Abstract
The pancreas is a gland in the abdomen that helps to produce hormones and digest food. The irregular development of tissues in the pancreas is termed as pancreatic cancer. Identification of pancreatic tumors early is significant for enhancing survival rate and providing appropriate treatment. Thus, an efficient Secretary Wolf Bird Optimization (SeWBO)_Efficient DenseNet is presented for pancreatic tumor detection using Computed Tomography (CT) scans. Firstly, the input pancreatic CT image is accumulated from a database and subjected to image preprocessing using a bilateral filter. After this, lesion is segmented by utilizing Parallel Reverse Attention Network (PraNet), and hyperparameters of PraNet are enhanced by using the proposed SeWBO. The SeWBO is designed by incorporating Wolf Bird Optimization (WBO) and the Secretary Bird Optimization Algorithm (SBOA). Then, features like Complete Local Binary Pattern (CLBP) with Discrete Wavelet Transformation (DWT), statistical features, and Shape Local Binary Texture (SLBT) are extracted. Finally, pancreatic tumor detection is performed by SeWBO_Efficient DenseNet. Here, Efficient DenseNet is developed by combining EfficientNet and DenseNet. Moreover, the proposed SeWBO_Efficient DenseNet achieves better True Negative Rate (TNR), accuracy, and True Positive Rate (TPR), of 93.596%, 94.635%, and 92.579%.
Similar content being viewed by others
Introduction
Pancreas is a crucial glandular organ at the bottom of the stomach, which plays an essential role in metabolism and digestion. The pancreas consists of two segments, such as endocrine and exocrine functions. In pancreas, the endocrine part generates a hormone, like glucagon and insulin, which are significant to regulate the blood sugar levels. Whereas, the exocrine part of the pancreas secretes and generates digestive enzymes, such as chymotrypsin and trypsin utilized to digest proteins, amylase to digest carbohydrates, and lipase is used to breakdown fats into the small intestine1. Pancreatic cancer is a malignant tumor, in which tissues grow abnormally and multiply out of control in the pancreas caused by cells mutation in its genes2. Moreover, pancreatic cancer is the most common tumor with rapid progression, high malignancy, and poor prognosis, Further, it is the seventh leading cause of cancer-related deaths worldwide3,4. The symptoms and signs of pancreatic tumor, comprise pain in the middle or upper abdomen, jaundice, loss of appetite, being always tired, and sudden weight loss for unknown reasons. Moreover, risk factors, such as alcohol consumption, smoking, diabetes, obesity, hypercholesterolemia, and pancreatic steatosis have been identified to cause pancreatic cancer4,5. Therefore, early detection of pancreatic cancer is essential to enhance patient survival and decrease the overall mortality rate6. On the other hand, pancreatic cancer exhibits a lack of particular clinical symptoms and becomes more complicated with early diagnosis and detection4.
Pancreatic cancer is considered as most fatal tumor, and screening programs are utilized for early diagnosis, risk stratification, treatment optimization, prognosis prediction, and research on early detection becomes necessary to move towards personalized medication7. Various image examination methods including CT, ultrasound scanning, Magnetic Resonance Imaging (MRI), and endoscopic investigation are utilized for detecting pancreatic cancer. Among the techniques mentioned above, CT images have relatively high resolution with less complication to humans and are utilized for early detection of pancreatic cancer, thus it is widely used in clinical settings. Accurate detection of pancreatic cancer earlier remains a challenge and is primarily reliant on image modalities. Nowadays, CT is more vital for the assessment and in treating pancreatic diseases. In addition, CT can be rapidly performed and provides detailed cross-sectional imaging for surrounding structures and the pancreas8. CT scans gather information about tumor size, morphology, and position effectively, thus they assist in the classification and diagnosis of pancreatic cancer compared to MRI and ultrasound imaging9. Furthermore, CT is an invaluable tool utilized for several pancreatic disease detection, such as chronic or acute pancreatitis and solid or cystic neoplastic lesions8. Due to the complexities during pancreatic cancer detection, the early diagnosis and treatment for pancreatic diseases are still unsatisfactory3. Besides, pancreatic cancer that occurs with regular contours and ill-defined margins is often unclear on CT and poses various challenges during detection at an early stage even for an experienced radiologist10.
Manual segmentation can generate less accurate outcomes, takes more time, and sometimes vulnerable to interobserver variability. Hence, to enhance the sensitivity of pancreatic cancer detection, an effective tool is required to support radiologists for proper treatment 11,12. Thus, an accurate, reliable, and objective technique is necessary for pancreatic cancer detection3. Therefore, a clinically applicable Computer-Aided Detection (CAD) tool is necessary for accurate classification and segmentation of pancreatic cancer with less human labor or annotation11. Moreover, Computer-Aided Diagnosis Systems (CADx) are used to produce accurate and automatic segmentation of pancreatic cancer involving thousands of abdominal medical scans for developing an essential correlation among organ volume, curvature, and anthropometric measures12. Nevertheless, establishing automatic algorithms for the segmentation and classification of pancreatic cancer is challenging because the pancreas shares boundaries with multiple organs and structures, and changes widely in shape and size, particularly in pancreatic cancer patients8. According to the severity of pancreatic tumors, generating CAD systems is crucial to detect cancerous from noncancerous cells. Subsequently, producing a advanced perceptive technique for pancreatic tumors is important. Convolutional Neural Network (CNN) models can address a wide variety of limitations related to pancreatic diseases effectively while classifying images by mining the features automatically from the images. Deep Learning (DL) is a branch of Machine Learning utilized to detect pancreatic cancer earlier by analyzing medical image data, especially CT scans13.
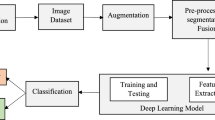
In this research, an optimization-enabled DL approach, called SeWBO_Efficient DenseNet is developed to detect the pancreatic cancer. Primarily, input CT images are accumulated and then forwarded to image pre-processing. Where the image is preprocessed by bilateral filter. Then, lesion segmentation is done utilizing PraNet, and hyperparameters of PraNet are adjusted to enhance the performance of lesion segmentation using SeWBO. Here, the SeWBO is developed by combining SBOA and WBO. Then, various features, such as SLBT, and CLBP with DWT, and statistical features are mined. Lastly, pancreatic tumor detection is executed by exploiting the newly devised SeWBO_Efficient DenseNet.
The primary contribution of this research is,
-
Devised SeWBO_Efficient DenseNet for pancreatic tumor detection: The SeWBO_Efficient DenseNet is established to perform pancreatic tumor detection using CT images. Here, Efficient DenseNet is developed by merging EfficientNet and DenseNet. Further, the Efficient DenseNet is structurally optimized by the SeWBO, which is attained by incorporating WBO and SBOA.
The rest of this research is organized as: The literature review of various models for pancreatic cancer detection is expounded in section "Motivation", the devised SeWBO_Efficient DenseNet technique is explained in segment 3, the outcomes of SeWBO_Efficient DenseNet are detailed in segment 4, and this work is concluded in segment 5.
Motivation
The literature review of different frameworks employed for detecting pancreatic cancer from CT images is depicted in this segment. Numerous approaches were proposed for pancreatic cancer detection, nevertheless, the ill-defined margin and irregular contours affect the performance of these methods, and the networks suffer from high computational complexity. Hence, there exists a need to establish a well-organized technique for pancreatic cancer detection.
Literature survey
Li et al.14 devised an Ensemble Learning-Support Vector Machine (EL-SVM) technique for preoperative diagnosis and staging of pancreatic cancer. This method solved the issues prevailing in preoperative pancreatic cancer diagnosis to a certain level. However, EL-SVM suffered from a high classification error after ensemble learning. Cao et al.15 developed a muti task CNN for detecting pancreatic cancer from CT images. This scheme was effectual in reducing false positives and was effective in performing detection at a large scale. Meanwhile, the technique was not validated in real-world practices. Dinesh et al.13 proposed CNN and YOLO model-based CNN (YCNN) approaches for the detection of pancreatic cancer based on pathological images. The learning rate of the CNN and YCNN was improved even in a variety of color variations. In contrast, this technique utilized a very small percentage of CT images, hence suffered from generalization issues. Vaiyapuri et al.16 Intelligent DL-Enabled Decision-Making Medical System- Pancreatic Tumor Classification (IDLDMS-PTC) method for pancreatic cancer classification with CT images. The IDLDMS-PTC method was exploited as an efficient instrument for a healthcare system and attained high accuracy with minimal parameters. However, this approach failed to consider techniques, such as deep instance segmentation to enhance outcomes.
Asadpour et al.17 developed a Cascaded multi-level method for pancreatic cancer analysis in CT images. Here, the Region of Interest (RoI) were attained by applying a threshold using Structured Forest Edge Detectors (SFED). Nevertheless, tumor shape region was not considered to enhance performance. Nakao et al.18 devised a Laplacian-based diffeomorphic shape matching (LDSM) technique for pancreatic cancer localization based on the shape features of multiple organs. Meanwhile, the technique was effective in maintaining a high stability rate during detection. In contrast, this approach was unsuccessful in representing non-linear deformation and complexity because of the influence of local shape errors and liver motion. Gandikota19 designed Tunicate Swarm Algorithm With Deep Learning-Based Pancreatic Cancer Segmentation And Classification (TSADL-PCSC). This method had good accuracy while classifying the pancreatic CT images. However, the method was futile in regarding deep ensemble techniques to enhance performance. Hooshangnejad et al.20 proposed Innovative DL for expeditious pancreatic cancer retinopathy in CT images. This scheme considerably advanced the workflow and reduced highly undesirable wait times. In contrast, this approach did not include high resolution and more data to attain high performance.
