Abstract
Background and objective: Identifying patients suitable for conversion therapy through early non-invasive screening is crucial for tailoring treatment in advanced gastric cancer (AGC). This study aimed to develop and validate a deep learning method, utilizing preoperative computed tomography (CT) images, to predict the response to conversion therapy in AGC patients. This retrospective study involved 140 patients. We utilized Progressive Distill (PD) methodology to construct a deep learning model for predicting clinical response to conversion therapy based on preoperative CT images. Patients in the training set (n = 112) and in the test set (n = 28) were sourced from The First Affiliated Hospital of Wenzhou Medical University between September 2017 and November 2023. Our PD models’ performance was compared with baseline models and those utilizing Knowledge Distillation (KD), with evaluation metrics including accuracy, sensitivity, specificity, receiver operating characteristic curves, areas under the receiver operating characteristic curve (AUCs), and heat maps. The PD model exhibited the best performance, demonstrating robust discrimination of clinical response to conversion therapy with an AUC of 0.99 and accuracy of 99.11% in the training set, and 0.87 AUC and 85.71% accuracy in the test set. Sensitivity and specificity were 97.44% and 100% respectively in the training set, 85.71% and 85.71% each in the test set, suggesting absence of discernible bias. The deep learning model of PD method accurately predicts clinical response to conversion therapy in AGC patients. Further investigation is warranted to assess its clinical utility alongside clinicopathological parameters.
Similar content being viewed by others
Introduction
Recent epidemiological data highlights gastric cancer (GC) as the fifth most common malignancy worldwide in terms of incidence, and the fourth leading cause of cancer-related mortality1. Due to the lack of obvious specificity in clinical symptoms of early gastric cancer, most GC patients are often diagnosed with progression. Advanced gastric cancer (AGC) accounts for 70–80% of all GC cases2. Owing to factors such as extensive distant metastasis, peritoneal metastasis, or local progression, some patients with AGC were unable to undergo radical resection surgery and can only undergo conservative treatment to delay disease progression. The prognosis of such patients is poor, with a 5-year overall survival rate ranging between 30%and 40%3. Better treatment for this type of patient still faces challenges in improving prognosis. Over recent years, advances in understanding of the occurrence, development and biological behavior characteristics of GC, as well as the development and application of new anticancer drugs, including chemotherapy drugs, molecularly targeted drugs and immunotherapy drugs, have reshaped the treatment concept and strategy of AGC. Various strategies are currently emerged to enhance survival rates, with conversion therapy proving effective and showing improved survival rates in patients with unresectable AGC4,5.
Conversion therapy is a treatment approach aimed at reducing initially unresectable or borderline resectable tumors, due to surgical technical and/or oncological reasons, can be reduced into a lower stage through active and effective.
chemotherapy and other comprehensive treatments. Its primary goal is to diminish the size of primary gastric lesions and effectively control metastatic lesions, facilitating R0 resection and enhancing long-term survival rates. However, due to tumor biology differences and heterogeneity, the precise implementation of conversion therapy remains a challenge, and not all patients derive benefit from it6. Histopathological examination, the current gold standard method for evaluating reactions, is only available postoperative, leading to delays in therapy adjustment. Therefore, a dependable method for early and individual prediction of treatment response is critically required for personalized therapy in AGC patients.
In the context of these challenges, artificial intelligence, particularly deep learning algorithms, has gained attention for its remarkable performance in image recognition tasks7. Deep learning has demonstrated remarkable capabilities in extracting meaningful patterns from medical imaging data, with successful applications spanning various medical domains including disease classification8,9, model interpretability research10,11,12and prognostic prediction13,14,15. Specifically in gastric cancer analysis, deep learning approaches have shown particular promise for risk stratification and treatment response prediction16,17,18.Deep convolutional neural network (CNN) models have demonstrated the ability to discern subtle details in medical images beyond human perception, offering automated and quantitative assessment. Combining the computed tomography (CT) signs of primary tumors with artificial intelligence to predict AGC patients’ response to conversion therapy and evaluate the feasibility of conversion therapy implementation may yield great diagnostic effects.
While deep learning has demonstrated remarkable success across diverse applications, the exclusive reliance on conventional CNN architectures often falls short of achieving optimal performance in complex clinical tasks. To address this limitation, we propose Progressive Distill (PD), a novel framework that synergistically integrates Knowl- edge Distillation (KD)19 with multi-iteration optimization. Unlike traditional KD methods that perform single- stage distillation, PD employs an iterative refinement process where intermediate student models progressively inherit and enhance discriminative features from teacher networks while incorporating stochastic noise (e.g., dropout and stochastic depth) to improve generalization. This hierarchical distillation mechanism not only mitigates overfitting—a critical challenge in medical imaging with limited datasets—but also enables the compression of knowledge from computationally intensive models into lightweight architectures without sacrificing accuracy.
