Abstract
We aimed to explore the role of circular RNA 0043256 (circ_0043256) in gastric cancer (GC) and its underlying mechanisms. The impact of circ_0043256 silencing on the proliferation, migration, apoptosis, and aerobic glycolysis of MKN-45 and AGS cells induced by CoCl2 was assessed through the utilization of CCK-8, wound healing assay, flow cytometry, and metabolic analysis. The interaction between circ_0043256 and miR-593-5p, as well as the involvement of the miR-593-5p/RRM2 axis in gastric cancer, were confirmed via luciferase assay, Western blot, and bioinformatics analysis. We found that circ_0043256 was up-regulated in GC tissues and CoCl2-treated MKN-45 and AGS cells. Silencing of circ_0043256 reversed CoCl2-induced proliferation, migration, and aerobic glycolysis in MKN-45 and AGS cells. Additionally, circ_0043256 silencing enhanced cell apoptosis and G2/M phase cell cycle arrest in response to CoCl2 treatment. Furthermore, the miR-593-5p/RRM2 axis was identified as a regulatory mechanism for circ_0043256 function in GC. Silencing of circ_0043256 and miR-593-5p mimic co-transfection significantly inhibited CoCl2-induced cellular responses in MKN-45 and AGS cells. A glycolysis inhibitor 2-DG further enhanced the inhibitory effect of circ_0043256 silencing on aerobic glycolysis of CoCl2-induced MKN-45 and AGS cells. Additionally, the inhibition of circ_0043256 resulted in a reduction in tumor volume and the expression of proliferation marker proteins in nude mice. Moreover, the suppression of circ_0043256 led to an increase in miR-593-5p expression and a decrease in RRM2 expression, ultimately causing a decrease in glycolytic-related proteins associated with the glycolytic pathway. Targeting this axis may offer a novel therapeutic approach for treating GC.
Similar content being viewed by others
Introduction
Gastric cancer (GC) is a prevalent digestive malignancy on a global scale, ranking fifth in terms of morbidity and fourth in terms of mortality among all tumors1,2. Despite advancements in the diagnosis and treatment of GC in recent years, the survival rates for patients remain low, particularly for those with advanced gastric cancer (AGC) where the 5-year overall survival (OS) rate is approximately 30% 3. The unfavorable prognosis for GC patients is linked to late-stage diagnosis, aggressive proliferation, and the invasive and metastatic capabilities of GC cells4,5,6. Consequently, the identification of reliable diagnostic biomarkers and promising therapeutic targets is crucial for enhancing the prognosis of patients with gastric cancer7.
Circular RNA (circRNA) is a single-stranded, closed RNA molecule generated through back splicing events. Its biogenesis primarily entails intron-paired splicing, loop formation driven by RNA-binding proteins, and loop formation driven by lassos. CircRNA has been implicated in the modulation of gene expression through its role as a competitive endogenous RNA (ceRNA) within the miRNA sponge network8,9. CircRNA plays a significant role in cancer development, as evidenced by its involvement in the modulation of proliferation, migration, invasion, and drug resistance in diverse cancer cell types. Specifically, in GC, circNRIP1 has been identified as as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway10. Additionally, circDLG1 was found to interact with miR-141-3p, functioning as a miRNA sponge to upregulate CXCL12 expression, thereby enhancing GC progression and resistance to anti-PD-1-based therapy11. Furthermore, circ_0067514 has been shown to inhibit GC progression and glycolysis through the miR-654-3p/LATS2 axis12.
Through data mining, we found that circ_0043256 was an obviously up-regulated circRNA in GC tissues. Previous research revealed that circ_0043256 promoted KLF2 expression by absorbing miR-1206 to suppress the tumorigenesis of lung cancer13. However, the functional roles of circ_0043256 in GC progression have not yet been examined. Based on prior research, it is hypothesized that circ_0043256 may play a role in promoting GC progression. This study aimed to investigate the impact of circ_0043256 knockdown on GC cell progression and aerobic glycolysis, as well as elucidated its molecular mechanism, with the goal of identifying circ_0043256 as a potential therapeutic target for GC.
Materials and methods
Clinical specimens
The study protocols were approved by the Experimental Animal Ethics Committee, West China Hospital, Sichuan University (20240617005). All methods were performed in accordance with the relevant guidelines and regulations. We collected GC tissues (n = 30) and adjacent non-tumor tissue samples (n = 30) from patients. None of the patients had received chemotherapy or radiotherapy before tumor excision. All specimens were collected after obtaining informed consent from all patients.
Cell culture and treatment
The control human gastric mucosal epithelial cells GES-1 (#icell-h062), the human GC cell lines MKN-45 (#iCell-h345), AGS (#iCell-h016), SNU-16 (#iCell-h430), and SNU-216 (#iCell-h526) were obtained from Saibaikang Biotechnology (China). The human GC cell lines HGC-27 (#CL-0107) and MKN-7 (#CL-0574) were purchased from Wuhan Pu-nuo-sai Life Technology Co. Ltd. (Wuhan, China). These cells were cultured in 1640 culture medium (#PM150110, Procell, USA) supplemented with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA, USA), and 100 µg/mL penicillin-streptomycin in a humidified incubator at 37 °C with 5% CO2. Cells in the logarithmic growth phase were utilized for subsequent experiments. Transient transfection of siRNA-circ_0043256 (si-circ_0043256), miR-593-5p mimic, and their respective negative controls was performed using Lipofectamine®2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. Subsequently, cells were exposed to different concentrations (50 µM, 100 µM, 200 µM, and 300 µM) of cobalt chloride (CoCl2; Sigma-Aldrich, USA), a chemical mimicking hypoxia conditions. Alternatively, cells were treated with 20 mM 2-deoxy-D-glucose (2-DG) for 30 min to inhibit glycolysis.
Real-time fluorescence quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted utilizing TRIzol® reagent (Thermo Fisher, USA), followed by the utilization of the SYBR Premix Ex Taq kit (Bao Biological Engineering, China) in accordance with the manufacturer’s instructions. The reverse transcription reaction was conducted under the following conditions: 95˚C for 10 min; 40 cycles of 95˚C for 5 s, 60˚C for 30 s, and 70 °C for 60 s. The relative gene expression level was quantified using the 2−△△Ct method with ABI software in Foster City, CA.
