Abstract
Recent advancements in large language models (LLMs) have significantly enhanced text generation across various sectors; however, their medical application faces critical challenges regarding both accuracy and real-time responsiveness. To address these dual challenges, we propose a novel two-step retrieval and ranking retrieval-augmented generation (RAG) framework that synergistically combines embedding search with Elasticsearch technology. Built upon a dynamically updated medical knowledge base incorporating expert-reviewed documents from leading healthcare institutions, our hybrid architecture employs ColBERTv2 for context-aware result ranking while maintaining computational efficiency. Experimental results show a 10% improvement in accuracy for complex medical queries compared to standalone LLM and single-search RAG variants, while acknowledging that latency challenges remain in emergency situations requiring sub-second responses in an experimental setting, which can be achieved in real-time using more powerful hardware in real-world deployments. This work establishes a new paradigm for reliable medical AI assistants that successfully balances accuracy and practical deployment considerations.
Similar content being viewed by others
Introduction
In the rapidly evolving healthcare ecosystem, the integration of big data analytics and Artificial Intelligence (AI) technologies is fundamentally transforming clinical practices. Notably, Large Language Models (LLMs)1,2, exemplified by ChatGPT3, have demonstrated significant potential across various healthcare domains, particularly in patient counseling and medical knowledge dissemination4. The emergence of specialized Medical LLMs5,6,7 has provided healthcare professionals with powerful tools for navigating complex clinical scenarios, offering valuable insights that facilitate early diagnosis and enhance patient guidance. However, the effective deployment of these models in medical settings necessitates rigorous validation processes and enhanced accuracy to ensure both the precision and comprehensibility of generated outputs.
This study investigates the development and evaluation of a medical-focused LLM architecture that incorporates Retrieval-Augmented Generation (RAG) technology8 and integrates external knowledge bases to significantly enhance the system’s information retrieval and response generation capabilities. While advanced language models (e.g., GPT3, BERT9, LLaMA10) have achieved remarkable progress in Natural Language Processing (NLP)11, demonstrating substantial capacity for storing factual knowledge during pre-training and excelling in various NLP tasks, they continue to face challenges in maintaining precision within knowledge-intensive domains such as medicine and open-domain question answering.
To address these limitations, Lewis et al. introduced the RAG framework8, which synergistically combines pre-trained parameterized memories (e.g., sequence-to-sequence models) with non-parameterized external knowledge sources (e.g., dense vector indexes derived from comprehensive databases such as Wikipedia). Although current large-scale pre-trained language models can store factual knowledge in their parameters and achieve state-of-the-art performance on downstream NLP tasks, their ability to access and accurately manipulate this knowledge remains limited, particularly in tasks requiring deep expertise. Additionally, providing traceable sources for modeling decisions and updating world knowledge are ongoing research challenges8. RAG models overcome these limitations by introducing a differentiable mechanism that enables access to explicitly non-parameterized memory. The researchers propose two forms of RAG models: one that performs conditional computation based on the same retrieval passages throughout the generation of sequences8, and another that allows different passages to be used for each generated token12. By utilizing neural retrieval and dense-representation techniques such as Dense Passage Retrieval (DPR)13, RAG enhances the performance of traditional sparse-retrieval methods, improving answer quality and contextual relevance. However, challenges such as capturing domain expertise more effectively, ensuring timely and accurate retrieval, and maintaining answer-source transparency and traceability remain areas for further research14,15. There are also many knowledge injection methods to enhance the generation of LLM16,17.
In the medical field, models like ChatDoctor18 integrate online and offline retrieval mechanisms to refine medical advice capabilities, surpassing the performance of general-purpose models like ChatGPT4. Zhongjing19, the first LLaMA-based large-scale model in the Chinese medical field, has demonstrated proficiency in handling complex conversations and spontaneous queries following intensive training and reinforcement learning from human feedback (RLHF). Despite these advancements, studies such as ChatGPT_USMLE20 highlight ongoing limitations in mastering detailed medical knowledge. Additionally, Wikichat21 illustrates how retrieval-augmented dialogue generation can effectively reduce misinformation, outperforming previous methods in both simulated and real-world user tests.
