. Tissue segmentation of computed tomography images using a Random Forest algorithm: A feasibility study. Phys. Med. Biol. 61, 6553–6569. https://doi.org/10.1088/0031-9155/61/17/6553 (2016).
Bedrikovetski, S. et al. Artificial intelligence for body composition and sarcopenia evaluation on computed tomography: A systematic review and meta-analysis. Eur. J. Radiol. 149, 110218. https://doi.org/10.1016/j.ejrad.2022.110218 (2022).
Herrod, P. J. J. et al. Quick and simple; psoas density measurement is an independent predictor of anastomotic leak and other complications after colorectal resection. Tech. Coloproctol. 23, 129–134. https://doi.org/10.1007/s10151-019-1928-0 (2019).
Chung, H. et al. Automated segmentation of muscle and adipose tissue on CT images for human body composition analysis. SPIE Proc. 7261, 72610K (2009).
Lee, S. B. et al. Automated segmentation of whole-body CT images for body composition analysis in pediatric patients using a deep neural network. Eur. Radiol. 32, 8463–8472. https://doi.org/10.1007/s00330-022-08829-w (2022).
Fu, Y. et al. Technical note: Automatic segmentation of CT images for ventral body composition analysis. Med. Phys. 47, 5723–5730. https://doi.org/10.1002/mp.14465 (2020).
Hu, P. et al. Automated characterization of body composition and frailty with clinically acquired CT. Comput. Methods Clin. Appl. Musculoskelet. Imaging 2017(10734), 25–35. https://doi.org/10.1007/978-3-319-74113-0_3 (2018).
Decazes, P., Tonnelet, D., Vera, P. & Gardin, I. Anthropometer3D: Automatic multi-slice segmentation software for the measurement of anthropometric parameters from CT of PET/CT. J. Digit. Imaging 32, 241–250. https://doi.org/10.1007/s10278-019-00178-3 (2019).
Koitka, S., Kroll, L., Malamutmann, E., Oezcelik, A. & Nensa, F. Fully automated body composition analysis in routine CT imaging using 3D semantic segmentation convolutional neural networks. Eur. Radiol. 31, 1795–1804. https://doi.org/10.1007/s00330-020-07147-3 (2021).
Mai, D. V. C. et al. A systematic review of automated segmentation of 3D computed-tomography scans for volumetric body composition analysis. J. Cachexia Sarcopenia Muscle 14, 1973–1986. https://doi.org/10.1002/jcsm.13310 (2023).
World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA 310, 2191-2194. https://doi.org/10.1001/jama.2013.281053 (2013).
Yao, Y., Rosasco, L. & Caponnetto, A. On early stopping in gradient descent learning. Constr. Approx. 26, 289–315. https://doi.org/10.1007/s00365-006-0663-2 (2007).
Shorten, C., Khoshgoftaar, T. M. & Furht, B. Text data augmentation for deep learning. J. Big Data 8, 101. https://doi.org/10.1186/s40537-021-00492-0 (2021).
He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. In Proc. of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, 770–778 (2016).
Acknowledgements
The Human Genome Center (University of Tokyo) provided supercomputing resources.
Funding
This work was supported by JSPS KAKENHI, Grant Number 21K15673JP.
Author information
Authors and Affiliations
Contributions
S.O. has wrote the main manuscript text and prepared all figures and tables. S.O. and T.K. analyzed data. T.K., M.S. and R.Y. reviewed, edited and revised manuscript. All contributed to the study conception and design, and contributed equally to this work and should be considered co-corresponding authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Onishi, S., Kuwahara, T., Tajika, M. et al. Artificial intelligence for body composition assessment focusing on sarcopenia. Sci Rep 15, 1324 (2025). https://doi.org/10.1038/s41598-024-83401-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-83401-8