Western blot analysis
Total protein extracted from MKN-45 and AGS cells underwent separation via 10% SDS polyacrylamide gel electrophoresis, followed by transfer to nitrocellulose membranes (Millipore, USA). The membranes were then subjected to blocking with 5% nonfat milk and incubation with specific protein antibodies or a rabbit anti-β-actin monoclonal antibody. Subsequently, the membranes were exposed to an HRP Goat anti-Rabbit IgG (1:20000; Boster, China; #BA1054) and the resulting net optical density was analyzed using the Image-pro Plus 6.0 gel Image processing system. Visualization of the bands was achieved through the use of the ECL system (Affinity Biosciences, USA) and β-actin was used as an internal control.
Dual luciferase assay
A portion of the RRM2 3′ UTR and circ_0043256 3′ UTR, which includes the predicted binding site for miR-593-5p, was amplified through PCR with specific primers. The resulting PCR product was then inserted into the pSI-Check2 vector, which contains the luciferase coding sequence. Cells were plated in 96-well plates at a density of 50-70% per well and transfected with either the RRM2 3′ UTR-wt, LOX 3′ UTR-mut, circ_0043256 3′ UTR-wt, or circ_0043256 3′ UTR-mut construct, along with either the miR-593-5p mimic or its normal control (NC). Luciferase activity was measured using the Dual-Luciferase system 24 h post-transfection.
Cell proliferation assay
The cell vitality of MKN-45 and AGS cells was monitored using a Cell Counting Kit-8 (CCK-8, Thermo Fisher Scientific) according to the manufacturer’s instructions. The cell vitality was measured using an enzyme-linked immune monitor at 450 nm.
Wound healing assay
The migration of MKN-45 and AGS cells was evaluated using the Wound Healing Assay methodology. Cells were cultured in a 6-well plate until reaching 90% confluence, at which point a sterile micropipette tip was used to create a wound in the cell monolayers. The wounded monolayers were then washed with phosphate buffered saline (PBS) to eliminate cellular debris. The distance between the wound edges was measured at three distinct locations, with a subsequent measurement taken 48 h post-wounding to assess cell migration.
Transwell assay
Transwell filters (BD Biosciences, USA) were filled with pre-chilled serum-free medium at 4 °C to establish the chamber. MKN-45 and AGS cells were suspended in serum-free medium (3.0 × 105/mL) and seeded into the upper chamber. A culture medium containing 10% FBS was added to the basolateral chamber and incubated at 37 °C for 24 h. Subsequently, the transwell cultures were rinsed twice with sterile PBS, fixed with methanol for 30 min, and stained with 0.1% crystal violet for 30 min at room temperature. Cell invasion was observed using an Olympus X51 inverted microscope (Japan).
Cell apoptosis assay
The cell apoptosis was measured by flow cytometry. MKN-45 and AGS cells were centrifuged at 250 × g for 5 min to collect the precipitate, which was then cooled with PBS at 4℃ and subjected to another round of centrifugation at 250 × g for 5 min. This process was repeated twice. After resuspending the cells in 500 µL binding buffer solution, they were treated with 5µL Annexin V-FTC solution. Following a dark reaction period of 5 min, the cells were supplemented with 5 ml PI solution and incubated for an additional 5 min. Apoptosis was subsequently detected by flow cytometry (Cytoflex, Beckman, USA).
Cell cycle assay
The cell cycle was assessed using flow cytometry. MKN-45 and AGS cells were washed with PBS, digested with pancreatic enzymes, and collected. Subsequently, 1 mL of pre-cooled 75% ethanol was added to the cells and left overnight for fixation. The cells were then centrifuged at 350 × g for 5 min, the supernatant was discarded, and the cells were washed with PBS at 4 ℃. Finally, the cells were resuspended in 500 µL of PI/RNase A staining solution and analyzed by flow cytometry after incubating for 30 min at room temperature in darkness.
Biochemical detection
The contents of ATP production (#A095-1-1), glucose consumption (#A154-1-1) and lactic acid (#A019-2-1) in MKN-45 and AGS cells or tumor tissues were measured with using biochemical kits (Nanjing Jiancheng, China), following the manufacturer’s instructions.
Animals
Twenty-four female nude mice (SPF grade, 4 weeks old) were obtained from Dashuo Animal Experiment Co., Ltd. (China). The mice were housed in a controlled environment with a temperature of 24 ± 1 °C, relative humidity of 55 ± 5%, and a 12-h light/dark cycle. They had ad libitum access to food and water. The animal experimental procedures were approved by the Experimental Animal Ethics Committee, West China Hospital, Sichuan University (20240617005) and conducted in accordance with established guidelines and regulations, as outlined in the ARRIVE guidelines for reporting animal experiments (https://arriveguidelines.org/).
In vivo tumorigenesis test
The BALB/c nude mice were randomly divided into 4 groups (n = 6), namely the nomoxic group, hypoxia group, adeno-associated virus (AAV)-shRNA-NC group, and AAV-shRNA-circ_0043256 group. The MKN-45 cells were transfected with AAV-shRNA-NC or shRNA-circ_0043256 after 48 h of hypoxia (5% O2, 5% CO2, 90% N2). Subsequently, the cells were cultured at a density of 3.0 × 105 cells/mL for 48 ho at a temperature of 37 °C and humidity level of 100%. Following digestion with pancreatic enzyme, a total of 1.0 × 108 cells per nude BALB/C mice were inoculated into the lateral subcutaneous skin of their lower extremities. In contrast, MKN-45 cells cultured under normal oxygen conditions were inoculated in the nomoxic group. The tumor volume of the mice was monitored weekly and euthanized after 28 days by inhaling carbon dioxide in a euthanasia chamber. Finally, photographs of the tumor were taken.