Hence, our study aims to establish a DL tool that empowers clinicians to stratify AGC patients for conversion therapy before treatment initiation, thereby addressing a pivotal gap in personalized oncology.
The key contributions of this study are:
-
Development of Progressive Distill (PD), a novel deep learning framework combining multi-iteration distillation and model noise.
-
First application of PD to predict clinical response to conversion therapy in AGC using preoperative CT.
-
Superior performance of PD over baseline CNNs, KD, and clinician assessments.
-
Demonstration of PD’s potential for non-invasive, personalized treatment planning in oncology.
Materials and methods
Ethical statement
This study received ethical approval from the Institutional Review Board of The First Affiliated Hospital of Wenzhou Medical University (Ethics approval No. 2024R043). Due to the retrospective nature of the study, the Institutional Review Board of The First Affiliated Hospital of Wenzhou Medical University waived the need of obtaining informed consent. All research involving human participants was conducted in accordance with the Declaration of Helsinki.
Data sets and study cohorts
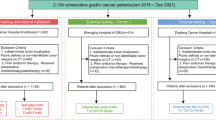
A cohort of 140 patients with histologically confirmed advanced-stage (cT3-4 N0/+M0/1) of GC, who underwent conversion therapy at a single hospital, was recruited from September 2017 to November 2022. The training set comprised 112 patients (mean age: 66 years; range: 39–81 years) consecutively treated at The First Affiliated Hospital of Wenzhou Medical University in Wenzhou, China. The test set consisted of 28 patients (mean age: 66 years; range: 46–81 years) who received treatment at the same hospital (Fig. 1). All baseline clinical characteristics (Table 1), including sex, age, CA199, CEA, and clinical T (cT), N (cN), and M (cM) stages according to the 8 th AJCC TNM staging system20, were extracted from medical records. CT images were sourced from the The First Affiliated Hospital of Wenzhou Medical University.
Conversion therapy protocols and response assessment
The chemotherapy regimens included SOX (Oxaliplatin plus S-1), XELOX (Oxaliplatin plus Capecitabine), FLOT (5-Fluorouracil Plus Leucovorin, Oxaliplatin, and Docetaxel), FLOFOX (Oxaliplatin plus Calcium Levofolinate,5- Fluorouracil), AS (Paclitaxel plus S-1), TP (Paclitaxel plus Cisplatin), DS (S-1 plus Docetaxel), DOS (Docetaxel plus S-1, Oxaliplatin). Trastuzumab was recommended for patients with HER2 positive cancers. The immunotherapy regi- mens included Camrelizumab, Pembrolizumab, Sintilimab, Tislelizumab, and Nivolumab, Penpulimab, Serplulimab. Clinicians determined the appropriate dose and treatment schedule. Within 1–3 weeks after the end of treatment, we evaluate the post-treatment efficacy of chemotherapy through comprehensive methods such as imaging examination and laparoscopic exploration. If the efficacy evaluation reaches complete remission, partial remission, or stable condition, and multidisciplinary team discussion suggest the possibility of R0 tumor resection, surgery would be performed after obtaining the patients’ informed consent, which is called conversion surgery.
The overall treatment response was assessed according to the Response Evaluation Criteria in Solid Tumors (RECIST version 1.1) guidelines21. These guidelines categorized responses into four levels:1. complete response: Defined as the disappearance of all target and non-target lesions. 2. partial response: Characterized by at least a 30% decrease in the sum of the lesion diameter (LD) of target lesions compared to the baseline sum LD. Non-target lesions may persist, and tumor marker levels may remain above normal limits. 3. progressive disease: Marked by at least a 20% increase in the sum of the LD of target lesions compared to the smallest sum LD recorded since treatment initiation. Additionally, the appearance of new lesions or unequivocal progression of existing non-target lesions may be observed.
4. stable disease: Neither meeting the criteria for partial response nor progressive disease. The sum of the LD of target.
lesions remains relatively stable compared to the smallest sum LD since treatment initiation, and non-target lesions show no significant change. The patients in our study were categorized into two groups based on treatment response: the good response (GR) group, comprising those with complete or partial response, and the poor response (PR) group, comprising those with progressive disease or stable disease.