With the introduction of several deep learning techniques, recent developments in pancreatic CT image segmentation have greatly increased segmentation accuracy. A multi-scale attention network was shown by Zhang et al.8, improving segmentation accuracy despite class imbalance issues brought on by a lack of annotated pancreatic CT images. A hybrid transformer-based segmentation approach was presented by Chen et al.11, which shown gains in recall and precision but had trouble managing tiny tumor areas.
A Tunicate Swarm Algorithm with deep learning was used by Gandikota et al.19; it produced good accuracy but needed practical validation. Hooshangnejad et al.20 presented a novel deep learning technique for quick segmentation that greatly shortened processing times; nevertheless, in order to increase robustness, their system needed a bigger and more varied dataset. Furthermore, a patch-based multi-resolution CNN model was proposed by Asadpour et al.17, which shown limitations in recognizing irregular tumor boundaries but improved segmentation resolution. A Laplacian-based diffeomorphic shape matching method was presented by Nakao et al.18, which had good segmentation stability but struggled with local shape defects and non-linear deformations. Last but not least, Placido et al.6 integrated multi-modal imaging approaches with radiomics and deep learning, which enhanced segmentation accuracy but increased computing load. With many approaches demonstrating advantages and disadvantages, these studies demonstrate the continuous progress in pancreas CT image segmentation. Even though segmentation accuracy has significantly increased as a result of deep learning, issues including high processing demands, class imbalance, and model generalizability still exist. The need for more accurate, flexible, and efficient methods to improve clinical applications in pancreatic cancer diagnosis is highlighted by the ongoing development of segmentation models.
Comparision Survey:
Author | Techniques used | Performance metrics | Limitations |
|---|---|---|---|
Li et al. (2023) | Ensemble Learning-Support Vector Machine (EL-SVM) | Accuracy: 85.2%, Sensitivity: 82.5%, Specificity: 87.4% | High classification error after ensemble learning |
Cao et al.15 | Multi-task CNN for pancreatic cancer detection | F1-score: 90.1%, Sensitivity: 91.3%, Specificity: 89.7% | Not validated in real-world clinical applications |
Dinesh et al.13 | CNN and YOLO-based CNN (YCNN) | Classification accuracy: 88.4%, Precision: 86.9% | Limited generalization due to a small dataset |
Vaiyapuri et al.16 | Intelligent DL-enabled decision-making system (IDLDMS-PTC) | Segmentation accuracy: 87.2%, Detection accuracy: 89.5% | Lack of deep instance segmentation techniques |
Asadpour et al.17 | Cascaded multi-level segmentation with SFED | Dice similarity coefficient: 82.7%, Jaccard index: 79.3% | Tumor shape region not considered |
Nakao et al.18 | Laplacian-based diffeomorphic shape matching (LDSM) | Detection accuracy: 85.6%, Precision: 83.2% | Struggles with non-linear deformation and liver motion |
Gandikota et al.19 | Tunicate Swarm Algorithm with Deep Learning (TSADL-PCSC) | Segmentation accuracy: 90.3%, Classification accuracy: 91.4% | Did not incorporate deep ensemble techniques |
Hooshangnejad et al.20 | DeepPERFECT: Deep Learning CT synthesis | Prediction accuracy: 88.9%, Sensitivity: 90.2% | Lack of high-resolution data affects performance |
Zhang et al.8 | Large-Scale Multi-Center CT and MRI segmentation | Pancreas segmentation Dice score: 89.4%, Accuracy: 91.0% | Limited multi-organ segmentation capability |
Ramaekers et al.21 | Computer-aided detection model utilizing secondary features | Pancreatic tumor detection sensitivity: 92.1%, Specificity: 90.5% | Secondary feature detection needs further validation |
Challenges
The challenges faced by Pancreatic cancer detection technique using CT images are as follows,
-
EL-SVM in14, delivered treatment options for various stages of pancreatic cancer patients, which had certain practicability and feasibility. Nevertheless, it was challenging for gathering various samples from several central institutions and to develop a classification approach with good performance.
-
A multi-task CNN was proposed for detecting pancreatic cancer from non-contrast CT images and this method attained exceptional results. Nevertheless, the technique missed certain cases when the contrast of the CT image was low.
-
In13, the YCNN was able to attain a consistent diagnosis of pancreatic cancer. However, it did not consider optimizing the performance of the YCNN by training the network using advanced algorithms.
-
The IDLDMS-PTC in16 was successful in detecting and classifying pancreatic cancer with minimum parameters, and reduced network size. However, the application of Emperor Penguin Optimizer (EHO) led to premature convergence, which affected performance.
-
Pancreatic cancer detection is challenging due to the difficulty in segmentation the cancer region accurately due to the ill-defined boundaries of the pancreatic region. In addition to this, the DL methods generally require high training time.
Developed secretary wolf bird optimization-based efficient DenseNet for pancreatic tumor detection
A pancreatic tumor occurs in the pancreas and develops abnormally and out of control. Recognizing pancreatic cancer at its starting stage is a major challenge, as the symptoms are visible only at a later stage. The most common symptoms of pancreatic cancer are weight loss, dark-colored urine, itching, and belly pain. Thus, an effective SeWBO_Efficient DenseNet approach is introduced to detect pancreatic cancer. Firstly, the input CT image is captured from the given database. After that, image pre-processing is done using a bilateral filter22 and then lesion segmentation is carried out by PraNet23. The hyperparameters of PraNet are adjusted by SeWBO and SeWBO is established by incorporating WBO24 and SBOA25. Then, in feature extraction, various features, such as CLBP26 with DWT27, SLBT28, and statistical features, including variance, standard deviation, mean, skewness, and kurtosis29 are extracted. Finally, pancreatic tumor is detected using SeWBO_Efficient DenseNet and Efficient DenseNet is enhanced by adjusting the hyperparameters using the SeWBO technique. Efficient DenseNet is developed by combining EfficientNet30 and DenseNet31. Figure 1 shows the schematic diagram of SeWBO_Efficient DenseNet for pancreatic tumor detection.
Image acquisition
Consider the CT pancreatic image as input, which is taken from the given database \(E\), and it is specified as,
Here, \(K_{r}\) specifies \(r^{th}\) pancreatic CT image input and \(s\) denotes the overall number of pancreatic CT images.
Representation of a set:
-
The equation defines E as a set of elements K, where each K represents a key parameter, variable.
-
It provides a structured way to denote multiple entities collectively, rather than listing them separately.
Image pre-processing
Image pre-processing is used to enhance image quality and decrease noise in the input image. Here, an input pancreatic CT image \(K_{r}\) is provided as input to the bilateral filter22 and a pre-processed image \(K_{a}\) is gained. A bilateral filter is regarded as a non-linear filter that depends on local image information. Moreover, the image’s spatial position information and pixel value are considered using bilateral filter for protecting the image edges during the process of denoising. In addition, a bilateral filter utilizes a weighted average of the neighborhood to substitute each noise pixel and is calculated as,
The summation across nearby pixels (d ∈ Ω) is represented by the Σ (Sigma) value. This indicates that within a certain region Ω, the equation is combining data from nearby pixels.
where, \(b\left( l \right)\) represents the output pixel value of the filter, \(A\left( d \right)\) denotes the pixels in the original input image, \(\Omega\) specifies the neighborhood of the pixel \(l\), \(c_{e}\) indicates normalization coefficient and is calculated as, \(c_{e} = \sum\nolimits_{d \in \Omega } {c_{f} \left( {l,d} \right)} \begin{array}{*{20}c} {c_{g} \left( {l,d} \right)} \\ \end{array}\), \(c_{f}\) stands for the spatial kernel and is expressed as \(c_{f} = \exp \left[ { - \frac{{\left| {l - d} \right|^{2} }}{{\sigma_{f}^{2} }}} \right]\), \(c_{g}\) represents range kernel and is expressed as \(c_{g} = \exp \left[ { - \frac{{\left| {A\left( l \right) - A\left( d \right)} \right|^{2} }}{{\sigma_{g}^{2} }}} \right]\), \(\sigma_{f}\) and \(\sigma_{g}\) are represents smoothening parameters.
Lesion segmentation
The process for outlining and identifying the boundaries of lesions in the pancreatic CT image is called lesion segmentation. In lesion segmentation, the pre-processed image \(K_{a}\) is considered as input and subjected to PraNet23 to attain the segmented lesion output \(K_{h}\).
Architecture of PraNet
The PraNet23 uses a parallel partial decoder to produce a high-level semantic global map and a cluster of reverse attention modules for accurate segmentation of pancreatic cancer from CT images.
-
(a)
Parallel Partial Decoder
During lesion segmentation, the high-level features are aggregated with the help of a parallel partial decoder component. Consider the pancreatic image \(K_{h}\) is fed as input with dimension \(c \times e\) and contains five feature levels , which is signified as \(\left\{ {i_{j} ,j = 1,.....,5} \right\}\) with a resolution of \(\left[ {{\raise0.7ex\hbox{$c$} \!\mathord{\left/ {\vphantom {c {2^{m - 1} }}}\right.\kern-0pt} \!\lower0.7ex\hbox{${2^{m - 1} }$}},{\raise0.7ex\hbox{$e$} \!\mathord{\left/ {\vphantom {e {2^{m - 1} }}}\right.\kern-0pt} \!\lower0.7ex\hbox{${2^{m - 1} }$}}} \right]\) are mined from Res2Net-based backbone network. Next, the \(i_{j}\) features are classified into high-level features \(i_{j} ,j = \left\{ {3,4,5} \right\}\) and low-level features \(\left\{ {i_{j} ,j = 1,2} \right\}\). After that, a partial decoder \(k_{n} \left( . \right)\) is established to combine the high-level feature with paralleled connections. Further, partial decoder feature is measured as \(\Upsilon = k_{n} \left( {i_{3} ,i_{4} ,i_{5} } \right)\) and finally, a global map \(S_{o}\) is attained.