This paper aims to advance existing RAG technology by reducing its hallucination rate and enhancing its interpretability for applications in the medical field, thereby enabling it to fulfill a unique role in this domain. Our proposed medical LLM represents an innovative integration of technologies, specifically built around Chroma, Elasticsearch, and ColBERTv2 models. The primary contributions of this work are as follows: (1) The implementation of dual retrieval mechanisms-combining word-term and semantic retrieval-to improve retrieval accuracy and mitigate the semantic gap between prompt words and retrieved text. (2) The introduction of double sorting and the incorporation of ColBERTv2 to enhance the precision of retrieval relevance judgment. (3) The pioneering application of RAG in the medical field, with results rigorously evaluated by professional physicians to ensure clinical relevance and accuracy.
Methods
All experiments were conducted on an Ubuntu 22.04 server equipped with an NVIDIA A40 GPU. The model used for inference is IvyGPT, a large language model (LLM) fine-tuned on medical data, which demonstrates sufficient performance to support this study.
IvyGPT
Wang et al. introduced IvyGPT22, a healthcare-specialized LLM based on the LLaMA architecture, designed to address the limitations of general-purpose LLMs like ChatGPT in healthcare applications, particularly concerning accuracy and professional suitability. IvyGPT was meticulously trained through supervised learning using high-quality medical Q&A datasets and further refined through reinforcement learning with human feedback (RLHF)23. This training enables IvyGPT to excel in conducting extended, informative dialogues and generating detailed, comprehensive treatment plans.
Utilizing the QLoRA training technique24, IvyGPT distinguishes itself as a state-of-the-art medical-oriented GPT model. Additionally, the research team contributed a novel dataset tailored to the Chinese healthcare context, on which IvyGPT demonstrates significant improvements over existing models. The design and empirical results of IvyGPT suggest its potential for broad applications in medical education, self-service patient assistance, and consultation services, significantly enhancing the precision and efficiency of medical diagnoses and treatment recommendations.
Retrieval-augmented generation
The core of Retrieval-Augmented Generation (RAG) lies in its integration of a pre-trained neural retriever with a pre-trained sequence-to-sequence (seq2seq) model, forming an end-to-end trainable probabilistic framework. This architecture not only enhances knowledge acquisition capabilities but also enables access to external knowledge resources without requiring additional training, thanks to its inherent pre-trained retrieval mechanism. Moreover, RAG extends beyond generative tasks and can be adapted to sequence classification tasks by treating the target category as a single-token target sequence.
In this study, we enhance the RAG model by implementing a dual-search strategy across Elasticsearch and Chroma, followed by passing the merged search results to ColBERTv2 for semantic ranking. This refinement significantly improves RAG’s retrieval capabilities.
Elasticsearch25,26 has become a critical technology in developing information retrieval systems for knowledge-centric chatbots. Its ability to handle vast repositories of large-scale text data with unparalleled retrieval speed and real-time information updates makes it particularly valuable. By integrating Elasticsearch, developers can efficiently create indexes and perform high-speed searches across extensive textual datasets, such as entire Wikipedia entries. This integration provides chatbots with precise and up-to-date knowledge support, enhancing both the accuracy and relevance of conversational content.
Chroma is an advanced vector similarity search technology designed to efficiently process large-scale text datasets. By leveraging a pre-trained text embedding model, Chroma transforms documents into compact, dense vectors. Its unique vector indexing architecture and optimized search algorithms enable precise identification of the most relevant documents. With its superior design, Chroma handles extensive datasets with minimal latency and high processing speeds, making it a versatile solution for various text retrieval applications.
ColBERTv227 is an advanced retrieval engine that employs multi-vector representations to capture the contextual semantic nuances of token-level features through cluster centroids. It innovatively reduces the storage requirements of the multi-vector system by utilizing residual representations, which encode the differences between the original vectors and their approximations, thereby improving retrieval accuracy while optimizing space efficiency. Building on the original ColBERTv2 model, this version enhances its supervisory mechanism by incorporating residual compression techniques and distillation insights from cross-encoder systems, further refining its performance.