CT imaging and data preprocessing
All patients underwent enhanced CT examination within 1–3 weeks before conversion therapy. Given the ability to distinguish GC from normal gastric tissues in portal venous phase (PVP) CT images, we have retained only the images captured during PVP phase. Additionally, we retained images captured from the moment the abdomen appeared to just before the liver disappeared, while discarding the remaining images. To address the limited amount of training data, we employed data augmentation techniques22,23. Addressing the constraint of limited training data within the dataset, we utilized data augmentation techniques for image processing. This methodology encompasses the application of various random geometric transformations, such as flipping, rotation, scaling, and shifting, to artificially augment the training image dataset. In addition, it can not only help to mitigate the impact of noise but also ensures that the model used focuses on GC tumors24. Subsequently, the remaining images underwent an initial filtering process with a window of [−215, 285] HU. Following this, the images underwent resizing to 256 × 256 pixels followed by random cropping to 224 × 224 pixels, ensuring a standardized distance scale. Previous research underscores the efficacy of data augmentation in preventing network overfitting and avoiding the memorization of specific details from training images25. These preprocessing procedures were executed through Python (version 3.9.16) with the PyTorch Transformer.
PD algorithm development and training
We developed a PD algorithm to predict a patient’s clinical response to conversion therapy based on CT images, as illustrated in Fig. 2. We use EfficientNets26 as baseline model of our method, due to its reputation as one of the most powerful CNNs, achieving the highest accuracy on the ImageNet top1 while demanding fewer computing resources compared to other models (21). Initially, all baseline models were trained without the utilization of any additional methods. To optimize model performance, we incorporated pretrained weights from ImageNet, as their efficacy has been demonstrated. To further boost the performance of baseline models, we introduced additional deep learning methods—PD, during model training.
The PD training template is depicted in Algorithm 1. Initially, we train a teacher network using cross entropy loss with label smoothing27. Subsequently, a student model is trained to minimize the combined loss, which includes:
(1) Kullback-Leibler divergence loss (KLDivLoss) using soft labels from teacher network and soft predictions from student network; (2) Cross entropy loss with label smoothing. This process iterates by replacing the student as a teacher and training a new student.
Knowledge Distillation. -The key objective of Knowledge Distillation (KD) is to minimize the discrepancy between the student network and the teacher network, assessed through the loss function. This facilitates the transfer of knowledge from the teacher network to the student network, enabling the latter to achieve performance comparable to or surpass the former.During the training process, a distillation loss is integrated to minimize the discrepancy between the softmax outputs generated by the teacher and student models. In this context, p denotes the true probability distribution, while z and r signify the outputs of the last fully-connected layer of the student and teacher models, respectively. In the equation, (p, softmax(z)) represents a negative cross-entropy loss measuring the difference between the true probability distribution p and the softmax output z. Therefore, the loss (p, softmax(z)) is changed to
where T represents the temperature hyper-parameter utilized to refine the softmax outputs and convey the knowledge of label distribution from the teacher’s predictions, α is a coefficient between 0 and 1.
Model Noise. –Utilizing conventional KD to distill knowledge to the student model may lead to premature overfitting, restricting the potential for performance improvement and causing a significant decline in performance after a few training epochs. To mitigate this and enhance the model’s robustness, we introduced difficult environments in terms of model noise to the student model. During student training, we use dropout28and stochastic depth29 as our model noise.
Multi-iteration. –The introduction of model noise often hinders students from reaching their maximum learning capability in a single distillation process. Hence, we incorporated multi-iteration to progressively distill knowledge. Furthermore, we want the student to better accommodate model noise by using the equal or larger student models to acquire knowledge, giving the student model enough capacity. Our method involves an iteration process comprised of three primary steps: (1) train a teacher model using standard training methods, (2) employing the teacher model to generate soft labels for distillation loss, and (3) training a student model based on the distillation loss with model noise. This iterative algorithm is repeated multiple times, with each iteration involving the student model acting as a new teacher to generate soft labels and subsequently training a new student model.
Cosine Learning Rate Decay. –To optimize training stability and convergence, we adopted the cosine annealing learning rate schedule, a dynamic optimization strategy proposed by Loshchilov and Hutter in their seminal work30. Unlike traditional stepwise decay, this method smoothly adjusts the learning rate ηt during training by following a cosine function, thereby avoiding abrupt changes that may destabilize gradient updates.The learning rate at batch t (excluding warmup stages) is defined as:
where T denotes the total number of training batches and η is the initial learning rate.
This approach offers two key advantages: (1) The gradual decay of ηt prevents sharp drops in learning rate, enabling steadier traversal of the loss landscape. (2) Cyclically “restarting” the learning rate (by resetting η after T batches) allows the optimizer to escape suboptimal local minima, improving generalization. In our study, cosine learning rate decay was applied during all model training process.