Immunohistochemistry (IHC) staining
The paraffin blocks were sectioned at a thickness of 4 μm, followed by IHC staining to identify protein expression within the tumor tissues. The levels of Ki-67 and PCNA protein expression were evaluated in accordance with the guidelines outlined in the IHC protocol.
Statistical analysis
The data in this study were presented as means ± standard deviation (SD), with each experiment conducted in triplicate. Statistical analysis was carried out using SPSS software (version 16.0, SPSS Inc., USA), utilizing a one-way analysis of variance (ANOVA). In cases where the variance was equal, the LSD method was employed for comparing between two groups, while the Dunnett t method was used when the variance was not equal. A P-value of less than 0.05 was deemed to be statistically significant.
Results
Circ_0043256 was up-regulated in GC tissues and CoCl2-induced MKN-45 and AGS cells
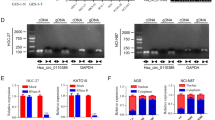
The expression analysis of hsa_circ_RNAs in GC tissues versus normal tissues was performed utilizing the GSE78092 dataset. This dataset was carefully curated from the Gene Expression Omnibus (GEO) database following an extensive screening process of circRNA sequencing data pertinent to GC. The findings revealed a significant up-regulation of hsa_circ_0043256 in GC tissues relative to normal tissues (Fig. 1A). Subsequently, we employed RT-qPCR to assess the expression levels of the top 25 up-regulated circ_RNAs in GC tissues and adjacent non-tumor tissues. As illustrated in Fig. 1B, circ_0043256 exhibited the most pronounced up-regulation in GC tissues. Consequently, we increased the sample size to include 30 GC tissue specimens to further evaluate the expression level of circ_0043256, obtaining consistent findings (Fig. 1C). Furthermore, the expression level of circ_0043256 was found to be elevated in various GC cell lines compared to normal gastric mucosa epithelial cells (Fig. 1D). This elevation was most pronounced in AGS and MKN-45 cells, which were subsequently selected for further experimental analysis (Fig. 1D).
Circ_0043256 was up-regulated in GC tissues and CoCl2-induced MKN-45 and AGS cells. (A) Expression of hsa_circ_0043256 in GC tissues and normal tissues showed by GSE78092 database. (B) The expression of the top 25 up-regulated circ_RNAs in tumor tissues and adjacent non-tumor tissues was assayed by RT-qPCR analysis. Numbers indicate mean. (C) The expression of circ_0043256 in tumor tissues and adjacent non-tumor tissues was measured by RT-qPCR analysis. ##P < 0.01 (vs. Control). (D) The expression of circ_0043256 in the control human gastric mucosal epithelial cells GES-1, the human GC cell lines MKN-45, AGS, SNU-16, SNU-216, HGC-27, and MKN-7 (#CL-0574) was tested by RT-qPCR analysis. *P < 0.05, **P < 0.01 (vs. GES-1). (C) The expression of circ_0043256 was measured by RT-qPCR analysis.**P < 0.01 (vs. siRNA-NC), ##P < 0.01 (vs. CoCl2 + siRNA-NC).
Circ_0043256 regulated CoCl2-induced proliferation, migration, apoptosis, and aerobic glycolysis of MKN-45 and AGS cells
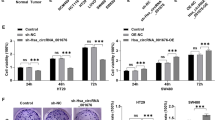
MKN-45 and AGS cell lines were subjected to varying concentrations of CoCl2 (50 µM, 100 µM, 200 µM, and 300 µM) to simulate hypoxic conditions. As illustrated in Fig. 2A,C, CoCl2 treatment resulted in the up-regulation of glycolysis-related proteins, specifically GLUT1 and HK2. Notably, the concentration of 100 µM CoCl2 was most efficacious in enhancing glycolytic activity in both MKN-45 and AGS cells (Fig. 2A,C). Furthermore, varying concentrations of CoCl2 were found to enhance the expression levels of circ_0043256 in MKN-45 and AGS cell lines. Notably, a concentration of 100 µM CoCl2 was the most effective in upregulating circ_0043256 expression in both cell types (Fig. 2D). These findings suggest that circ_0043256 may contribute to the hypoxia-induced progression of MKN-45 and AGS cells. Then, MKN-45 and AGS cells were transfected with si-circ_0043256 and treated with 100 µM CoCl2. Treatment with CoCl2 enhanced the expression of circ_0043256, whereas circ_0043256 level were reduced in cells treated with both CoCl2 and si-circ_0043256 (Fig. 2E). CCK-8 detection and Wound Healing assay demonstrated that treatment with CoCl2 markedly enhanced cell proliferation and migration, which was effectively reversed by silencing circ_0043256 (Fig. 2F,J). Flow cytometry analysis revealed that circ_0043256 silencing increased the rate of cell apoptosis and induced G2/M phase cell cycle arrest of MKN-45 and AGS cells (Fig. 2K,P). Conversely, CoCl2 treatment reduced the apoptosis rate and G2/M phase cell cycle arrest, both of which were significantly counteracted by silencing circ_0043256 (Fig. 2K,P).
Circ_0043256 regulated CoCl2-induced proliferation, migration, and apoptosis of MKN-45 and AGS cells. MKN-45 and AGS cells were exposed to different concentrations (50 µM, 100 µM, 200 µM, and 300 µM) of CoCl2 to induce a hypoxic environment. (A–C) Western blot analysis was performed to detect the expression levels of glycolytic related proteins HK2 and GLUT1 in MKN-45 and AGS cells induced by different concentrations of CoCl2. β-actin is a loading control. (D) The expression of si-circ_0043256 was quantified using RT-qPCR analysis. *P < 0.05 (vs. 0 µM), **P < 0.01 (vs. 0 µM). (E) MKN-45 and AGS cells were transfected with si-circ_0043256 and treated with CoCl2. The expression level of si-circ_0043256 was determined through RT-qPCR analysis (F and G) The proliferation of MKN-45 and AGS cells were evaluated by CCK-8 assay. (H–J) MKN-45 and AGS cell migration was tested by Wound-healing assay. Experiments were terminated after scratching for 48 h. (K-M) Flow cytometry was used to detect apoptosis and measure the rate of apoptosis. (N-P) The cell cycle of MKN-45 and AGS cells was examined by flow cytometry. *P < 0.05 (vs. siRNA-NC), **P < 0.01 (vs. siRNA-NC), ##P < 0.01 (vs. CoCl2 + siRNA-NC).