-
(b)
Reverse Attention framework
The global map \(S_{o}\) has the ability to capture rough position of pancreatic cells without any structural information. Therefore, a principal strategy is required to erase foreground object in pancreatic images for discriminating the cancer regions. Thus, reverse attention is utilized to aggregate features from high-level and low-level and features from all levels in three parallel high-level features. Moreover, the outcome reverse attention feature \(F_{j}\) is attained by multiplying high-level side-output feature \(\left\{ {i_{j} ,j = 3,4,5} \right\}\) using reverse attention weight \(B_{j}\) by an element-wise operation \(\odot\) and is expressed as,
$$F_{j} = i_{j} \odot B_{j}$$(3)where, \(B_{j}\) represents reverse attention weight and is computed as,
$$B_{j} = \Theta \left[ {\sigma \left( {\text{M}\left( {G_{j} + 1} \right)} \right)} \right]$$(4)After applying a series of operations, the final computed value or feature transformation at index j is represented as Bj.
Theta Function (Θ): This usually denotes a non-linear transformation or activation function that regulates the scaling or Thresholding of the values that have been processed.
σ (Sigma Function): This typically indicates a sigmoid function that adds non-linearity and normalizes values, like softmax.
M(Gj + 1): Gj is an index j input feature or function.
-
Adding 1 could be a scaling factor to change the range of values or a bias term to avoid zero values.
-
M(⋅) denotes a transformation function, which could be a convolution operation or weight multiplication.
where, \(\sigma \left( . \right)\) symbolizes sigmoid function, \(\Theta \left( . \right)\) denotes reverse operation detracting the input from matrix \(H\), and \({\text{M}}\left( . \right)\) epitomizes up-sampling operation. Figure 2 represents the PraNet architecture, and the lesion-segmented output attained is denoted as \(K_{h}\).
Training of PraNet using proposed SeWBO
The PraNet23 is trained by the proposed SeWBO, which is developed by incorporating SBOA25 and WBO24. SBOA is a population-based metaheuristic approach, which is formulated on the basis of survival instincts of the secretary bird. The secretary bird’s snake-hunting behavior that forms the basis of the exploration phase and the escaping strategy forms the idea behind the exploitation phase. Moreover, SBOA enhances efficiency and minimizes computational cost. WBO is formulated by considering the relationship among the ravens and wolves. The ravens are extremely intelligent in finding preys and they seek the help of wolves in hunting the prey by sending signals. The WBO is flexible and is suited for high dimensional problems. Further, they are effective in enhancing the convergence rate. The inclusion of WBO in SBOA enhances the adaptability of the SeWBO technique and assists in avoiding premature convergence. The phases followed by SeWBO are discussed beneath.
-
(i)
Initialization: The initial process in the implementation of SeWBO is the arbitrary initialization of the location of a secretary bird in a search space and is expressed as,
$$C_{m,n} = lb_{n} + a \times \left( {ub_{n} - lb_{n} } \right),m = 1,2,.....,D,n = 1,2,.....,Dim$$(5)wherein, \(c_{m}\) designates position of \(m^{th}\) secretary bird, \(lb_{n}\) indicates lower bound, \(ub_{n}\) stipulates the upper bound, and \(a\) signifies a random number among \([0,1][0,1]\). Furthermore, the optimization begins with a candidate solution \(C\), which are randomly produced within lower-bound and upper-bound constraints for a specified problem. Furthermore, the candidate solution \(C\) is defined as,
$$C = \left[ {\begin{array}{*{20}c} {c_{1,1} } & {c_{1,2} } & \cdots & {c_{1,n} } & \cdots & {c_{1,Dim} } \\ {c_{2,1} } & {c_{2,2} } & \cdots & {c_{2,n} } & \cdots & {c_{2,Dim} } \\ \vdots & \vdots & \ddots & \vdots & \ddots & \vdots \\ {c_{m,1} } & {c_{m,2} } & \cdots & {c_{m,n} } & \cdots & {c_{m,Dim} } \\ \vdots & \vdots & \ddots & \vdots & \ddots & \vdots \\ {c_{D,1} } & {c_{D,2} } & \cdots & {c_{D,m} } & \cdots & {c_{D,Dim} } \\ \end{array} } \right]_{D \times Dim}$$(6)where, amount of group members is symbolized as \(D\), \(C\) represents secretary bird group, \(c_{m}\) specifies \(m^{th}\) secretary bird, \(c_{m,n}\) denotes \(m^{th}\) secretary bird in the \(n^{th}\) dimension, and variable dimension is indicated as \(Dim\).
-
(ii)
Fitness Function: Here, optimization algorithm is utilized to find the ideal hyperparameters of the PraNet and hence Mean Square Error (MSE) is considered as fitness, which is articulated as,
$$P = \frac{1}{o}\sum\limits_{t = 1}^{o} {\left( {K_{h}^{t} - \widehat{{K_{h}^{t} }}} \right)}^{2}$$(7)where, \(P\) specifies MSE, \(o\) represents a number of observations, \(K_{h}^{t}\) stands for actual output of PraNet, and \(\widehat{{K_{h}^{t} }}\) denotes expected value. The secretary bird has two diverse natural behaviors, and they are exploited to update SeWBO members and the behavior of secretary bird’s hunting plan and escape strategy are detailed below.
-
(iii)
Exploration Stage: When feeding on snakes, the secretary bird’s hunting behavior is alienated into three phases, such as searching for prey, hunting of prey, and consumption of prey. Depending on the biological statistics of a secretary bird hunting steps and the overall hunting procedure is classified into three-time durations at each phase, including \(p < \frac{1}{3}I\), \(\frac{1}{3}I < p < \frac{2}{3}I\), and \(\frac{2}{3}I < p < I\), are based on the above-mentioned phases with \(p\). Here, \(p\) represents the present iteration number, and \(I\) designates the entire iteration number, and the behaviors are explained as follows.
-
(a)
Searching for Prey: The secretary bird’s hunting process is initiated with searching for its possible prey, particularly snakes. Generally, these birds have sharp vision, which is used to find the hidden snakes in tall grass. In addition, the secretary bird uses its long legs for gently sweeping the ground by paying attention to the environments. The long necks and legs help them to avoid snake attacks and maintain a safe distance from snakes. Further, differential evolution is used here, which exploits the changes between the individuals for improving the algorithm diversity by creating global search capabilities and new solutions. Thus, a differential mutation process is employed to help avoid local optima. The when searching for prey, the secretary bird’s updating location is mathematically given by,
$$when\begin{array}{*{20}c} {} \\ \end{array} p < \frac{1}{3}I,c_{{_{m,n} }}^{{new\begin{array}{*{20}c} {} \\ \end{array} \begin{array}{*{20}c} {J_{1} } \\ \end{array} }} = c_{m,n} + \left( {c_{rand\_1} - c_{rand\_2} } \right) \times L_{1}$$(8)where, \(c_{m}^{{new\begin{array}{*{20}c} {J1} \\ \end{array} }}\) stands for \(m^{th}\) secretary bird’s new state at the first stage, and \(c_{m,n}^{{new\begin{array}{*{20}c} {J1} \\ \end{array} }}\) its value of \(n^{th}\) dimension, \(L_{1}\) specifies a arbitrarily created array of size \(1 \times Dim\) in the interval \(\left[ {0,1} \right]\), and \(Dim\) denotes dimensionality of solution space.
-
(b)
Consumption of Prey: A secretary bird engages in a strategy for hunting after detecting the snake. The agile footwork and the tricks of secretary bird is employed around a snake instead of diving immediately into fight. Every sign of a snake is observed by secretary bird from a high vantage point. Moreover, the secretary bird utilizes their keen judgment by analyzing the movement of snake to provoke the snake. Additionally, a Brownian motion is established to simulate the actions of secretary bird. Moreover, the secretary bird’s leg surface is enclosed with thick keratin scales makes it hard for the snake to trap in its body. At this stage, \(c_{best}\) concept and Brownian motion \(E^{*}\) is employed, and it is defined as,
$$E* = random(1,Dim)$$(9)Here, \(random(1,Dim)\) signifies an arbitrarily formed array with dimension size \(1 \times Dim\). When consuming the prey, the secretary bird’s updated position is modeled by,
$$while\begin{array}{*{20}c} {} \\ \end{array} \frac{1}{3}I < p < \frac{2}{3}I,c_{m,n}^{{new\begin{array}{*{20}c} {} \\ \end{array} J1}} = c_{best} + \exp \left[ {\left( \frac{p}{I} \right) \wedge 4} \right] \times \left( {E* - 0.5} \right) \times \left( {c_{best} - c_{m,n} } \right)$$(10)where, \(c_{best}\) specifies the current best value.
By regulating the exponential term’s growth rate, exponent 4 has a major impact on the behavior of the equation.
By raising \(\left( \frac{p}{I} \right)\) to the power (4) increases its non-linearity.