Label Smoothing. –Label smoothing is a regularization technique employed during training to prevent the model from becoming overly confident and excessively reliant on the training data. It involves replacing the hard targets (one-hot encoded labels) with smoothed targets that distribute some probability mass to other classes, and alteration transforms the formulation of the true probability to:
where β denotes a constant, and K is the number of labels. In this study, label smoothing was applied to cross entropy loss.
Model testing and statistical analysis
The test set (n = 28) was constituted by 20% of the dataset sourced from The First Affiliated Hospital of Wenzhou Medical University, and the training set (n = 112) of the prediction model was conducted with the remaining 80% of the dataset. Parameters gleaned from internal evaluation were applied during the training process. The trained models outputted the predicted probability of all images belonging to each patient in the test set being classified as class 1 according to CT images, then calculates the average of the 10 highest probabilities as the prediction result.
All models underwent performance evaluation utilizing receiver operating characteristic (ROC) analysis, with the area under the ROC curve (AUC) computed and compared across various models and methodologies. Furthermore,
predictive accuracy, specificity, and sensitivity were evaluated. The model-predicted clinical response scores were dichotomized into two scores—low or high, with an optimal threshold selected based on the Youden index within the test set, aiming to maximize both sensitivity and specificity. To assess the predictive proficiency of readers on the test set, three clinicians with diverse experience levels (3, 20, and 40 years, respectively) independently interpreted images from 28 patients in the test set. Each clinician provided binary predictions for clinical response (GR or PR) for each patient.
Results
Patient characteristics
Among the patients who underwent conversion therapy for AGC, the following were excluded(n = 164): those whose tumors metastasized outside the abdominal cavity(n = 19), and those whose CT scans without PVP stage(n = 5). Finally, 140 patients were included (Fig. 1).
As depicted in Table 1, the rates of GR in the training and test sets stood at 65.17% and 50.00%, respectively. Notably, there were no discernible differences in sex, age, cN stage, cT stage, or cM stage prior to starting conversion therapy between the GR and PR groups in two cohorts.
Performance of PD
To optimize model performance, we implemented additional deep learning methods based on the baseline models. The corresponding performance outcomes and comparisons in the training set and test set are summarized in Table 2, and the AUCs in test set are shown in Fig. 3. We first report the performance of baseline models. In the test set, the baseline models exhibited AUCs and accuracies as follows: ResNet50 (0.67, 67.86%), ResNet101 (0.69, 71.43%), ResNext101 (0.77, 75.0%), DenseNet121 (0.66, 71.43%), DenseNet201 (0.74, 75.0%), Vgg11 (0.69, 67.86%), Vgg16.
(0.72, 75.0%), EfficientNetB0 (0.68, 71.43%), EfficientNetB3 (0.69, 71.43%), and EfficientNetB7 (0.74, 75.00%). Additionally, our exploratory experiments with 3DResNet-50 and a hybrid CNN + Transformer model (integrating Transformer modules into EfficientNetB7 for global-local feature fusion) achieved test AUCs of 0.69 and 0.75, respectively, with comparable accuracies (67.86% and 75.00%), though their performance did not surpass the baseline 2D models in overall generalizability.
Then we report the performance of KD. When both teachers and students use the same model, students exhibit only marginal improvements in AUC. EfficientNetB0 experienced an AUC increase from 0.68 to 0.72, while maintaining a consistent accuracy of 71.43%. Likewise, EfficientNetB3 demonstrated an improvement in AUC from 0.69 to 0.72, with no change in accuracy at 71.43%. Notably, EfficientNetB7 showcased an AUC increase from 0.74 to 0.77, while maintaining a steady accuracy of 75%. By employing a smaller student model for iteration compared to the teacher model (EfficientNetB7 to B3), it is feasible to attain performance that equals or exceeds that of the larger model, EfficientNetB3 can also achieve an AUC and an accuracy of 0.80 and 82.14%, which is much better than teacher EfficientNetB7.
Moreover, PD outperforms KD. In the following, we report the performance of models with PD. The best model in our study resulted from two iterations of placing the student back as the new teacher, exclusively using EfficientNetB7. The model performance, with an AUC and accuracy, improved from 0.72 to 75.0% to 0.78 and 78.57% in the first iteration and then to 0.87 and 85.71% in the second iteration. As for iterating by solely using EfficientNetB3, the best results were also achieved after two iterations, with an AUC and an accuracy of 0.76 and 75.0%. Iterating from a smaller model to a larger model (EfficientNetB0 to B3 to B7) is feasible to shorten the training time while achieving excellent performance for the larger model. EfficientNetB7 can also achieve an AUC and an accuracy of 0.84 and 82.14% in half of the time compared to iterating with three EfficientNetB7. However, when iterating from a larger model to a smaller model (EfficientNetB7 to B3), albeit not to the same extent as conventional KD, the student model’s performance still improves. The AUC and accuracy, which were achieved by EfficientNetB7 at 0.78 and 75.0%, respectively, increased to 0.81 and 78.57% by EfficientNetB3. Experimental results indicate that larger models benefit positively from the inclusion of model noise during training, whereas smaller models experience negative effects when model noise is added. These findings will be explored further in the Ablation Study.