In terms of cellular metabolism, the silencing of si-circ_0043256 significantly impeded ATP production, glucose consumption, and lactic acid content (Fig. 3A,B). However, under CoCl2 treatment, the inhibition caused by si-circ_0043256 silencing on ATP production, glucose consumption, and lactic acid production capacity was notably counteracted (Fig. 3A,B). In addition, the expression levels of key proteins associated with aerobic glycolysis were assessed by Western blot analysis. The results demonstrated that si-circ_0043256 significantly suppressed the expression levels of HIF-1α, HK2, ENO1, LDHA, GLUT1 and PKM2 (Fig. 3C,F). Treatment with CoCl2 enhanced the expression of these proteins, whereas their levels were reduced in cells treated with both CoCl2 and si-circ_0043256 (Fig. 3C,F). These findings suggest that CoCl2 treatment enhanced aerobic glycolysis by up-regulating the expression of these pivotal proteins, while silencing circ_0043256 partially inhibited this effect.
Circ_0043256 regulated CoCl2-induced aerobic glycolysis of MKN-45 and AGS cells. MKN-45 and AGS cells were transfected with si-circ_0043256 and treated with CoCl2. (A and B) The ATP production, glucose consumption, and lactic acid content in CoCl2-induced MKN-45 and AGS cells were detected by commercial kits. (C and D) The protein levels of HIF-1α, HK2, ENO1, LDHA, GLUT1, and PKM2 in AGS cells were determined via Western Blot analysis. β-actin is a loading control. (E and F) The protein levels of HIF-1α, HK2, ENO1, LDHA, GLUT1, and PKM2 in MKN-45 cells were measured by Western Blot analysis. β-actin is a loading control. *P < 0.05 (vs. siRNA-NC), **P < 0.01 (vs. siRNA-NC), #P < 0.05 (vs. CoCl2 + siRNA-NC), ##P < 0.01 (vs. CoCl2 + siRNA-NC).
Circ_0043256 targeted miR-593-5p/RRM2axis
The targeting relationship between circ_0043256 and miR-593-5p was predicted using circMine database (http://www.biomedical-web.com/circmine/circRNA-miRNA_prediction, Fig. 4A). We found that miR-593-5p might interact with circ_0043256 3’ UTR (Fig. 4A). As shown in Fig. 4B, miR-593-5p mimic transfection decreased the luciferase activity of circ_0043256-WT reporter in 293T cells. As shown in Fig. 4C, the expression of miR-593-5p was down-regulated in GC tissues. In MKN-45 and AGS cells, treatment with CoCl2 reduced the expression of miR-593-5p, whereas miR-593-5p level were reduced by the transfection of si-circ_0043256 (Fig. 4D). Furthermore, by using bioinformatics predictions TargetScanHuman 8.0 (https://www.targetscan.org/), we identified that miR-593-5p could bind with RRM2 (Fig. 4E). miR-593-5p mimic suppressed the luciferase activity of the RRM2 wild type (WT) reporter, but not that of mutant (mut) reporter in 293T cells (Fig. 4F). As shown in Fig. 4G, the mRNA level of RRM2 was up-regulated in GC tissues. In MKN-45 and AGS cells, Treatment with CoCl2 enhanced the expression of RRM2, whereas RRM2 level were reduced in cells treated with both CoCl2 and si-circ_0043256 (Fig. 4H,I).
Circ_0043256 targeted miR-593-5p/RRM2axis. (A) The targeting relationship between circ_0043256 and miR-593-5p was predicted using circMine database (http://www.biomedical-web.com/circmine/circRNA-miRNA_prediction). Diagram of the predicted miR-593-5p binding site in the 3’UTR sequences of circ_0043256. (C) The luciferase activity in 293T cells transfected with miR-593-5p mimic or normal control (NC) mimic. **P < 0.01 (vs. NC mimic). (D) The expression of miR-593-5p was measured by RT-qPCR analysis. (E) miRNA target prediction software (TargetScanHuman 8.0, https://www.targetscan.org/) was used to identify potential miR-593-5p target genes. (F) The luciferase activity in 293T cells transfected with miR-593-5p mimic or NC mimic. **P < 0.01 (vs. NC mimic). (G) The mRNA expression of RRM2 was measured by RT-qPCR analysis. (H) The protein levels of RRM2 were measured by Western Blot analysis. β-actin is a loading control. *P < 0.05 (vs. Con), **P < 0.01 (vs. Con),#P < 0.05 (vs. CoCl2 + siRNA-NC), ##P < 0.01 (vs. CoCl2 + siRNA-NC).
Circ_0043256 regulated CoCl2-induced proliferation, migration, apoptosis, and aerobic glycolysis of MKN-45 and AGS cells by targeting the miR-593-5p/RRM2 axis
MKN-45 and AGS cells were transfected with si-circ_0043256 and/or miR-593-5p mimic to investigate their effects on CoCl2-induced cellular responses. RT-qPCR analysis revealed that circ_0043256 knockdown led to a significant increase in miR-593-5p levels in both MKN-45 and AGS cells, with further enhancement observed upon co-transfection with the miR-593-5p mimic (Fig. 5A). CCK-8 and Wound Healing assays demonstrated that circ_0043256 silencing resulted in a notable inhibition of cell proliferation and migration in CoCl2-induced MKN-45 and AGS cells, which was further attenuated by co-transfection with the miR-593-5p mimic (Fig. 5B,F). Flow cytometry analysis demonstrated that the downregulation of circ_0043256 resulted in a heightened level of cellular apoptosis and prompted G2/M phase cell cycle arrest in CoCl2-induced MKN-45 and AGS cells. These effects were further augmented by the co-transfection of miR-593-5p mimic, as illustrated in Fig. 5J,L.