-
(c)
Hunting of Prey: If a snake is tired, a secretary bird perceives an opportunity to move using their high-powered leg muscles for attack. Further, a leg-kicking approach is initiated by a secretary bird, where it rises its leg and distributes correct kicks with their sharp talons and targets the head of the snake. Besides, some snakes are too long therefore, it is difficult to kill rapidly. Here, the secretary bird carries a snake into sky and releases it and this causes an immediate death. Thus, a levy flight strategy is presented for enhancing the global search capability of the optimizer, which is performed by enhancing the convergence rate. In addition, a nonlinear perturbation factor is developed for making the model more energetic, adaptive, and flexible by avoiding premature convergence. A nonlinear perturbation factor is designated as \(\left( {1 - \frac{p}{I}} \right)\left( {2 \times \frac{p}{I}} \right)\). Moreover, the secretary bird’s updating position while hunting the prey is given by,
$$while\begin{array}{*{20}c} {} \\ \end{array} p > \frac{2}{3}I,c_{m,n}^{{new\begin{array}{*{20}c} {} \\ \end{array} J1}} = c_{best} + \left[ {\left( {1 - \frac{p}{I}} \right) \wedge \left( {2 \times \frac{p}{I}} \right)} \right] \times c_{m,n} \times M$$(11)where, weighted levy flight is implied as \(M\).
-
(a)
-
(iv)
Exploitation Stage: Secretary bird’s natural enemies are big predators, which include jackals, foxes, eagles, and hawks which steal food or kill them. There are mainly two kinds of evasion strategies to safeguard themselves. The first strategy is run or fly away, in which the secretary bird utilizes its long leg enabling them to fly at high speed to escape from danger. Furthermore, Camouflage is considered as the second strategy, in which secretary bird uses the structures or colors in their surroundings to make it difficult for the predators to identify them. These two conditions occur with equal possibility and they are:
-
(A)
\(N_{1}\): Camouflage by environment.
-
(B)
\(N_{2}\): Run or fly away.
In a first strategy, a secretary bird identifies a predator and they look for suitable camouflage surroundings. If there is no safe and suitable camouflage, then the secretary bird will choose to fly or rapidly run to escape. At this stage, a dynamic perturbation factor is introduced and signified as \(\left( {1 - \frac{p}{I}} \right)^{2}\). This dynamic perturbation factor assists the algorithm to balance between exploitation and exploration. Finally, the secretary bird’s position is updated,
$$c_{m,n}^{new,J2} = \left\{ {\begin{array}{*{20}l} {N_{1} :c_{best} + \left( {2 \times E* - 1} \right) \times \left( {1 - \frac{p}{I}} \right)^{2} \times c_{m,n} ,} & {if\,q < q_{j} } \\ {N_{2} \begin{array}{*{20}l} {:c_{m,n} + L_{2} \times \left( {c_{random} O \times c_{m,n} } \right),} \\ \end{array} } & {else} \\ \end{array} } \right.$$(12)where, \(q_{j} = 0.5,\) \(E*\) represents Brownian motion, \(q\) represents the random number, \(L_{2}\) signifies the a random array of normal distribution with size \(\left( {1 \times Dim} \right)\). \(c_{random}\) denotes random candidate solution, and \(O\) indicates random selection among the integer \([1,2]\). When a condition \(N_{2}\) met equation, the above equation becomes,
$$c_{m,n} \left( {p + 1} \right) = c_{m,n} \left( p \right) + L_{2} *\left[ {c_{random} - O*c_{m,n} \left( p \right)} \right]$$(13)$$c_{m,n} \left( {p + 1} \right) = c_{m,n} \left( p \right) + L_{2} *c_{random} - L_{2} O*c_{m,n} \left( p \right)$$(14)$$c_{m,n} \left( {p + 1} \right) = c_{m,n} \left( p \right)\left[ {1 - L_{2} O} \right] + L_{2} *c_{random}$$(15)The WBO24, is a nature-inspired optimization algorithm utilized for identifying the behavior of birds while searching for food. Moreover, the inclusion of WBO in SBOA enhances the adaptability of the SeWBO technique and assists in avoiding premature convergence. From WBO,
$$c_{m,n} \left( {p + 1} \right) = c_{m,n} \left( p \right) + q_{1} \begin{array}{*{20}c} * \\ \end{array} c_{m,n}^{J} (p) - q_{2} *pcp$$(16)$$c_{m,n} \left( p \right) = c_{m,n} \left( {p + 1} \right) - q_{1} *c_{m,n}^{J} \left( p \right) - q_{2} *pcp$$(17)Substituting Eq. (17) in Eq. (13),
$$c_{m,n} \left( {p + 1} \right) = \left[ {c_{m,n} \left( {p + 1} \right) - q_{1} *c_{m,n}^{J} \left( p \right) - q_{2} *pcp} \right]\left[ {1 - L_{2} O} \right] + L_{2} *c_{random}$$(18)$$c_{m,n} \left( {p + 1} \right) - c_{m,n} \left( {p + 1} \right)\left[ {1 - L_{2} O} \right] = L_{2} *c_{random} - \left( {q_{1} \times c_{m,n}^{J} \left( p \right) + q_{2} *pcp} \right)\left[ {1 - L_{2} O} \right]$$(19)$$c_{m,n} \left( {p + 1} \right)\left[ {1 - 1 + L_{2} O} \right] = L_{2} *c_{random} - \left( {q_{1} \times c_{m,n}^{J} \left( p \right) + q_{2} *pcp} \right)\left[ {1 - L_{2} O} \right]$$(20)$$c_{m,n} \left( {p + 1} \right) = \frac{1}{{L_{2} O}}\left[ {L_{2} *c_{random} - \left( {q_{1} *c_{m,n}^{J} \left( p \right) + q_{2} *pcp} \right)\left( {1 - L_{2} O} \right)} \right]$$(21)The Eq. (21) denotes the updated equation of the SeWBO algorithm. Further, in the above equation, \(q_{1}\) and \(q_{2}\) designates two random numbers ranging in [0,1], \(c_{m,n}^{J} \left( p \right)\) is the hungriest prey, and \(pcp\) is the prey’s center point.
Furthermore, the random selection is indicated as,
$$O = round\left( {1 + random\left( {1,1} \right)} \right)$$(22)Here, \(random(1,1)\) represents an arbitrarily generated random between \(\left[ {0,1} \right]\).
-
(A)
-
(v)
Evaluation of Feasibility: The SeWBO members location is updated using the above-mentioned steps in every iteration. When the updated location is better than other locations, then the updated position is taken as the new updated location.
-
(vi)
Termination: The above steps are done repeatedly till attain the best outcomes. The new location of SeWBO members gives the best solution for a specified issue. The pseudocode for SeWBO is demonstrated in Algorithm 1.
The Secretary Bird Optimization Algorithm (SBOA) draws inspiration from the unique hunting techniques of secretary birds. These remarkable birds exhibit a keen ability to find their prey through thoughtful movements and precise positioning. Similarly, SBOA leverages this instinctual behavior to tackle various optimization challenges effectively.
Thus, the SeWBO effectively enhances the efficiency of lesion segmentation by finding the ideal hyperparameters of PraNet and the segmented output \(K_{h}\) is attained.
Feature extraction
Here, the lesion-segmented image \(K_{h}\) is considered as input for the extraction of features, such as DWT27 with CLBP26, SLBT28, and statistical features29, like mean, variance, standard deviation, kurtosis, and skewness. The final feature vector gained is signified as \(Q\).
DWT with CLBP features
The DWT with CLBP feature is excerpted by first applying DWT to split the image \(K_{h}\) into multiple bands first, followed by the extraction of CLBP features. The obtained CLBP features are combined to get the DWT with CLBP feature.
DWT27 is utilized to analyze the image at several scales. In addition, DWT is used to extract the co-efficient wavelets from CT images. During classification, frequency information of signal function is localized by wavelet. The DWT converts the images into four sub bands including Low–High, Low-Low, High-Low, and High-High. Here, the low-level bands are alone considered. The distinct frequency components with resolution matched with scale in DWT are expressed as,
where, \(z_{{y_{1} ,y_{2} }}\) represents a signal’s component attribute, \(w\left( x \right)\) corresponds to the wavelet function, \(a_{1} \left( x \right)\) and \(b_{1} \left( x \right)\) indicates the high-pass and low-pass filter coefficient, \(y_{2}\) stands for translation factor, and \(y_{1}\) stipulates wavelet scale. Further, Low–High, Low-Low, and High-Low bands obtained are subjected to the extraction of the CLBP feature.
CLBP26 is used to extract the local texture of the input image. For a given pixel \(d_{e}\) and its neighbor pixel \(d_{f},\) the difference among \(d_{f}\) and \(d_{e} :g_{f} = d_{f} - d_{e}\) is computed. Here, \(g_{f}\) is classified into \(g_{f} = h_{f} *j_{f}\).
where, \(h_{f}\) represents sign component, \(j_{f}\) indicates magnitude component. The CLBP is measured utilizing \(d_{e}\), \(j_{f}\), and \(h_{f}\) individually. The magnitude component is given by,
wherein, amount of neighboring points is indicated as \(n\), \(k\) implies the radius and \(e\) signifies the \(j_{f}^{th}\) mean value from the entire image. Here, the sign component is signified as,
Further, the information about the central pixel is expressed as,
where, \(e_{m}\) stands for image’s mean value. The histogram’s size corresponds to \(CLBP\_j_{n,k}\) is epitomized as \(n + 2\), the dimension of \(CLBP\_h_{n,k}\) is also \(n + 2\), and \(CLBP\_p_{n,k}\) is 2. To achieve better classification accuracy, simple concatenation of \(CLBP\_j_{n,k}\),\(CLBP\_h_{n,k}\), and \(CLBP\_p_{n,k}\) is utilized. The output gained from DWT with CLBP features is specified as \(R_{1}\).