System design
The design of our system is fundamentally motivated by a profound consideration of domain-specific knowledge representation. In highly specialized fields such as rare diseases and cutting-edge medical research, relevant terminologies often exhibit extremely low frequency or complete absence in model training corpora. This data sparsity poses significant challenges for traditional vector databases (e.g., Chroma) in converting these specialized terms into semantic vectors, because the limited understanding of domain knowledge in pre-trained models and the out-of-distribution nature of these terms may lead to vector representations that do not accurately capture professional semantics. To address this, we innovatively incorporate Elasticsearch’s term-based search mechanism, which ensures that even the most recently emerged, hard-to-vectorize professional terms can be effectively retrieved and utilized to enhance the model’s generation capabilities.
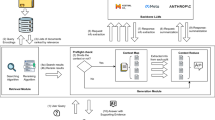
The entire system in Fig. 1 is thoughtfully structured into 5 interconnected modules: Knowledge Base, Question, Retrieval, Generation, and Check. The Check module is responsible for validating the generated answer against the retrieved results to ensure consistency and accuracy. In unison, these 5 modules collaborate effectively to generate potentially accurate responses to every user inquiry, guaranteeing a fluid and efficient solution-finding process.
The Question module expertly handles the receipt and processing of healthcare-related queries from users through an interactive chat interface. The system permits users to type in free-form text and provides real-time feedback as the user types, ensuring a responsive and dynamic interaction. Once the user submits their question, it is transformed into a vector representation.
In the Retrieval stage, the vector representation of the user’s question is sought in both the Chroma and Elasticsearch databases. The retrieval process inherently involves computing similarities between vectors, employing differing methodologies: Chroma leverages dense vector embeddings for semantic similarity search, while Elasticsearch utilizes traditional term-based search with support for structured data. Given their respective merits and limitations, the system adopts a hybrid approach to combine the strengths of both methods. Subsequently, the hybrid search results from both databases are collectively passed to the ColBERTv2 model. ColBERTv2 employs multi-vector representations and residual compression techniques to capture fine-grained semantic nuances, enabling it to re-rank the hybrid results and select the top five most semantically aligned matches.These top five selections are then inserted as prompts into IvyGPT, which generates the corresponding answer.
Finally, the system’s UI presents the generated answer to the user, accompanied by the first five search results underneath. This arrangement allows the user to evaluate if the search outputs correspond with the actual generated answer, thereby deciding whether to accept or reject the answer produced by IvyGPT.
The external medical files utilized in this system have been sourced from a collaborating Hospital, a distinguished Grade 3A comprehensive healthcare facility that combines multiple facets of medical service delivery, emergency care, health maintenance, rehabilitation, scientific research, and educational instruction.
The knowledge base houses an extensive array of medical subspecialties, extending from Orthopedics, which encompasses diagnosing and treating orthopedic trauma, hand and foot injuries, joint dysfunctions, bone pathologies, and the resultant symptoms such as pain, swelling, and limited mobility; to Pediatrics, which specializes in handling pediatric emergencies like respiratory distress, circulatory collapse, shock, and a variety of other life-threatening conditions; and Ophthalmology, which delves deeply into diagnosing and managing a broad spectrum of eye disorders, including eyelid abnormalities, surface ocular diseases, conjunctivitis, keratitis, and conditions affecting the sclera. These medical files are preprocessed and converted into vector representations, which are stored in both the Chroma and Elasticsearch vector databases for efficient retrieval. Figure 2 illustrates some of the diseases and their presenting symptoms that may occur in orthopedics.
Moreover, the database features a thorough and meticulous Hospital Departmental Guide, which meticulously delineates the functions and areas of expertise for each department within the institution, specifies their floor-by-floor location arrangements, and offers detailed descriptions of the symptoms and unique attributes of the diseases they specialize in treating. This guide is also indexed and stored in the vector databases, enabling the system to retrieve relevant department information when needed.