Circ_0043256 regulated CoCl2-induced proliferation, migration, and apoptosis of MKN-45 and AGS cells by targeting the miR-593-5p/RRM2 axis. MKN-45 and AGS cells were transfected with si-circ_0043256 and/or miR-593-5p mimic to investigate their effects on CoCl2-induced cellular responses. (A) The expression of miR-593-5p was quantified using RT-qPCR analysis. (B and C) Cell proliferation in MKN-45 and AGS cells was assessed by the CCK-8 assay. (D-F) Wound-healing assay was performed to evaluate cell migration in MKN-45 and AGS cells. Experiments were terminated 48 h after scratching. (G-I) Flow cytometry was employed to detect apoptosis and determine the apoptotic rate. (J-L) Cell cycle analysis of MKN-45 and AGS cells was conducted using flow cytometry. **P < 0.01 (vs. siRNA-NC), ##P < 0.01 (vs. CoCl2 + siRNA-NC), &P < 0.05 (vs. CoCl2 + si-circ_0043256 + mimic-NC), &&P < 0.01 (vs. CoCl2 + si-circ_0043256 + mimic-NC).
The silencing of si-circ_0043256 significantly decreased ATP production, glucose consumption and lactic acid content in CoCl2-induced MKN-45 and AGS cells, as illustrated in Fig. 6A,B. Additionally, the suppression of ATP production, glucose consumption, and lactic acid production capacity resulting from si-circ_0043256 silencing was further exacerbated by co-transfection with the miR-593-5p mimic (Fig. 6A,B). Furthermore, our findings indicated that si-circ_0043256 effectively down-regulated the expression of HIF-1α, HK2, ENO1, LDHA, GLUT1, and PKM2 in CoCl2-induced MKN-45 and AGS cells. This down-regulation was further attenuated by co-transfection with a miR-593-5p mimic (Fig. 6C,F).
Circ_0043256 regulated CoCl2-induced aerobic glycolysis of MKN-45 and AGS cells by targeting the miR-593-5p/RRM2 axis. MKN-45 and AGS cells were transfected with si-circ_0043256 and/or miR-593-5p mimic to investigate their effects on CoCl2-induced aerobic glycolysis. (A and B) The ATP production, glucose consumption, and lactic acid content in CoCl2-induced MKN-45 and AGS cells were assessed using commercially available kits. (C and D) The protein levels of HIF-1α, HK2, ENO1, LDHA, GLUT1, and PKM2 in AGS cells were determined by Western Blot analysis with β-actin serving as a loading control. (E and F) The protein levels of HIF-1α, HK2, ENO1, LDHA, GLUT1, and PKM2 in MKN-45 cells were measured via Western Blot analysis. β-actin is a loading control. **P < 0.01 (vs. siRNA-NC), #P < 0.05 (vs. CoCl2 + siRNA-NC), ##P < 0.01 (vs. CoCl2 + siRNA-NC), &P < 0.05 (vs. CoCl2 + si-circ_0043256 + mimic-NC), &&P < 0.01 (vs. CoCl2 + si-circ_0043256 + mimic-NC).
DG further enhanced the inhibitory effect of circ_0043256 silencing on cell progression of CoCl2-induced MKN-45 and AGS cells
The MKN-45 and AGS cells were transfected with si-circ_0043256 and treated with 2-DG. CCK-8 and Wound Healing assays demonstrated that silencing of circ_0043256 resulted in a significant inhibition of cell proliferation and migration in CoCl2-induced MKN-45 and AGS cells, which was further attenuated by co-treatment with 2-DG (Fig. 7A,E). Flow cytometry analysis revealed that silencing of circ_0043256 led to an increased level of cellular apoptosis and induced G2/M phase cell cycle arrest in CoCl2-induced MKN-45 and AGS cells. These effects were further enhanced by co-treatment with 2-DG, as depicted in (Fig. 7F,K).
2-DG further enhanced the inhibitory effect of circ_0043256 silencing on cell progression of CoCl2-induced MKN-45 and AGS cell. The MKN-45 and AGS cells were transfected with si-circ_0043256 and treated with 2-DG. (A and B) Cell proliferation in MKN-45 and AGS cells was assessed by the CCK-8 assay. (C–E) Wound-healing assay was performed to evaluate cell migration in MKN-45 and AGS cells. Experiments were terminated 48 h after scratching. (F–H) Flow cytometry was employed to detect apoptosis and determine the apoptotic rate. (I–K) Cell cycle analysis of MKN-45 and AGS cells was tested using flow cytometry. **P < 0.01 (vs. siRNA-NC), ##P < 0.01 (vs. CoCl2 + siRNA-NC), &P < 0.05 (vs. CoCl2 + 2-DG + siRNA-NC), &&P < 0.01 (vs. CoCl2 + 2-DG + siRNA-NC).
As shown in Fig. 8A,B. the knockdown of si-circ_0043256 significantly reduced ATP production, glucose consumption, and lactic acid content in CoCl2-induced MKN-45 and AGS cells. Moreover, the inhibition of ATP production, glucose consumption, and lactic acid production capacity resulting from si-circ_0043256 knockdown was further exacerbated by co-treatment with 2-DG (Fig. 8A,B). Furthermore, our findings demonstrated that si-circ_0043256 effectively down-regulated the protein expression of HIF-1α, HK2, ENO1, LDHA, GLUT1, and PKM2 in CoCl2-induced MKN-45 and AGS cells. This down-regulation was further attenuated by co-treatment with 2-DG (Fig. 8C,F).
2-DG further enhanced the inhibitory effect of circ_0043256 silencing on aerobic glycolysis of CoCl2-induced MKN-45 and AGS cells. The MKN-45 and AGS cells were transfected with si-circ_0043256 and treated with 2-DG. (A and B) The ATP production, glucose consumption, and lactic acid content in CoCl2-induced MKN-45 and AGS cells were assessed using commercially available kits. (C and D) The protein levels of HIF-1α, HK2, ENO1, LDHA, GLUT1, and PKM2 in AGS cells were determined by Western Blot analysis with β-actin serving as a loading control. (E and F) The protein levels of HIF-1α, HK2, ENO1, LDHA, GLUT1,and PKM2 in MKN-45 cells were measured via Western Blot analysis. β-actin is a loading control. *P < 0.05 (vs. siRNA-NC), **P < 0.01 (vs. siRNA-NC), ##P < 0.01 (vs. CoCl2 + siRNA-NC), &&P < 0.01 (vs. CoCl2 + 2-DG + siRNA-NC).