SLBT features
SBLT28 features combine both texture and shape information in the segmented image \(K_{h}\). Let us consider, \(S = \left\{ {S_{1} ,S_{2} ,.....,S_{T} } \right\}\) signifies a training set of \(T\) images with \(U = \left\{ {U_{1} ,U_{2} ,.....,U_{T} } \right\}\) as their shape landmark points, which are used to obtain shape variations and shape vector \(U\) in a training set is given by,
where, \(\overline{U}\) specifies mean value, \(W_{v}\) consists of eigenvectors of biggest eigenvalues \(\left( {\lambda_{v} } \right)\), \(X_{v}\) signifies shape or weight model parameters, and \(v\) indicates shape in \(X_{v}\).
This formula represents a linear transformation that maps the shape vector U to a new feature space using PCA. It helps in reducing dimensionality and extracting the most significant shape variations.
where, \(\overline{U}\) specifies mean value, \(W_{v}\) consists of eigenvectors of biggest eigenvalues \(\left( {\lambda_{v} } \right)\), \(X_{v}\) signifies shape or weight model parameters, and \(v\) indicates shape in \(X_{v}\). Considering a \(3 \times 3\) window with a center pixel \(\left( {r_{v} ,s_{v} } \right)\) intensity value be \(t_{v}\) and local texture as \(z = w\left( {t_{x} } \right)\) where, \(t_{x} \left( {x = 0,1,2,3,4,5,6,7} \right)\) corresponds to eight neighboring pixel’s grey values. These neighboring pixels are thresholded with a center value \(t_{v}\) by \(w\left( {z\left( {t_{0} - t_{x} } \right),.....,z\left( {t_{7} - t_{x} } \right)} \right)\) and here \(z\left( a \right)\) is modelled as,
The Local Binary Pattern (LBP) at a center pixel \(t_{v}\) is gained using Eq. (32) and consider \(A = \left[ {A_{1} ,A_{2} ,.....,A_{T} } \right]\) be an LBP feature histogram of overall tuning set images. Likewise, texture model is carried out in the same manner as shape modeling with weight or texture model parameter \(X_{c}\) and \(c\) represents texture in \(X_{c}\), \(W_{c}\) specifies eigenvectors, and \(\overline{A}\) stands for a mean vector. the texture model parameter \(X_{c}\) is defined as,
The texture and shape parameters vectors are combined together and a shape texture parameter controlling local, texture, and global shape is attained by combining parameter vectors and is expressed as,
where, \(E\) represents shape feature parameter, \(W_{Vc}\) specifies eigenvector, \(\overline{{X_{Vc} }}\) stands for mean vector, and \(C_{V}\) denotes a diagonal matrix of weight for every shape parameter. The overall feature attained from SLBT is denoted as \(R_{2}\).
Statistical features
The first-order statistical measurements are considered as the statistical features29, and they are skewness, kurtosis, mean, Standard Deviation, and variance. Statistical features are quantitative measures utilized for modeling, classification, or analysis.
-
(a)
Mean: Mean \(\left( {S_{1} } \right)\) is measured by summing entire values in the image \(K_{h}\) and dividing by overall grey level count and is defined by,
$$S_{1} = \sum\limits_{d* = 0}^{{B_{1} - 1}} {d*C_{1} \left( {d*} \right)}$$(36)where, \(B_{1}\) denotes a total number of grey levels, \(C_{1} \left( {d*} \right)\) represents probability of \(d*\) happening, \(d*\) stands for grey level in the image.
Mean (S1) is 127.35.
-
(b)
Variance: A statistical measure that quantifies deviation of an image’s grey level from the mean grey level is called variance \(\left( {S_{2} } \right)\), and it is specified as,
$$S_{2} = \sum\limits_{d* = 0}^{{B_{1} - 1}} {\left( {d* - S_{1} } \right)^{2} *C_{1} \left( {d*} \right)}$$(37)Variance: 1024.67
-
(c)
Standard Deviation: The dispersion of values about the mean is represented as Standard Deviation \(\left( {S_{3} } \right)\), and it is formulated as,
$$S_{3} = \sqrt {\sum\limits_{d* = 0}^{{B_{1} - 1}} {\left( {d* - S_{1} } \right)^{2} *C_{1} \left( {d*} \right)} }$$(38)Standard Deviation: 32.00
-
(d)
Kurtosis: The fourth normalized moment is also called as Kurtosis \(\left( {S_{4} } \right)\), and it is measured as leveling of a distribution related to a normal distribution. Here, kurtosis is signified as,
$$S_{4} = S_{1}^{ - 4} \left[ {\sum\limits_{d* - 0}^{{B_{1} - 1}} {\left( {d* - S_{1} } \right)^{4} *C_{1} \left( {d*} \right)} } \right]$$(39)Kurtosis: 2.85
-
(e)
Skewness: The degree of asymmetry of a distribution of a particular feature around a mean value is referred as skewness \(\left( {S_{5} } \right)\) and defined by,
$$S_{5} = S_{1}^{ - 3} \left[ {\sum\limits_{d* = 0}^{{B_{1} - 1}} {\left( {d* - S_{1} } \right)^{3} *C_{1} \left( {d*} \right)} } \right]$$(40)Skewness: 0.12
The overall statistical features \(R_{3}\) is given by,
where, \(S_{1}\) stands for mean, \(S_{2}\) represents variance, \(S_{3}\) signifies Standard Deviation, \(S_{4}\) denotes kurtosis, and \(S_{5}\) indicates skewness. The final output obtained by feature extraction \(Q\) is expressed as,
Where, \(R_{1}\) denotes DWT with CLBP features, \(R_{2}\) denotes SLBT features, and \(R_{3}\) represents statistical features.
Pancreatic tumor detection
Early detection of pancreatic tumors is crucial to increasing the survival rate and early pancreatic cancer may be treatable with surgery. Here, feature \(Q\) is taken as input and passed to Efficient DenseNet. The Efficient DenseNet is developed by merging EfficientNet30 and DenseNet31. Further, the Efficient DenseNet is structurally optimized with the help of the SeWBO algorithm, which is developed by the combination of SBOA25 and WBO24.
Efficient DenseNet
He Efficient DenseNet is a hybrid DL framework, which is formulated to detect the pancreatic cancer. It is made up of two DL models, such as the EfficientNet30 and DenseNet31. The input image \(K_{r}\) is given to EfficientNet, and it obtains an outcome \(D_{1}\). The output \(D_{1}\) is combined with the feature vector \(Q\) in the Efficient DenseNet layer on the basis of Fractional Calculus to establish regression modeling to attain output \(D_{2}\). Thereafter, the final DenseNet model, when applied with the \(D_{2}\), produces the final detected outcome \(D_{3}\). Figure 3 portrays the architecture of Efficient DenseNet.
-
EfficientNet
EfficientNet30 is a CNN established to optimize the efficiency and performance of image classification tasks. During classification, EfficientNet enhances parameter efficiency, reduces overfitting issues, and attains high accuracy. Here, the image \(K_{r}\) is subjected to EfficientNet as input. Moreover, an activation function is used with fully connected, batch normalization, and softmax layers. In a convolution (Conv) layer, neurons are utilized to mine the features using Conv kernels. Furthermore, each neuron is convoluted with same dimension in a previous layer and converts into a large receptive field, and it indicates a learned feature vector of an image from a neuronal vector. Generally, Global Positioning Pooling (GAP) is utilized to compute a simple average with weights to input images lacking of applying any specific attention to a particular receptive region. The output \(D_{1}\) obtained from EfficientNet is given by,
$$D_{1} = J_{1} \Theta i_{2}$$(43)where,\(J_{1}\) indicates attention weight ranging from \(\left[ {0,1} \right]\), \(\Theta\) stands for element-wise multiplication, and \(i_{2}\) denotes receptive region.
-
EfficientNet DenseNet layer
Here, the EfficientNet output \(D_{1}\) and feature vector \(Q\) is provided as input. Here, both inputs are combined using regression modeling and fusion is performed based on Fractional Calculus32. Fractional Calculus (FC) is employed to decrease information loss and improve performance. The output gained by Efficient DenseNet is \(D_{2}\) and is expressed as,
$$D_{2} = \eta *\sum\limits_{{d_{2} = 1}}^{2} {\sum\limits_{{e_{2} = 1}}^{a^{\prime}} {\sum\limits_{{f_{2} = 1}}^{b^{\prime}} {F_{{d_{2} e_{2} f_{2} }} G_{{d_{2} e_{2} f_{2} }} + \frac{1}{2}} } } \eta *\sum\limits_{{d_{2} = 1}}^{5} {R_{3} G_{{d_{2} }} + \frac{1}{6}} \left( {1 - \eta *} \right)D_{1}$$(44)where, \(\eta\) stands for constant value, \(R_{3}\) indicates statistical features, \(G\) signifies weight value, \(F_{2} = \left\{ {R_{1} ,R_{2} } \right\}\) specifies the combination of DWT with CLBP and SLBT features with dimension \(a^{\prime} \times b^{\prime}\), and \(D_{1}\) denotes EfficientNet output.