The performance of the PD models for predicting clinical response was also compared with those of skilled clinicians. As illustrated in Table 3, clinicians with 3, 20, and 40 years of experience exhibited varying accuracies for clinical response, achieving 92.86%, 82.14%, and 78.57%, respectively. Their sensitivities and specificities likewise displayed variability across experience levels, with the most experienced clinician surprisingly performing less effectively than others. In contrast, the PD model demonstrated an accuracy of 85.71% for clinical response, either comparable to or significantly higher than the accuracy achieved by clinicians with 20 and 40 years of experience, as well as the average performance of all three clinicians.
ROC curves of baseline models, KD and PD for clinical response prediction in test set. The EfficientNetB7 model with PD achieved a high accuracy and substantially improved upon baseline models and KD. Sensitivity and specificity for clinician predictions are reported, both as an average and for the individual clinicians with 3 (C3), 20 (C20), and 40 (C40) years of experience. ROC = receiver operating characteristic, KD = Knowledge Distillation, PD = Progressive Distill.
The confusion matrix graph illustrates category predictions made by our best model for patients in the test set with clinical responses of complete response, partial response, progressive disease, and stable disease. It demonstrates that our model exhibits strong predictive capabilities and shows no discernible bias (Fig. 4A). Furthermore, the decision curve analysis graphically indicates that the PD provides a larger net benefit compared to other models within the pertinent threshold range in the test set (Fig. 4B).
The heat maps, generated using the gradient-weighted activation mapping method (Fig. 5), showcasing the areas that PD model is most concerned about for these images, underscores the deep learning network’s emphasis on the most predictive image features related to clinical response.
Figure 6 illustrates the changes in model performance in the test set with increasing iterations for models trained using PD iterates with equal models. In the second iteration, both EfficientNetB7 and EfficientNetB3 performed the best. EfficientNetB7 showed a substantial improvement, while EfficientNetB3 also experienced improvement, albeit not as remarkable as EfficientNetB7. However, after the second iteration, the performance of both models started to decline.
Ablation study
To further validate the effectiveness of the designs in our model, we conducted several ablation studies. The findings are presented in Table 4, highlighting the pivotal role of model noise and multi-iteration in enhancing the performance of the student model surpass that of the teacher.Conventional KD only results in marginal improvements in AUC for the model. However, incorporating model noise in addition to KD brings substantial positive gains for the large model (EfficientNetB7), while the small model (EfficientNetB3) experiences only slight gains. Increasing the number of iterations in the preceding steps leads to enhanced performance for both the large and small models.
Discussion
Conversion therapy stands as a significant treatment for unresectable GC patients; however, its scheme and effectiveness vary among patients. The absence of a reliable preoperative method to predict clinical responses results in treatment failures for some individuals, preventing them from undergoing radical surgery due to disease progression16. Hence, the creation of a precise predictive model to evaluate the effectiveness of conversion therapy before its commencement holds immense importance in the meticulous management of patients with AGC. Within this retrospective investigation encompassing 140 patients, we developed and validated a deep learning method capable of accurately predicting the clinical response of conversion therapy for AGC patients based on preoperative PVP CT images. Furthermore, PD demonstrated superior performance compared to both baseline models and KD.
As early studies31,32provided preliminary evidence of the safety and efficacy of conversion therapy, subsequent high-quality investigations33,34,35 have further substantiated its feasibility. Yoshida et al.6,36 introduced a systematic classification of conversion therapy for AGC, offering guidance for clinical practice based on the presence of visible peritoneal metastasis. Their further findings based on this classification showed that patients with stage IV GC who underwent conversion surgery had relatively longer survival. Additionally, Li et al.37 demonstrated the efficacy and feasibility of immune checkpoint inhibitor (ICI)- and antiangiogenesis-based conversion therapy in patients with AGC. Despite the availability of various approaches to evaluate the clinical response to conversion therapy, they often rely on subjective visual assessment of experienced experts or lack non-invasive assessment options.