The silencing of circ_0043256 inhibited tumor growth and glycolysis through the miR-593-5p/RRM2 axis
The silencing of circ_0043256 decreased tumor volume in nude mice (Fig. 9A,B). Meanwhile, the expression of a proliferation marker protein Ki67 and PCNA was decreased by silencing circ_0043256 (Fig. 9C,D). Our study further confirmed that the silencing of circ_0043256 inhibited the level of circ_0043256 and promoted the level of miR-593-5p in tumor tissues. IHC stain of the tumor tissues demonstrated that the positive rate of RRM2 in the tumor tissues was greatly decreased in the AVV-shRNA-circ_0043256-treated group compared with the control group (Fig. 9G,H). In addition, the production of ATP, glucose consumption, and lactic acid production capacity of tumor tissues were suppressed by circ_0043256 silencing (Fig. 8I). Western Blot analysis showed that the silencing of circ_0043256 decreased the expressions of HIF-1α, HK2, ENO1, LDHA, GLUT1, and PKM2 in tumor tissues compared with the control group (Fig. 9J,K).
The silencing of circ_0043256 inhibited tumor growth and glycolysis through the miR-593-5p/RRM2 axis. (A and B) Compared with the control group, circ_0043256 silencing inhibited the tumor weight. (C and D) Western blot was used to assay determined Ki67 and PCNA. β-actin is a loading control. (E) The expression of circ_0043256 was performed through RT-qPCR analysis. (F) The expression of miR-593-5p was quantified using RT-qPCR analysis. (G and H) IHC stain of Ki67 and PCNA, magnification is 400×. (I–K) Western blot was used to assay determined HIF-1α, HK2, ENO1, LDHA, GLUT1, and PKM2 expression in tumor tissues. **P < 0.05 (vs. Normoxic), #P < 0.05 (vs. AAV-shRNA-NC), ##P < 0.01 (vs. AAV-shRNA-NC).
Discussion
Numerous studies have confirmed the aberrant expression of circRNA in various tumor types, suggesting their potential utility in cancer diagnosis, monitoring, prognosis, and predicting therapeutic response14,15. The identification of multiple circRNAs in GC offers novel insights into the molecular pathways underlying GC pathogenesis16. In the early stage of this study, data mining showed that circ_0043256 was significantly up-regulated in GC tissues, indicating its potential use as an independent prognostic biomarker for GC.
In recent years, it has been widely acknowledged that cancer cells demonstrate a distinct metabolic phenotype characterized by heightened glucose uptake and lactate release to facilitate their malignant biological activities17. Numerous investigations have highlighted the close association between circRNA and the modulation of the Warburg effect, which sustains the proliferation and viability of cancer cells18,19,20. A representative study showed that circ_LDLR drove the proliferation and aerobic glycolysis of GC cells by targeting CHD1 with miR-449b-5p21. Moreover, the suppression of circ_VPS33B attenuated the Warburg effect and inhibited tumor growth by modulating the miR-873-5p/HNRNPK axis in infiltrative GC22. In accordance with prior research on circRNAs, our study demonstrated that circ_0043256 silencing acts as a tumor suppressor gene by inhibiting tumor aggressiveness and influencing cellular energy metabolism. circ_0043256 silencing regulated metabolism by downregulating glycolytic gene expression and enzyme activity, thus inhibiting glycolysis in vitro and in vivo. In addition, the glycolytic inhibitor 2-DG further reduced the glycolysis of GC cells silenced by circ_0043256.
MicroRNAs (miRNAs) are a class of endogenous noncoding RNAs typically consisting of approximately 22 nucleotides in length, known to modulate protein expression by inhibiting mRNA translation23. Previous research has established the significant involvement of miRNAs in the progression of various diseases at the cellular level24,25. Given their association with the initiation and development of numerous cancers, miRNAs have been recognized as oncogenes or tumor-suppressing genes that play a role in tumorigenesis26,27. The study confirmed that circRNA is able to bind to specific miRNA functional sequences, thereby inhibiting miRNA regulation of mRNA and indirectly influencing mRNA expression to regulate the cancer progression28,29. Specifically, our study showed that miR-593-5p, a validated tumor-suppressing gene in human glioma30 and lung adenocarcinoma31, was found to bind to circ_0043256 and exhibit low expression in GC cells. Similarly, miR-593-5p was frequently downregulated in GC patients and GC cell lines. miR-593-5p was significantly correlated with tumor size and distant metastasis in GC patients32. Additionally, we found that circ_0043256 could negatively regulate miR-593-5p expression. Moreover, miR-593-5p up-regulation could retard GC progression.
Ribonucleotide Reductase M2 (RRM2) serves as a pivotal subunit of the enzyme Ribonucleotide Reductase (RNR) essential for DNA synthesis and repair processes33,34. Specifically, RRM2 facilitates the conversion of ribonucleotides to deoxyribonucleotides, the fundamental components of DNA. RRM2 plays a critical role in regulating the cell cycle and isabnormally expressed in diverse cancer types, rendering it a promising target for anticancer interventions33,35,36. Prior research demonstrated that the overexpression of RRM2 in GC cells enhanced their invasiveness through the activation of the AKT/NF-κB signaling pathway37. Overexpression of RRM2 exhibited a significant association with tumor invasion, survivin expression, and DNA methyltransferase levels in GC patients38. Conversely, RRM2 silencing hindered the proliferation of GC cell lines MKN-1, MKN-7, and SNU-719, indicating a potential role of RRM2 in the advancement of GC38. Serum RRM2 has been identified as a promising diagnostic biomarker for liver cancer, as it plays a crucial role in maintaining intracellular glutathione levels by preventing the degradation of glutathione synthetase39. This mechanism ultimately contributes to the anti-iron death function observed in liver cancer cells39. Moreover, inhibition of RRM2 had the potential to augment the anticancer efficacy of chemotherapy, serving as a potential indicator of chemosensitivity and prognosis40. Similarly, this study showed that the expression of RRM2 in gastric cancer tissues was significantly increased, and circ_0043256 silencing inhibited the expression of RRM2 in GC cells by up-regulating the expression of miR-593-5p.