-
DenseNet
DenseNet31 is a DL framework employed for classification tasks that comprises trainable parameters. In addition, DenseNet has robust feature propagation and can resolve disappearing gradient problems and achieve a high accuracy with a reduced number of trainable parameters. DenseNet consists of a max-pooling, dense, and a Conv layer. Here, the DenseNet is applied with the outcome \(D_{2}\) of the Efficient DenseNet layer.
-
(i)
Conv Layer
In the Conv layer, a feature map denotes a number of exposed features at various locations. In addition, the filters in the Conv layer are smaller when contrasted to a given input and are utilized to extract feature maps.
-
(ii)
Max Pooling
In order to lessen the feature map’s magnitude, this layer is used. Moreover, a pooling filter is slid above the feature map and thus the highest value over the area is obtained as the output.
-
(iii)
Dense Layer
Dense layer is fully connected with the prior layer, and all the neuron is interconnected with those in the previous layer. The outcome from the former layer is subjected as input to all neurons and a matrix–vector multiplication is effectuated to attain the output. The outcomes attained from DenseNet \(D_{3}\) is expressed as,
$$D_{3} = \chi \left( {\varpi_{uv}^{\ell } } \right)$$(45)where,
$$\varpi_{u,v}^{\ell } = \sum\limits_{i = 0}^{\alpha - 1} {\sum\limits_{j = 0}^{\alpha - 1} {\wp_{ij} D_{{2} {(u + i)(v + j)}}^{\ell - 1} } }$$(46)where, \(\wp_{ij}\) is the weight, \(\alpha \times \alpha\) is the filter size,\(\varpi^{\ell }\) specifies the outcome of \(\ell^{th}\) layer,\(D_{2}\) characterizes Efficient DenseNet outcome and \(D_{2}^{*}\) denotes non-linear units.
Training of efficient DenseNet utilizing proposed SeWBO
The tuning process of Efficient DenseNet is executed using the SeWBO, which is elucidated in section "Training of PraNet using proposed SeWBO". Here, SeWBO is developed by merging SBOA25 and WBO24. However, the fitness parameter differs and is expressed as,
where, \(P\) specifies MSE, \(o\) represents a number of observations, \(D_{3}^{t}\) stands for the actual output of Efficient DenseNet, and \(\widehat{{D_{3}^{t} }}\) denotes expected value.
Integration of Secretary Wolf Bird Optimization with DenseNet.
-
Efficient Dense Net is used with Secretary Wolf Bird Optimization (SeWBO) to enhance classification accuracy and optimize hyperparameters for pancreatic tumor identification from CT scans. Hyperparameter tweaking, feature selection, and network architecture optimization are all part of the integration process.
There are three main stages to the integration process:
-
-
1.
Preprocessing and Feature Extraction
-
A bilateral filter is used to remove noise from CT images during preprocessing.
PraNet is used to segregate tumor regions, and SeWBO is used to adjust its parameters.
-
Discrete Wavelet Transform (DWT), Shape Local Binary Texture (SLBT), Complete Local Binary Pattern (CLBP), and Statistical Features (Mean, Variance, Kurtosis, etc.) are among the features that were extracted.
-
-
2.
Efficient DenseNet Hyperparameter Optimization with SeWBO
-
SeWBO is applied to tune Dense Net’s hyper parameters like:
-
Learning rate
-
Number of Dense layers
-
Growth rate
-
Batch size
-
Filter size
-
-
SeWBO ensures fast convergence and avoids local minima.
-
-
3.
Classification using Efficient DenseNet
-
The existence of pancreatic tumors is classified using the optimal Efficient DenseNet model.
-
SeWBO’s hyperparameter adjustment enhances the sensitivity and accuracy of the DenseNet architecture.
-
-
1.
Procedure for Integration
Step 1: Feature extraction and image preprocessing
-
A bilateral filter is used to preprocess the input CT images I:
$$I^{\prime} = BilateralFilter(I) \,$$The segmented lesion SSS is extracted using PraNet:
$$S = PraNet(I^{\prime})$$ -
Feature extraction is performed using:
-
CLBP with DWT
-
SLBT features
-
Statistical features
-
The extracted feature vector is denoted as:
Step 2: Hyper parameter tuning using SeWBO
Hyper parameters Tuned by SeWBO
Hyper parameter | Search Space |
|---|---|
Learning Rate (α) | 10−5 to 10−2 |
Growth Rate (γ) | 8 to 48 |
Number of Dense Layers (L) | 2 to 8 |
Batch Size (B) | 8, 16, 32, 64 |
Filter Size (F) | 3 × 3, 5 × 5 |
Hyper parameter Optimization for Efficient Dense Net (Classification)
Parameter | Optimized value |
|---|---|
Learning Rate | 0.0001 |
Batch Size | 32 |
Number of Epochs | 100 |
Weight Decay | 0.0001 |
Dropout Rate | 0.3 |
Activation Function | Swish |
Pooling Layer | Global Average Pooling (GAP) |
Fully Connected Layers | 512, 256, 128 (with Batch Normalization) |
Optimizer | AdamW |
Loss Function | Binary Cross-Entropy (BCE) |
Hyper parameter Optimization for PraNet Segmentation
Parameter | Optimized value |
|---|---|
Learning Rate | 0.001 |
Batch Size | 16 |
Number of Epochs | 50 |
Weight Decay | 0.0005 |
Momentum | 0.9 |
Activation Function | ReLU |
Loss Function | Cross-Entropy Loss + Dice Loss |
Optimizer | Adam |
SeWBO searches for the optimal set of parameters to minimize classification error.
Mean Squared Error (MSE) is used as the fitness function:
For Efficient DenseNet training, the parameters with the highest performance are chosen.
Step 3: Effective DenseNet Model for Categorization Improvements to the DenseNet Architecture
-
Dense Net and Efficient Net are combined to create Efficient Dense Net.
-
Fractional Calculus Regression Modeling for Fusion:
$$\begin{aligned} O_{EffDense}& = W_{1} \cdot F_{DWT} + W_{2} \cdot F_{CLBP} + W_{3} \cdot F_{SLBT} \\&+ W_{4} \cdot F_{Stat} + W_{5} \cdot EfficientNet(I^{\prime})\end{aligned}$$
Results and discussion
The effectiveness of SeWBO_Efficient DenseNet for detecting of pancreatic cancer is evaluated and its outcomes, dataset description, and evaluation measures are discussed in this section.
Experimental setup
The SeWBO_Efficient DenseNet for identifying the pancreatic tumor is effectively implemented by using the PYTHON tool.
Dataset description
Medical Segmentation Decathlon Dataset33 is taken from, “http://medicaldecathlon.com/”, accessed on July 2024 that comprises 420 portal-venous phase CT scans of patients undergoing resection of pancreatic masses. The pancreatic mass and pancreatic parenchyma are corresponding to the target RoI. Among the images, a total of 282 images are taken for tuning and the testing set contains 139 images. The 512 × 512 pixel resolution of each CT scan ensures a high level of detail in the examination of pancreatic tumors.
Evaluation metrics
The accuracy, TNR, TPR, segmentation accuracy, and Receiver Operating Characteristics (ROC) are utilized for analyzing the proposed SeWBO_Efficient DenseNet to recognize the pancreatic cancer, and they are discussed in the segment.
Accuracy
The ratio of precise detections to the overall amount of detection made by SeWBO_Efficient DenseNet is called accuracy \(\left( {K_{1} } \right)\), which is given by,
where, \(Q_{1}\) designates True Negative, \(P_{1}\) indicates True Positive, \(S_{1}\) designates False Positive, and \(T_{1}\) represents False Negative.
TPR
The proportion of appropriately recognized positive occurrences to the entire amount of actual positive samples is termed TPR \(\left( {L_{1} } \right)\) and defined by,
TNR
The proportion of appropriately recognized negative occurrences from the entire quantity of negative samples is called TNR \(\left( {M_{1} } \right)\) and is expressed as,
Segmentation accuracy
Segmentation accuracy \(\left( {N_{1} } \right)\) measures a proportion of accurately classified pixels in the segmented image, which is represented by,
where, \(\beta\) typifies overall count of correctly classified pixels and \(\gamma\) is the pixel count.
ROC
ROC is a graphical representation, which is used for evaluating the binary classification performance. Moreover, ROC is employed for measuring the trade-off among False Positive Rate (FPR) and TPR at numerous threshold settings.
Image output
The image outcomes of SeWBO_Efficient DenseNet to identify the pancreatic cancer by considering abnormal and normal tissues are exhibited in this section.
Image samples for abnormal tissues
The image outcomes of SeWBO_Efficient DenseNet for pancreatic cancer detection for abnormal tissues are exhibited in Fig. 4. The input and pre-processed image is depicted in Fig. 4a and b. The segmented image is designated in Fig. 4c. In Fig. 4d and e the DWT with CLBP feature SLBT feature image are portrayed.
Image results for normal tissues
Here, Fig. 5 demonstrates the image outputs of SeWBO_Efficient DenseNet for normal tissues. Figure 5a and b signifies the input and pre-processed image. In Fig. 5c, the segmented image is exhibited. Figure 5d and e shows DWT with CLBP feature, and SLBT feature image.
Performance analysis
The efficacy of SeWBO_Efficient DenseNet for pancreatic tumor identification is assessed with changing layer size and batch size and is demonstrated as follows.