Results
Experiments
To more comprehensively evaluate and substantiate the effectiveness of the proposed system, the experimental design comprises a two-fold approach. The first component involves an ablation experiment, aimed at discerning the individual contributions and significance of each integrated module within the system. This phase systematically removes or alters components of the Chroma + Elasticsearch + ColBERTv2 architecture to assess the impact on overall performance, thereby elucidating the inherent value and functionality of each constituent element. The three systems tested are listed in Table 1.
The second part of the experiment focuses on a comparative analysis, comparing the system with RAG to the system without RAG. This comparison aims to demonstrate the importance of RAG in Medical LLMs operations through both quantitative and qualitative methods. By examining the efficacy of the system in handling complex medical scenarios and generating accurate and rich responses, a robust assessment of the system’s superiority in real-world medical applications is made. System 3 and System 4, and SELF-RAG in Table 2 are the subjects of this experiment.
To further validate our system’s performance, we conducted a series of rigorous evaluations focusing on complex clinical scenarios. Specifically, we designed diagnostic tests for complicated disease cases, establishing multiple simulated scenarios with intricate clinical manifestations. Each test case was carefully constructed to include complex symptom combinations and challenging diagnostic conditions, thereby thoroughly assessing the system’s robustness and practical utility in real-world medical applications. We used 20 complex cases to test our system to evaluate whether it can correctly guide patients to the designated floor and designated department for treatment. As shown in Table 3, we present two examples. Our system can correctly complete this task in most cases. In the first example, the model explains high blood pressure and the vascular system, considers other possible diagnoses, and addresses symptoms such as headaches and physical manifestations. In the second case, the model should first consider specific aneurysm or nuclear disease, and the next is the problem of thyroid function.
Evaluation
Assessing the performance of LLMs with RAG has posed a significant challenge, the generation quality of the model is difficult to evaluate using quantitative standards. Among the myriad approaches taken by previous researchers, some have subjected the models to human-like examinations29, while others employed crowdsourcing to gauge the quality of the models’ responses21. We believe that using crowdsourced human inspections will be biased due to the lack of professionalism among non-experts, so we asked professional doctors to participate in our evaluation.
In contrast, this present study employs a questionnaire-based methodology. Given that LLM engage in conversations with humans, the subjective perceptions of users during these interactions hold paramount importance. To this end, the experiment translates human subjects’ subjective impressions into numerical scores across various dimensions, thereby providing a metric for evaluating the models’ performance. Five key dimensions were identified for the evaluation: (1) Relevance, (2) Accuracy, (3) Anthropomorphism, (4) Speed, and (5) Usefulness.
To enhance credibility, this questionnaire departs from previous practices by selecting a heterogeneous sample of 100 testers, comprising practicing doctors from collaborating hospital, medical students, and university professors specializing in medicine. This diversity enriches the reliability of the collected data.
Nonetheless, the questionnaire is not entirely subjective. It establishes clear boundaries for the types of questions and expected answers, briefing testers about these limitations before they embark on the evaluation. Specifically, the medical inquiries are restricted to common, non-life-threatening ailments, as our system is intended to support patients in making informed decisions and to facilitate expedited access to medical services.
Questionares
For each experiment, a pair of custom-designed questionnaires was crafted to capture distinct aspects of the systems’ performance. Questionnaire 1, predominantly emphasizes the assessment of the systems’ search capabilities. It discriminates among the three systems being evaluated based on their unique searching proficiencies alone, thus allowing for a targeted evaluation of this crucial functionality. In contrast, Questionnaire 2 shifts its focus to delve deeper into the level of medical specialization embedded within the systems undergoing testing.
Questionnaire 1 for Ablation Experiment:

Questionnaire 2 for Comparison Experiment:

Results analysis
Figure 3 illustrates the outcomes of the administered Questionnaire 1. From Fig. 3, it is evident that System 3 consistently surpasses Systems 1 and 2 in the dimensions of Relevance and Usefulness, showcasing a notable advantage. We believe that this is because when Elasticsearch performs keyword search, the original text with slightly different semantics from the declarative sentence causes the traditional RAG method to deviate in the relevance calculation when asking questions using interrogative sentences, while the auxiliary meta-search can find relevant content as much as possible. Concerning Accuracy, System 3 marginally edges out Systems 1 and 2, suggesting that all three systems possess a commendable ability to deliver relatively accurate medical information. However, when it comes to Speed, System 3 exhibits a slight decrease in performance compared to its predecessors, which can be attributed to its integration of dual search models, leading to a somewhat prolonged response time.