Although conversion therapy can effectively treat AGC, positive results may not be universal for all individuals. In the study conducted by Yamaguchi et al.36, 43 patients underwent conversion therapy were classified into four categories as described by Yoshida et al.6. The median survival times (MSTs) differed by 13 months between patients in category 1 and category 4. Similar results were reported by Chen et al.38, where the MSTs differed by 30 months between patients in Category 3 and Category 4. These results underscore the importance of developing a non-invasive screening method to identify the subset of individuals who would benefit from it. In our study, patients were classified into two groups based on their clinical response: the good response (GR) group, including patients with complete response and partial response, and the poor response (PR) group, including patients with stable disease and progressive disease, as defined according to the response assessment criteria proposed in RECIST version 1.121.
CT images and heat maps of four AGC patients in four categories of clinical responses. (a) Images in a 69-year-old man with a clinical response of complete response. (b) Images in a 69-year-old man with a clinical response of partial response. (c) Images in a 59-year-old man with a clinical response of stable disease. (d) Images in a 67-year-old man with a clinical response of progressive disease. CT = computed tomography, AGC = advanced gastric cancer.
Our study identifies four notable advantages: (1) Compared to previous assessment methods, with the use of deep learning models and preoperative CT images allows us to predict clinical responses non-invasively, which is critically required for personalized therapy in AGC patients. (2) Traditional methods of evaluating medical images often rely on human visual interpretation, which can be subjective and limited by the observer’s experience and expertise. In contrast, deep learning algorithms analyze images systematically, processing each pixel to derive comprehensive insights. This approach allows for a more objective and consistent assessment of medical images, potentially leading to more accurate diagnoses and treatment decisions. (3) Applying the KD can enhance the model’s performance, surpassing the baseline model and enabling the small model to outperform the large model, resulting in a fast-running model with satisfactory performance and fewer parameters. (4) Through a progressive approach combined with KD, the model’s performance steadily improves, ultimately surpassing both the baseline model and the KD. The best-performing model obtained using the PD achieved an AUC of 0.87, an accuracy rate of 85.71%, a sensitivity of 85.71%, and a specificity of 85.71%.
Model performance of PD with an increase in the number of iterations. (A) The accuracies of EfficientNetB3, EfficientNetB7 for clinical response. (B) The AUCs of EfficientNetB3, EfficientNetB7 for clinical response. PD = Progressive Distill, AUC = area under the receiver operating characteristic curve.
Comparisons between our models and predictions made by clinicians offer valuable insights. Interestingly, the clinician with the most experience exhibited the lowest accuracy in predicting clinical response, emphasizing the inherent challenges and subjective nature of human-based predictions. Clinicians face the critical task of determining whether a patient’s survival can be prolonged through life-saving surgery, highlighting the weight of their decisions.Our models demonstrated superior accuracy compared to clinician predictions, suggesting potential enhancements in prognosticating clinical responses over qualitative assessments. Upon thorough validation, our model could provide quantitative preoperative information non-invasively, empowering clinicians to make rapid, reproducible, and more precise decisions in guiding the care of patients with AGC.
Our study has limitations. First, our sample was collected from a single center and our model may not perform well on CT images from other hospitals. In future studies, we aim to conduct a multicenter study to minimize variations between hospitals and enhance the robustness of our model. Additionally, this study was retrospective, and the outcomes were influenced by the composition of dataset, which was limited in size. Prior to actual clinical application, significant advancements are required through larger and prospective studies. Due to the retrospective nature of this study, clinicians were not involved in patient classification using our PD model, this will be considered in our future work. To address clinical utility, we are pursuing ethical approvals for deploying the PD model as a real-time decision-support tool in radiology workflows. Future work will involve collaborations with multidisciplinary clinical teams to measure its impact on treatment planning and patient outcomes through prospective trials. What’s more, our current framework focuses exclusively on CT imaging features without integrating clinical parameters such as age, family history, or biochemical markers. While this streamlined approach establishes baseline efficacy for CT-based prediction, future work should explore multimodal fusion architectures to leverage complementary clinical and imaging biomarkers. Though focused on 2D tumor analysis for clinical alignment, we explored 3D CNNs and CNN + Transformer hybrids for volumetric and global-local feature modeling. These showed modest gains over 2D baselines but encountered overfitting, computational burdens, and generalizability issues, potentially from data limitations. Future work should develop efficient 3D architectures and improved fusion methods to optimize clinical- computational tradeoffs.
Conclusion
This study developed and validated a deep learning model based on CT scans for early prediction of clinical response to conversion therapy in AGC patients. The introduced PD model demonstrated encouraging predictive capabilities, offering significant insights for individualized treatment approaches in AGC patients. These findings emphasize the need for prospective validation in forthcoming randomized trials to assess the clinical applicability of our imaging model in conjunction with clinicopathological criteria, facilitating personalized treatment strategies.
Data availability
The datasets generated and/or analysed during the current study are not publicly available due to hospital ethics committee regulations and potential involvement of patient privacy, but are available from the corresponding author on reasonable request.