Based on layer size
The investigation of SeWBO_Efficient DenseNet for pancreatic tumor detection based on numerous layer sizes is represented in Fig. 6. The performance appraisal of SeWBO_Efficient DenseNet regrading accuracy is designated in Fig. 6a). The accuracy calculated by SeWBO_Efficient DenseNet using a training data of 60% for 2 layers is 77.957%, 4 layers is 79.042%, 6 layers is 80.854%, and 8 layers is 81.367%. The valuation of SeWBO_Efficient DenseNet with TPR is displayed in Fig. 6b). the TPR gained by SeWBO_Efficient DenseNet for 2 layers is 82.621%, 4 layers is 84.38%, 6 layers is 85.201%, and 8 layers is 86.795% when training data is 70%. Figure 6c) depicts the performance analysis of SeWBO_Efficient DenseNet regarding TNR. With 2, 4, 6, and 8 layers, the SeWBO_Efficient DenseNet achieves the TNR of 83.126%, 84.542%, 85.679%, and 86.928% when assuming training data 80%.
Performance concerning batch size
Figure 7 shows the assessment of SeWBO_Efficient DenseNet by concerning various batch sizes. The estimation of SeWBO_Efficient DenseNet with accuracy is represented in Fig. 7a). The accuracy gained by SeWBO_Efficient DenseNet for a batch size of 8 is 90.054%, a batch size of 16 is 91.201%, a batch size of 32 is 92.856%, and a batch size of 64 is 93.596% when considering training data 90%. The valuation of SeWBO_Efficient DenseNet regarding TPR is designated in Fig. 7b). With 8, 16, 32, and 64 batch sizes, the TPR attained by SeWBO_Efficient DenseNet is 80.401%, 81.854%, 83.010%, and 84.079% when considering training data 60%. Figure 7c) deliberates, the valuation of SeWBO_Efficient DenseNet regarding TNR. When the training data is 70%, the TNR achieved by SeWBO_Efficient DenseNet for a batch size of 8 is 78.645%, a batch size of 16 is 79.875%, a batch size of 32 is 81.205%, and a batch size of 64 is 82.304%.
Algorithmic techniques
The performance of SeWBO_Efficient DenseNet is analyzed by associating it with prevailing algorithmic approaches, such as Flamingo Search Algorithm (FSA) + Efficient DenseNet30,31,34, Gannet Optimization Algorithm (GOA) + Efficient DenseNet30,31,35, SBOA + Efficient DenseNet25,30,31, and WBO + Efficient DenseNet24,30,31.
Algorithmic analysis
The analysis of Efficient DenseNet by changing solution size is depicted in Fig. 8. The investigation of SeWBO + Efficient DenseNet by exploiting accuracy is stipulated in Fig. 8a). When a solution size is 5, the accuracy achieved by existing algorithmic approaches, including FSA + Efficient DenseNet is 85.012%, GOA + Efficient DenseNet is 88.210%, SBOA + Efficient DenseNet is 88.613%, and WBO + Efficient DenseNet is 89.647%, and the proposed SeWBO + Efficient DenseNet is 90.648%. Figure 8b) exhibits the examination of SeWBO + Efficient DenseNet concerning TPR. The TPR recorded by SeWBO + Efficient DenseNet is 93.310% and existing algorithmic techniques, like GOA + Efficient DenseNet is 87.201%, WBO + Efficient DenseNet is 91.346%, FSA + Efficient DenseNet is 85.499%, and SBOA + Efficient DenseNet is 90.042%, when a solution size is 10. The examination of SeWBO + Efficient DenseNet with TNR is shown in Fig. 8c). When the solution size is 15, the TNR gained by several algorithmic approaches, such as FSA + Efficient DenseNet is 86.318%, SBOA + Efficient DenseNet is 89.643%, GOA + Efficient DenseNet is 87.449%, WBO + Efficient DenseNet is 90.544% and the proposed SeWBO + Efficient DenseNet is 92.010%.
Comparative approaches
The comparative assessment is performed to measure the effectiveness of the SeWBO_Efficient DenseNet technique with respect to prevailing approaches, namely EL-SVM14, Multitask CNN15, YCNN13, IDLDMS-PTC16, and Efficient DenseNet.
Comparative analysis
The comparative appraisal of SeWBO_Efficient DenseNet to identify the pancreatic cancer is established concerning variosu performance metrics based on training data.
Based on training data
Figure 9 displays the estimation of SeWBO_Efficient DenseNet with respect to varying training data. The valuation of SeWBO_Efficient DenseNet concerning accuracy is represented in Fig. 9a). For training data 60%, the accuracy measured by SeWBO_Efficient DenseNet is 81.367% and the prevailing methods, including Efficient DenseNet is 73.542%, IDLDMS-PTC is 69.012%, YCNN is 66.549%, Multitask CNN is 67.810%, and EL-SVM is 65.145%. Here, the accuracy of SeWBO_Efficient DenseNet is enhanced by 11.08%, which is better than Multitask CNN. Figure 9b), represents, the comparative appraisal of SeWBO_Efficient DenseNet with respect to TPR. The TPR quantified by conventional techniques, namely EL-SVM is 69.813%, YCNN is 77.010%, Efficient DenseNet is 83.167%, Multitask CNN is 75.491%, and IDLDMS-PTC is 83.145% and the proposed SeWBO_Efficient DenseNet is 86.795% when training data is 70%. Now, the TPR of SeWBO_Efficient DenseNet is enhanced by 4.73% than EL-SVM. In Fig. 9c), the comparative assessment of SeWBO_Efficient DenseNet based on TNR is designated. The TNR obatined by SeWBO_Efficient DenseNet is 86.928% and the prevailing techniques, namely Multitask CNN, IDLDMS-PTC, EL-SVM, YCNN, and Efficient DenseNet is 72.154%, 78.645%, 70.348%, 74.857%, and 82.346% for training data 80%. Likewise, the TNR figured by SeWBO_Efficient DenseNet is 7.18% better than EL-SVM.
ROC analysis
The ROC analysis of SeWBO_Efficient DenseNet utilized to detect the pancreatic cancer is designated in Fig. 10. The TPR measured by SeWBO_Efficient DenseNet is 89.642% and the prevailing methods, namely Multitask CNN is 77.104%, EL-SVM is 73.124%, IDLDMS-PTC is 82.124%, YCNN is 80.164%, and Efficient DenseNet is 85.010%, for FPR of 80%.
Segmentation analysis
In order to evaluate the performance of SeWBO_PraNet is segmenting the lesions, it is compared with other methods, such as Deep Convolutional Network (DCN)36, 3D U-Net21, the multi-layer up-sampling structure37 based on segmentation accuracy. Figure 11 deliberates the segmentation analysis of SeWBO_PraNet for pancreatic cancer detection regarding training data. The segmentation accuracy gained by the prevailing frameworks, namely DCN is 72.454%, 3D U-Net is 78.645%, the multi-layer up-sampling structure is 82.105%, and SeWBO_PraNet is 85.042%. The performance of the SeWBO_PraNet is enhanced by 7.5% than the 3D-UNet.
Comparative discussion
Table 1 exhibits the comparative discussion of the developed SeWBO_Efficient DenseNet employed to detect pancreatic tumor. The SeWBO_Efficient DenseNet is assessed with a learning set of 90% with accuracy, TNR, and TPR, and the values figured by several models are delineated in the below table. The accuracy attained by the SeWBO_Efficient DenseNet is 93.596% and other techniques including Multitask CNN, EL-SVM, IDLDMS-PTC, YCNN, and Efficient DenseNet is 83.542%, 81.014%, 87.542%, 85.642%, and 89.240%. The TPR attained by the prevailing techniques, like IDLDMS-PTC is 90.434%, Multitask CNN is 85.246%, Efficient DenseNet is 92.382%, YCNN is 92.382%, and EL-SVM is 80.134% and the proposed SeWBO_Efficient DenseNet is 94.635%. Likewise, the TNR measured by the proposed SeWBO_Efficient DenseNet is 92.579%, and prevailing frameworks, namely Efficient DenseNet is 89.316%, YCNN is 83.612%, EL-SVM is 75.844%, IDLDMS-PTC is 86.391%, and Multitask CNN is 79.642%. In addition, Efficient DenseNet enhances parameter efficiency, and feature propagation, decreases computational cost, versatility, and better scalability. Moreover, SeWBO has better adaptability, effective exploitation and exploration, dynamic adjustment, and adaptability. Thus, the proposed SeWBO_Efficient DenseNet effectively detects pancreatic cancer from the CT images.
Conclusion
A kind of cancer which creates in the pancreas is termed as pancreatic cancer. Early recognition of pancreatic cancer is important for improving survivability by providing proper treatments. Therefore, an efficient SeWBO_Efficient DenseNet is presented for pancreatic tumor identification. Primarily, the CT image gathered is considered as an input. After that, image preprocessing is executed by utilizing a bilateral filter. Following this, lesion is segmented using PraNet, which is tuned by SeWBO. Here, the SeWBO is engineered by fusing SBOA and WBO. Then, in feature extraction, various features including DWT with CLBP, statistical, and SLBT features are mined. At last, pancreatic tumor detection is performed by using Efficient DenseNet trained by the SeWBO model, where Efficient DenseNet is developed by integrating EfficientNet and DenseNet. Additionally, the SeWBO_Efficient DenseNet attains better accuracy, TPR, and TNR of 93.596%, 94.635%, and 92.579% respectively. In the future, image augmentation methods can be used to address class imbalance and overfitting issues. Another possibility worth pursuing is the usage of transfer learning to minimize the training time and achieve efficient performance with fewer samples. One limitation of the proposed SeWBO_Efficient Dense Net model is its high computational cost, requiring powerful hardware (such as NVIDIA RTX 3090 or A100 GPUs) for training and inference. This may limit its deployment in real-time clinical applications, especially in resource-constrained environments such as small hospitals or remote healthcare centers. Future work will focus on optimizing the model’s efficiency, potentially by reducing the computational complexity through model pruning, quantization, or lightweight Transformer-based architectures.