Regarding the cumulative scores garnered from Questionnaire 1, both System 1 and System 3 managed to attain satisfactory scores exceeding the benchmark of 60, with System 3 notably achieving a commendable high score of 70. Conversely, System 2 fell short of the passing threshold. This outcome underscores the superior performance of Chroma, equipped with its semantic search functionality, in accomplishing this particular task.
In the illustrations depicted in Fig. 4, System 3 outperforms System 4 across nearly all metrics, except for Speed. Here, System 4 takes a considerable lead as it lacks the RAG feature. Nevertheless, the scores in the remaining categories clearly show that LLM enhanced with RAG capabilities provides superior overall performance, higher accuracy, and greater practicality, especially in fields such as medicine that require precise expertise, where the necessity to reduce hallucination rates and improve accuracy is higher. The shortcomings in speed can be corrected through hardware upgrades and optimizations. Although the dual search slows down the whole system, it is still slightly faster than SELF-RAG, and the proposed pipeline is only slightly inferior to SELF-RAG in terms of Anthropomorphism. This may be due to the influence of training and fine-tuning of the large language model. Since our medical language model has been fine-tuned on professional medical knowledge, it has certain professional capabilities. However, the distribution of professional data is quite different from that of the open domain, which affects its score in anthropomorphism.
We quantified all our tests that are shown in Table 4, measuring the performance of each system by MRR and MAP, and added SELF-RAG for comparison. Our proposed pipeline achieved the best results in both indicators. SELF-RAG was second only to our pipeline in MRR. We believe this is because the model with 13B parameters in SELF-RAG is not a professional medical model and does not include word-meta retrieval. In an environment containing a large number of professional domain words, it is difficult to query content that has not been correctly vectorized by the database.
Discussion
The study reveals the revolutionary effect of incorporating RAG technology within a Medical LLM Guidance System. The fusion of Chroma’s vector similarity search, Elasticsearch’s real-time indexing capabilities, and a continuously updated medical knowledge corpus forms a hybrid architecture that markedly enhances the precision and pertinence of generated responses. Experimental findings indicate that the strategic blend of Chroma and Elasticsearch technologies optimizes information retrieval and synthesis. Finally uses ColBertv2 to re-rank the screening results. This approach greatly improves the accuracy of existing RAGs, thereby laying a robust groundwork for dependable AI-powered decision-making in the realm of healthcare.
While the Medical LLM with RAG demonstrates substantial benefits and improvements in medical knowledge retrieval and dialogue generation, several limitations should be acknowledged. The reliance on a dual search mechanism involving both Chroma and Elasticsearch may contribute to a decrease in response speed, though this trade-off is offset by the significant gains in accuracy and richness of provided information. Moreover, despite the inclusion of vetted medical documents, there remains a need for continuous monitoring and updating of the knowledge base to account for the latest advancements and changes in medical science.
In our medical database, which contains a vast collection of specialized terminology such as ’palpitations’, ’skin rash’, and ’erosion’, we encounter a significant challenge when processing patient self-reports. These reports often include layman’s descriptions like ’irregular heartbeat’, ’blistering’, or ’skin damage’, which cannot be effectively matched with professional medical terms using Elasticsearch’s token-based search alone. To address this limitation, we integrate Chroma for semantic vector matching, which enables more accurate mapping between colloquial expressions and technical medical terminology.
However, in professional contexts, such as physician-curated medical records, we observe that inputs often fall outside the training data distribution of both the LLM and the vectorization model used by Chroma. In such cases, when the matching content exists within our database, token-based search proves to be more accurate than vector search for identifying highly similar content. Therefore, we propose a hybrid approach that combines Chroma’s vector retrieval for supplementary content matching beyond keywords with Elasticsearch’s token-based search, resulting in enhanced overall performance and more comprehensive search capabilities.