References
Sung, H. et al. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J. Clin. 71 (3), 209–249. https://doi.org/10.3322/caac.21660 (2021).
Hu, C., Terashima, M. & Cheng, X. Conversion therapy for stage Iv gastric cancer. Sci. Bull. 68 (7), 653–656. https://doi.org/10.1016/j.scib.2023.03.011 (2023).
Machlowska, J., Baj, J., Sitarz, M., Maciejewski, R. & Sitarz, R. Gastric cancer: epidemiology, risk factors, classification, genomic characteristics and treatment strategies. Int. J. Mol. Sci. 21 (11), 4012. https://doi.org/10.3390/ijms21114012 (2020).
Kinoshita, J. et al. Efficacy of conversion gastrectomy following docetaxel, cisplatin, and s-1 therapy in potentially resectable stage Iv gastric cancer. Eur. J. Surg. Oncol. (EJSO). 41 (10), 1354–1360. https://doi.org/10.1016/j.ejso.2015.04.021 (2015).
Mieno, H. et al. Conversion surgery after combination chemotherapy of docetaxel, cisplatin and s-1 (dcs) for far-advanced gastric cancer. Surg. Today. 47, 1249–1258. https://doi.org/10.1007/s00595-017-1512-z (2017).
Yoshida, K., Yamaguchi, K., Okumura, N., Tanahashi, T. & Kodera, Y. Is conversion therapy possible in stage Iv gastric cancer: the proposal of new biological categories of classification. Gastric cancer. 19, 329–338. https://doi.org/10.1007/s10120-015-0575-z (2016).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521 (7553), 436–444. https://doi.org/10.1038/Nature14539 (2015).
Ullah, N., Khan, J. A., De Falco, I. & Sannino, G. Bridging clinical gaps: multi-dataset integration for reliable multi-class lung disease classification with deepcrinet and occlusion sensitivity, in: 2024 IEEE Symposium on Computers and Communications (ISCC), IEEE, pp. 1–6, (2024). https://doi.org/10.1109/ISCC61673.2024.10733651
Ullah, N. et al. A lightweight deep learning-based model for tomato leaf disease classification. Computers Mater. Continua. 77 (3), 3969–3992. https://doi.org/10.32604/cmc.2023.041819 (2023).
Ullah, N. et al. Tumordetnet: A unified deep learning model for brain tumor detection and classification. Plos One. 18 (9), e0291200. https://doi.org/10.1371/journal.pone.0291200 (2023).
Ullah, N., Khan, J. A., De Falco, I. & Sannino, G. Explainable artificial intelligence: importance, use domains, stages, output shapes, and challenges. ACM Comput. Surveys. 57 (4), 1–36. https://doi.org/10.1145/3705724 (2024).
Ullah, N., Hassan, M., Khan, J. A., Anwar, M. S. & Aurangzeb, K. Enhancing explainability in brain tumor detection: A novel Deepebtdnet model with lime on mri images. Int. J. Imaging Syst. Technol. 34 (1), e23012. https://doi.org/10.1002/ima.23012 (2024).
Xie, T. et al. Self-supervised contrastive learning using Ct images for pd-1/pd-l1 expression prediction in hepatocellular carcinoma. Front. Oncol. 13, 1103521. https://doi.org/10.3389/fonc.2023.1103521 (2023).
Wei, Y. et al. Novel computed-tomography-based transformer models for the noninvasive prediction of pd-1 in pre-operative settings. Cancers 15 (3), 658. https://doi.org/10.3390/cancers15030658 (2023).
Chen, S. et al. Reinforcement learning based diagnosis and prediction for covid-19 by optimizing a mixed cost function from Ct images. IEEE J. Biomedical Health Inf. 26 (11), 5344–5354. https://doi.org/10.1109/JBHI.2022.3197666 (2022).
Hu, C. et al. Deep learning radio-clinical signatures for predicting neoadjuvant chemotherapy response and prognosis from pretreatment Ct images of locally advanced gastric cancer patients. Int. J. Surg. 109 (7), 1980–1992. https://doi.org/10.1097/JS9.0000000000000432 (2023).
Cui, Y. et al. A ct-based deep learning radiomics nomogram for predicting the response to neoadjuvant chemotherapy in patients with locally advanced gastric cancer: A multicenter cohort study. EClinicalMedicine 46 https://doi.org/10.1016/j.eclinm.2022.101348 (2022).
Jiang, Y. et al. Predicting peritoneal recurrence and disease-free survival from Ct images in gastric cancer with multitask deep learning: a retrospective study. Lancet Digit. Health. 4 (5), e340–e350. https://doi.org/10.1016/S2589-7500(22)00040-1 (2022).