Data availability
The Medical Segmentation Decathlon dataset used in this study is available to public and from “http://medicaldecathlon.com/”, accessed on July 20, 2024, that comprises 420 portal-venous phase CT scans of patients undergoing resection of pancreatic masses.
References
Zhang, Z., Yao, L., Keles, E., Velichko, Y. & Bagci, U. Deep learning algorithms for pancreas segmentation from radiology scans: A review. Adv. Clin. Radiol. 5(1), 31–52 (2023).
Sehmi, M. N. M., Fauzi, M. F. A., Ahmad, W. S. H. M. W. & Chan, E. W. L. Pancreatic cancer grading in pathological images using deep learning convolutional neural networks. F1000Research 10, 1057 (2021).
Zhang, Y., Wang, S., Qu, S. & Zhang, H. Support vector machine combined with magnetic resonance imaging for accurate diagnosis of paediatric pancreatic cancer. IET Image Proc. 14(7), 1233–1239 (2020).
Chi, H. et al. Proposing new early detection indicators for pancreatic cancer: Combining machine learning and neural networks for serum miRNA-based diagnostic model. Front. Oncol. 13, 1244578 (2023).
Sekaran, K., Chandana, P., Krishna, N. M. & Kadry, S. Deep learning convolutional neural network (CNN) with Gaussian mixture model for predicting pancreatic cancer. Multimed Tools Appl 79(15), 10233–10247 (2020).
Placido, D. et al. A deep learning algorithm to predict risk of pancreatic cancer from disease trajectories. Nat. Med. 29(5), 1113–1122 (2023).
Preuss, K. et al. Using quantitative imaging for personalized medicine in pancreatic cancer: A review of radiomics and deep learning applications. Cancers 14(7), 1654 (2022).
Zhang, Z., Keles, E., Durak, G., Taktak, Y., Susladkar, O., Gorade, V., Jha, D., Ormeci, A. C., Medetalibeyoglu, A., Yao, L. & Wang, B. Large-scale multi-center CT and MRI segmentation of pancreas with deep learning. arXiv preprint arXiv:2405.12367 (2024).
Zhang, Z., Li, S., Wang, Z. & Lu, Y. A novel and efficient tumor detection framework for pancreatic cancer via CT images. In Proceedings of 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), 1160–1164 (IEEE, 2020).
Liu, K. L. et al. Deep learning to distinguish pancreatic cancer tissue from non-cancerous pancreatic tissue: A retrospective study with cross-racial external validation. Lancet Digit. Health 2(6), e303–e313 (2020).
Chen, P. T. et al. Pancreatic cancer detection on CT scans with deep learning: A nationwide population-based study. Radiology 306(1), 172–182 (2023).
Asaturyan, H., Gligorievski, A. & Villarini, B. Morphological and multi-level geometrical descriptor analysis in CT and MRI volumes for automatic pancreas segmentation. Comput. Med. Imaging Graph. 75, 1–13 (2019).
Dinesh, M. G., Bacanin, N., Askar, S. S. & Abouhawwash, M. Diagnostic ability of deep learning in detection of pancreatic tumour. Sci. Rep. 13(1), 9725 (2023).
Li, M. et al. Computer-aided diagnosis and staging of pancreatic cancer based on CT images. IEEE Access 8, 141705–141718 (2020).
Cao, K. et al. Large-scale pancreatic cancer detection via non-contrast CT and deep learning. Nat. Med. 29(12), 3033–3043 (2023).
Vaiyapuri, T., Dutta, A. K., Punithavathi, I. H., Duraipandy, P., Alotaibi, S. S., Alsolai, H., Mohamed, A. & Mahgoub, H. Intelligent deep-learning-enabled decision-making medical system for pancreatic tumor classification on CT images. In Healthcare Vol. 10(4), 677 (MDPI, 2023).
Asadpour, V. et al. Pancreatic cancer tumor analysis in CT images using patch-based multi-resolution convolutional neural network. Biomed. Signal Process. Control 68, 102652 (2021).
Nakao, M., Nakamura, M., Mizowaki, T. & Matsuda, T. Statistical deformation reconstruction using multi-organ shape features for pancreatic cancer localization. Med. Image Anal. 67, 101829 (2021).
Gandikota, H. P. CT scan pancreatic cancer segmentation and classification using deep learning and the tunicate swarm algorithm. PLoS ONE 18(11), e0292785 (2023).
Hooshangnejad, H., Chen, Q., Feng, X., Zhang, R. & Ding, K. deepPERFECT: novel deep learning CT synthesis method for expeditious pancreatic cancer radiotherapy. Cancers 15(11), 3061 (2023).
Ramaekers, M. et al. Improved pancreatic cancer detection and localization on CT scans: A computer-aided detection model utilizing secondary features. Cancers 16(13), 2403 (2024).
Wang, T., Feng, H., Li, S. & Yang, Y. Medical image denoising using bilateral filter and the K-SVD algorithm. In Proceedings of Journal of Physics: Conference Series, Vol. 1229(1), 012007 (IOP Publishing, 2019).
Fan, D. P., Ji, G. P., Zhou, T., Chen, G., Fu, H., Shen, J. & Shao, L. Pranet: Parallel reverse attention network for polyp segmentation. In Proceedings of International Conference on Medical Image Computing and Computer-Assisted Intervention, 263–273, (Springer International Publishing, Cham, 2020).
Azizi, M. et al. Wolf-Bird Optimizer (WBO): A novel metaheuristic algorithm for Building Information Modeling-based resource tradeoff. J. Eng. Res. https://doi.org/10.1016/j.jer.2023.11.024 (2023).
Fu, Y., Liu, D., Chen, J. & He, L. Secretary bird optimization algorithm: A new metaheuristic for solving global optimization problems. Artif. Intell. Rev. 57(5), 1–102 (2024).
Guermoui, M. & Mekhalfi, M. L. A sparse representation of complete local binary pattern histogram for human face recognition. arXiv preprint arXiv:1605.09584 (2016).
Varuna Shree, N. & Kumar, T. N. R. Identification and classification of brain tumor MRI images with feature extraction using DWT and probabilistic neural network. Brain Inform. 5(1), 23–30 (2018).
Lakshmiprabha, N. S. & Majumder, S. Face recognition system invariant to plastic surgery. In Proceedings of 12th International Conference on Intelligent Systems Design and Applications (ISDA), 258–263 (IEEE, 2012).
Lessa, V. & Marengoni, M. Applying artificial neural network for the classification of breast cancer using infrared thermographic images. In Proceedings of Computer Vision and Graphics: International Conference, ICCVG, Warsaw, Poland, 19–21, Proceedings Vol. 8, 429–438 (Springer International Publishing, 2016).
Alhichri, H., Alswayed, A. S., Bazi, Y., Ammour, N. & Alajlan, N. A. Classification of remote sensing images using EfficientNet-B3 CNN model with attention. IEEE Access 9, 14078–14094 (2021).
Vulli, A. et al. Fine-tuned DenseNet-169 for breast cancer metastasis prediction using FastAI and 1-cycle policy. Sensors 22(8), 2988 (2022).
Bhaladhare, P. R. & Jinwala, D. C. A clustering approach for the l-diversity model in privacy preserving data mining using fractional calculus-bacterial foraging optimization algorithm. Adv. Comput. Eng. 2014(1), 396529 (2014).
Medical Segmentation Decathlon dataset is taken from, “http://medicaldecathlon.com/”, accessed on July 2024.
Zhiheng, W. & Jianhua, L. Flamingo search algorithm: a new swarm intelligence optimization algorithm. IEEE Access 9, 88564–88582 (2021).
Pan, J. S., Zhang, L. G., Wang, R. B., Snášel, V. & Chu, S. C. Gannet optimization algorithm: A new metaheuristic algorithm for solving engineering optimization problems. Math. Comput. Simul. 202, 343–373 (2022).
Roth, H. R., Farag, A., Lu, L., Turkbey, E. B. & Summers, R. M. Deep convolutional networks for pancreas segmentation in CT imaging. In Medical Imaging 2015: Image Processing, Vol. 9413, 378–385 (SPIE, 2015).
Fu, M. et al. Hierarchical combinatorial deep learning architecture for pancreas segmentation of medical computed tomography cancer images. BMC Syst. Biol. 12, 119–127 (2018).
Author information
Authors and Affiliations
Contributions
MEKALA SANDHYA was responsible for conceptualization, methodology, and data analysis. Phani Kumar S conducted data collection and visualization and prepared the initial draft of the manuscript. Both authors contributed to the review and editing of the manuscript and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing interests as defined by Nature Research, or any other interests that might be perceived to influence the results and/or discussion reported in this paper.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mekala, S., S, P. Enhancing pancreatic cancer detection in CT images through secretary wolf bird optimization and deep learning. Sci Rep 15, 19787 (2025). https://doi.org/10.1038/s41598-025-00512-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-00512-6
Keywords
This article is cited by
-
Secretary bird optimization algorithm incorporating independent thinking mechanism and sine-square step length for feature selection
Scientific Reports (2026)
-
HBoEQN: Hybrid Attention-Based Quantum ReLU-Applied Lightweight Deep Learning Model for Pancreatic Cancer Detection Using Computed Tomography
Biomedical Materials & Devices (2025)