The proposed system exhibits two primary limitations: temporal efficiency and the necessity of high-quality database content. Due to the implementation of dual retrieval mechanisms (lexical and vector-based) and subsequent two-stage ranking, the system incurs higher computational overhead compared to conventional approaches. This increased processing time is a trade-off for enhanced retrieval accuracy and comprehensive results.
Furthermore, the quality of the database is a critical factor in determining the system’s output reliability. In our experiments, when utilizing professionally curated medical data from hospital sources-such as physician-annotated records and department-level information-the system demonstrated high precision in recommending appropriate medical departments and their corresponding physical locations within the hospital. However, when integrating data scraped from medical forums into the database, the system’s performance degraded significantly. For instance, even for common symptoms like “fever,” the system retrieved highly improbable disease-related snippets (e.g., cancer), which were irrelevant to the query context. This highlights the importance of maintaining a high-quality, professionally vetted database to ensure the system’s practical utility and reliability in real-world medical scenarios.
Future directions will explore three optimization pathways: 1) Hardware-accelerated retrieval pipelines 2) Lightweight neural ranking models 3) Context-aware search pruning algorithms. These developments aim to preserve accuracy gains while achieving emergency-ready performance, ultimately bridging the gap between algorithmic innovation and clinical implementation requirements.
Conclusion
Our two-stage retrieval and ranking RAG framework represents a significant advancement in medical QA systems, addressing the dual challenges of accuracy and responsiveness in medical AI applications. By synergistically integrating embedding search with Elasticsearch technology and leveraging ColBERTv2 for context-aware ranking, our approach achieves a 10% improvement in accuracy over standalone LLMs and single-search RAG variants. This performance gain underscores the effectiveness of hybrid retrieval architectures in handling complex medical inquiries, particularly when built upon a dynamically updated medical knowledge base curated from expert-reviewed documents.
In the experimental environment, the response time is slightly longer than other systems due to equipment limitations, but better hardware in actual deployment can mitigate this issue and achieve real-time response. Our work establishes a robust paradigm for reliable medical AI assistants, successfully balancing accuracy with practical deployment considerations.
Data availability
Some of the data that support the fndings of this study are available from the corresponding author upon reasonable request. Some of the public ones are available through https://github.com/WangRongsheng/awesome-LLM-resourses
References
Hadi, M. U. et al. Large language models: A comprehensive survey of its applications, challenges, limitations, and future prospects. https://doi.org/10.36227/techrxiv.23589741.v8 (2025).
Brown, T. et al. Language models are few-shot learners. Adv. Neural. Inf. Process. Syst. 33, 1877–1901 (2020).
Achiam, J. et al. Gpt-4 technical report. arXiv preprint arXiv:2303.08774 (2023).
Wu, T. et al. A brief overview of chatgpt: The history, status quo and potential future development. IEEE/CAA J. Autom. Sin. 10, 1122–1136 (2023).
Thirunavukarasu, A. J. et al. Large language models in medicine. Nat. Med. 29, 1930–1940 (2023).
Li, J. et al. Instruction fine-tuning of large language models for traditional Chinese medicine. In China Conference on Knowledge Graph and Semantic Computing, 419–430 (Springer, 2024).
Wang, H. et al. Knowledge-tuning large language models with structured medical knowledge bases for trustworthy response generation in Chinese. ACM Trans. Knowl. Discov. Data 19, 1–17 (2025).
Lewis, P. et al. Retrieval-augmented generation for knowledge-intensive nlp tasks. Adv. Neural. Inf. Process. Syst. 33, 9459–9474 (2020).
Devlin, J., Chang, M.-W., Lee, K. & Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, volume 1 (Long and Short Papers), 4171–4186 (2019).
Touvron, H. et al. Llama 2: Open foundation and fine-tuned chat models. arXiv preprint arXiv:2307.09288 (2023).
Chowdhary, K. & Chowdhary, K. Natural language processing. In Fundamentals of Artificial Intelligence, 603–649 (2020).
Borgeaud, S. et al. Improving language models by retrieving from trillions of tokens. In International Conference on Machine Learning, 2206–2240 (PMLR, 2022).