In summary, our findings indicated that the suppression of circ_0043256 effectively hindered the proliferation, migration, and glycolysis induced by CoCl2 in GC cells by modulating the miR-593-5p/RRM2 pathway. These results suggest that circ_0043256 has the potential to be utilized as a therapeutic target for gastric cancer.
Data availability
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
References
Smyth, E. C., Nilsson, M., Grabsch, H. I., van Grieken, N. C. & Lordick, F. Gastric cancer. Lancet (London, England) 29 (10251), 635–648. https://doi.org/10.1016/s0140-6736(20)31288-5 (2020).
Yang, T. & Guo, L. Advancing gastric cancer treatment: Nanotechnology innovations and future prospects. Cell. Biology Toxicol. 20 (1), 101. https://doi.org/10.1007/s10565-024-09943-9 (2024).
Beeharry, M. K., Liu, W. T., Yan, M. & Zhu, Z. G. New blood markers detection technology: A leap in the diagnosis of gastric cancer. World J. Gastroenterol. 21 (3), 1202–1212. https://doi.org/10.3748/wjg.v22.i3.1202 (2016).
Khalayleh, H., Kim, Y. W., Yoon, H. M. & Ryu, K. W. Assessment of Lymph Node Metastasis in patients with gastric cancer to identify those suitable for middle segmental gastrectomy. JAMA Netw. Open. 1 (3), e211840. https://doi.org/10.1001/jamanetworkopen.2021.1840 (2021).
Takahashi, K. & Yoshikawa, T. Different risks of nodal metastasis by tumor location in remnant gastric cancer after curative gastrectomy for gastric cancer. Gastric Cancer 23 (1), 195–201. https://doi.org/10.1007/s10120-019-00989-x (2020).
Kim, K. T. & Lee, J. E. Deciphering metastatic route-specific signals and their microenvironment interactions in peritoneal metastasis of gastric cancer. Cancer Commun. (Lond.) 44 (4), 514–517. https://doi.org/10.1002/cac2.12533 (2024).
Chalabi, M. Stomach cancer gets a triple punch of therapy. Nature 600 (7890), 608–609. https://doi.org/10.1038/d41586-021-03458-7 (2021).
Patop, I. L., Wüst, S. & Kadener, S. Past, present, and future of circRNAs. EMBO J. 15 (16), e100836. https://doi.org/10.15252/embj.2018100836 (2019).
Zhou, W. Y. et al. Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 14 (1), 172. https://doi.org/10.1186/s12943-020-01286-3 (2020).
Zhang, X. et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Molecular Cancer 4 (1), 20. https://doi.org/10.1186/s12943-018-0935-5 (2019).
Chen, D. L. et al. The circular RNA circDLG1 promotes gastric cancer progression and anti-PD-1 resistance through the regulation of CXCL12 by sponging miR-141-3p. Mol. Cancer 15 (1), 166. https://doi.org/10.1186/s12943-021-01475-8 (2021).
Jiang, S. F. & Li, R. R. hsa_circ_0067514 suppresses gastric cancer progression and glycolysis via miR-654-3p/LATS2 axis. Neoplasma 69 (5), 1079–1091. https://doi.org/10.4149/neo_2022_220225N209 (2022).
Zhou, Y., Liu, H., Wang, R. & Zhang, M. Circ_0043256 upregulates KLF2 expression by absorbing miR-1206 to suppress the tumorigenesis of lung cancer. Thorac Cancer 14 (7), 683–699. https://doi.org/10.1111/1759-7714.14794 (2023).
Chen, L. & Shan, G. CircRNA in cancer: Fundamental mechanism and clinical potential. Cancer Lett. . 505, 49–57. https://doi.org/10.1016/j.canlet.2021.02.004 (2021).
Kristensen, L. S., Jakobsen, T., Hager, H. & Kjems, J. The emerging roles of circRNAs in cancer and oncology. Nat. Reviews Clin. Oncol. 19 (3), 188–206. https://doi.org/10.1038/s41571-021-00585-y (2022).
Li, R. et al. CircRNA: A rising star in gastric cancer. Cell. Mol. life Sciences: CMLS . 77 (9), 1661–1680. https://doi.org/10.1007/s00018-019-03345-5 (2020).
Yu, T. et al. CircRNAs in cancer metabolism: A review. J. Hematol. Oncol. 4 (1), 90. https://doi.org/10.1186/s13045-019-0776-8 (2019).
Wang, Y., Chen, X. & Yang, Y. CircRNA-regulated glucose metabolism in ovarian cancer: An emerging landscape for therapeutic intervention. Clin. Transl. Oncol; 26(3):584–596. https://doi.org/10.1007/s12094-023-03285-4 (2024).
Huang, Y., Zheng, S., Lin, Y. & Ke, L. Circular RNA circ-ERBB2 elevates the Warburg Effect and facilitates triple-negative breast cancer growth by the MicroRNA 136-5p/Pyruvate dehydrogenase kinase 4 Axis. Mol. Cell. Biol.. 24 (10), e0060920. https://doi.org/10.1128/mcb.00609-20 (2021).
Wang, X. et al. Hsa_circ_0052611 and mir-767-5p guide the warburg effect, migration, and invasion of BRCA cells through modulating SCAI. J. Bioenergetics Biomembr. 55 (5), 381–396. https://doi.org/10.1007/s10863-023-09985-4 (2023).