Karpukhin, V. et al. Dense passage retrieval for open-domain question answering. In EMNLP, Vol. 1, 6769–6781 (2020).
Fan, W. et al. A survey on rag meeting llms: Towards retrieval-augmented large language models. In Proceedings of the 30th ACM SIGKDD Conference on Knowledge Discovery and Data Mining, 6491–6501 (2024).
Jeong, S., Baek, J., Cho, S., Hwang, S. J. & Park, J. Adaptive-rag: Learning to adapt retrieval-augmented large language models through question complexity. In NAACL (2024).
Yasunaga, M. et al. Deep bidirectional language-knowledge graph pretraining. In Neural Information Processing Systems (NeurIPS) (2022).
Cheng, Y., Li, K. & Kang, Z. Emkg: Efficient matchings for knowledge graph integration in stance detection. In 2024 International Joint Conference on Neural Networks (IJCNN), 1–8. https://doi.org/10.1109/IJCNN60899.2024.10651163 (2024).
Li, Y. et al. ChatDoctor: A Medical Chat Model Fine-Tuned on a Large Language Model Meta-AI (LLaMA) Using Medical Domain Knowledge. Cureus 15(6), e40895. https://doi.org/10.7759/cureus.40895 (2023).
Yang, S. et al. Zhongjing: Enhancing the Chinese medical capabilities of large language model through expert feedback and real-world multi-turn dialogue. In In Proceedings of the AAAI Conference on Artificial Intelligence, Vol. 38, 19368–19376 (2024).
Gilson, A. et al. How does ChatGPT perform on the united states medical licensing examination (USMLE)? The implications of large language models for medical education and knowledge assessment. JMIR Med. Educ. 9, e45312 (2023).
Semnani, S., Yao, V., Zhang, H. C. & Lam, M. Wikichat: Stopping the hallucination of large language model chatbots by few-shot grounding on Wikipedia. In The 2023 Conference on Empirical Methods in Natural Language Processing (2023).
Wang, R. et al. Ivygpt: Interactive Chinese pathway language model in medical domain. In CAAI International Conference on Artificial Intelligence, 378–382 (Springer, 2023).
Ouyang, L. et al. Training language models to follow instructions with human feedback. Adv. Neural. Inf. Process. Syst. 35, 27730–27744 (2022).
Dettmers, T., Pagnoni, A., Holtzman, A. & Zettlemoyer, L. Qlora: Efficient finetuning of quantized llms. In Advances in Neural Information Processing Systems, Vol. 36 (2024).
Elasticsearch, B. Elasticsearch. In software], version, Vol. 6 (2018).
Chen, D. et al. Real-time or near real-time persisting daily healthcare data into HDFS and elasticsearch index inside a big data platform. IEEE Trans. Ind. Inf. 13, 595–606 (2016).
Santhanam, K., Khattab, O., Saad-Falcon, J., Potts, C. & Zaharia, M. A. Colbertv2: Effective and efficient retrieval via lightweight late interaction. In North American Chapter of the Association for Computational Linguistics (2021).
Asai, A., Wu, Z., Wang, Y., Sil, A. & Hajishirzi, H. Self-rag: Self-reflective retrieval augmented generation. In NeurIPS 2023 Workshop on Instruction Tuning and Instruction Following (2023).
Zhang, H. et al. HuatuoGPT, towards taming language model to be a doctor. In Bouamor, H., Pino, J. & Bali, K. (eds.) Findings of the Association for Computational Linguistics: EMNLP 2023, 10859–10885 (Association for Computational Linguistics, Singapore, 2023). https://doi.org/10.18653/v1/2023.findings-emnlp.725.
Acknowledgements
This work is supported by Science and Technology Development Fund of Macao (0041/2023/RIB2) and Macao Polytechnic University Grant (RP/FCA-15/2022).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, Q., Zuo, H., Su, R. et al. Dual retrieving and ranking medical large language model with retrieval augmented generation. Sci Rep 15, 18062 (2025). https://doi.org/10.1038/s41598-025-00724-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-00724-w