Zeng, F. & Zhao, J. CircRNA LDLR promotes proliferation and aerobic glycolysis of gastric cancer cells by targeting CHD1 with miR-449b-5p. Turk J Biol.; 48(1):46–58. (2024). https://doi.org/10.55730/1300-0152.2681
Lu, Y. et al. Inhibition of circRNA circVPS33B reduces Warburg Effect and Tumor Growth through regulating the miR-873-5p/HNRNPK Axis in Infiltrative Gastric Cancer. Onco Targets Ther.. 14, 3095–3108. https://doi.org/10.2147/ott.s292575 (2021).
Buhagiar, A. F. & Kleaveland, B. To kill a microRNA: Emerging concepts in target-directed microRNA degradation. Nucleic Acids Res. 28 (4), 1558–1574. https://doi.org/10.1093/nar/gkae003 (2024).
Pozniak, T. & Shcharbin, D. Circulating microRNAs in Medicine. Int. J. Mol. Sci.https://doi.org/10.3390/ijms23073996 (2022).
Plotkin, L. I. & Wallace, J. M. MicroRNAs and osteocytes. Bone 150, 115994. https://doi.org/10.1016/j.bone.2021.115994 (2021).
Zhang, C. et al. Overview of MicroRNAs as diagnostic and prognostic biomarkers for high-incidence cancers in 2021. Int. J. Mol. Sci. 23 (19). https://doi.org/10.3390/ijms231911389 (2022).
Jothimani, G., Bhatiya, M., Pathak, S., Paul, S. & Banerjee, A. Tumor suppressor microRNAs in gastrointestinal cancers: A Mini-review. Recent. Adv. Inflamm. Allergy Drug Discov.. 16 (1), 5–15. https://doi.org/10.2174/2772270816666220606112727 (2022).
Li, H. et al. Circular RNA circRNA_000203 aggravates cardiac hypertrophy via suppressing miR-26b-5p and mir-140-3p binding to Gata4. Cardiovasc. Res. . 1 (7), 1323–1334. https://doi.org/10.1093/cvr/cvz215 (2020).
Zhao, X., Zhong, Y., Wang, X., Shen, J. & An, W. Advances in circular RNA and its applications. Int. J. Med. Sci. 19 (6), 975–985. https://doi.org/10.7150/ijms.71840 (2022).
Zhang, R. et al. Long noncoding RNA ENST00000413528 sponges microRNA-593-5p to modulate human glioma growth via polo-like kinase. CNS Neurosci Ther. 25(8):842–854. (2019). https://doi.org/10.1111/cns.13121
Zhang, H. B. et al. MiR-593-5p inhibited proliferation and migration of lung adenocarcinoma by targeting ICAM-1. Eur. Rev. Med. Pharmacol. Sci. 24(8):4298–4305. (2020). https://doi.org/10.26355/eurrev_202004_21010
Yu, H. et al. Regulation of cell proliferation and metastasis by microRNA-593-5p in human gastric cancer. OncoTargets Therapy. 11, 7429–7440. https://doi.org/10.2147/ott.s178151 (2018).
Zuo, Z. et al. Ribonucleotide reductase M2 (RRM2): Regulation, function and targeting strategy in human cancer. Genes Dis. 11 (1), 218–233. https://doi.org/10.1016/j.gendis.2022.11.022 (2024).
Shah, K. N., Wilson, E. A., Malla, R., Elford, H. L. & Faridi, J. S. Targeting Ribonucleotide reductase M2 and NF-κB activation with Didox to circumvent tamoxifen resistance in breast Cancer. Mol. cancer Ther.. 14 (11), 2411–2421. https://doi.org/10.1158/1535-7163.mct-14-0689 (2015).
Shi, S. C., Zhang, Y. & Wang, T. High RRM2 expression has poor prognosis in specific types of breast cancer. 17(3):e0265195. (2022). https://doi.org/10.1371/journal.pone.0265195
Jiang, H. et al. RRM2 mediates the anti-tumor effect of the natural product pectolinarigenin on glioblastoma through promoting CDK1 protein degradation by increasing autophagic flux. Front. Oncol. 12, 887294. https://doi.org/10.3389/fonc.2022.887294 (2022).
Zhong, Z., Cao, Y., Yang, S. & Zhang, S. Overexpression of RRM2 in gastric cancer cell promotes their invasiveness via AKT/NF-κB signaling pathway. Die Pharmazie. 71 (5), 280–284 (2016).
Morikawa, T. et al. Expression of ribonucleotide reductase M2 subunit in gastric cancer and effects of RRM2 inhibition in vitro. Hum. Pathol. 41 (12), 1742–1748. https://doi.org/10.1016/j.humpath.2010.06.001 (2010).
Yang, Y. et al. RRM2 protects against ferroptosis and is a tumor biomarker for liver cancer. Cancer Cell Int. 7 (1), 587. https://doi.org/10.1186/s12935-020-01689-8 (2020).
Zhan, Y. et al. Inhibiting RRM2 to enhance the anticancer activity of chemotherapy. Biomed. Pharmacotherapy = Biomedecine Pharmacotherapie 133, 110996. https://doi.org/10.1016/j.biopha.2020.110996 (2021).
Funding
This study was supported by a grant from the Zhanjiang City science and technology plan project (2022A01174) and Clinical study in Affiliated Hospital of Guangdong Medical University (LCYJ2018A001).
Author information
Authors and Affiliations
Contributions
Wenting Ou and Caijin Zhou conceived and designed the experiments. Wenting Ou, Rongjian Tan, Jiawei Zhai, Lijun Sun, Zhenhao Quan and Xianjin Huang performed the experiments. Wenting Ou, Rongjian Tan, Jiawei Zhai, Feipeng Xu, and Qingwen Xu analyzed the data. Caijin Zhou contributed to the reagents and materials. Caijin Zhou wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Approval of the research protocol by an institutional reviewer board
The study protocols were approved by the Experimental Animal Ethics Committee, West China Hospital, Sichuan University (20240617005).
Informed consent
All specimens were collected after obtaining informed consent from all patients.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ou, W., Tan, R., Zhai, J. et al. Silencing circ_0043256 inhibited CoCl2-induced proliferation, migration, and aerobic glycolysis in gastric cancer cells. Sci Rep 15, 171 (2025). https://doi.org/10.1038/s41598-024-84548-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-84548-0