Hinton, G., Vinyals, O. & Dean, J. Distilling the knowledge in a neural network. ArXiv Preprint arXiv. https://doi.org/10.48550/arXiv.1503.02531 (2015).
Sano, T. et al. Proposal of a new stage grouping of gastric cancer for Tnm classification: international gastric cancer association staging project. Gastric cancer. 20, 217–225. https://doi.org/10.1007/s10120-016-0601-9 (2017).
Eisenhauer, E. A. et al. New response evaluation criteria in solid tumours: revised recist guideline (version 1.1). Eur. J. Cancer. 45 (2), 228–247. https://doi.org/10.1016/j.ejca.2008.10.026 (2009).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. Imagenet classification with deep convolutional neural networks. Adv. Neural. Inf. Process. Syst. 25 https://doi.org/10.1145/3065386 (2012).
Xie, T. et al. Cut-thumbnail: A novel data augmentation for convolutional neural network, in: Proceedings of the 29th ACM International Conference on Multimedia, 2021, pp. 1627–1635. https://doi.org/10.1145/3474085. 3475302.
Roth, H. R. et al. Improving computer-aided detection using convolutional neural networks and random view aggregation. IEEE Trans. Med. Imaging. 35 (5), 1170–1181. https://doi.org/10.1109/TMI.2015.2482920 (2015).
Kayalibay, B., Jensen, G. & van der Smagt, P. Cnn-based segmentation of medical imaging data, arxiv Preprint arxiv:1701.03056https: // (2017). https://doi.org/10.48550/arXiv.1701.03056
Tan, M. & Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks, in: International conference on machine learning, PMLR, pp. 6105–6114, (2019). https://doi.org/10.48550/arXiv.1905.11946
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J. & Wojna, Z. Rethinking the inception architecture for computer vision, in: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 2818–2826, (2016). https://doi.org/10.48550/arXiv.1512.00567
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I. & Salakhutdinov, R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15 (1), 1929–1958. https://doi.org/10.5555/2627435.2670313 (2014).
Huang, G., Sun, Y., Liu, Z., Sedra, D. & Weinberger, K. Q. Deep networks with stochastic depth, in: Computer Vision–ECCV 2016: 14th European Conference, Amsterdam, The Netherlands, October 11–14, 2016, Proceedings, Part IV 14, Springer, pp. 646–661, https: (2016). https://doi.org/10.48550/arXiv.1603.09382
Loshchilov, I. & Hutter, F. Stochastic gradient descent with warm restarts. ArXiv Preprint arXiv. 160803983. https://doi.org/10.48550/arXiv.1608.03983 (2016).
Nakajima, T. et al. Combined intensive chemotherapy and radical surgery for incurable gastric cancer. Ann. Surg. Oncol. 4, 203–208. https://doi.org/10.1007/BF02306611 (1997).
Yano, M. et al. Neoadjuvant chemotherapy followed by salvage surgery: effect on survival of patients with primary noncurative gastric cancer. World J. Surg. 26, 1155–1159. https://doi.org/10.1007/s00268-002-6362-0 (2002).
Yoshikawa, T. et al. Phase Ii study of neoadjuvant chemotherapy and extended surgery for locally advanced gastric cancer. J. Br. Surg. 96 (9), 1015–1022. https://doi.org/10.1002/bjs.6665 (2009).
Tsuburaya, A. et al. Neoadjuvant chemotherapy with s-1 and cisplatin followed by d2 gastrectomy with para-aortic lymph node dissection for gastric cancer with extensive lymph node metastasis. J. Br. Surg. 101 (6), 653–660. https://doi.org/10.1002/bjs.9484 (2014).
Satoh, S. et al. Phase Ii trial of combined treatment consisting of preoperative s-1 plus cisplatin followed by gastrectomy and postoperative s-1 for stage Iv gastric cancer. Gastric cancer. 15, 61–69. https://doi.org/10.1007/s10120-011-0066-9 (2012).
Yamaguchi, K. et al. The long-term survival of stage Iv gastric cancer patients with conversion therapy. Gastric Cancer. 21, 315–323. https://doi.org/10.1007/s10120-017-0738-1 (2018).
Li, S. et al. Neoadjuvant therapy with immune checkpoint Blockade, antiangiogenesis, and chemotherapy for locally advanced gastric cancer. Nat. Commun. 14 (1), 8. https://doi.org/10.1038/s41467-022-35431-x (2023).
Chen, G. M. et al. Surgical outcome and long-term survival of conversion surgery for advanced gastric cancer. Ann. Surg. Oncol. 27, 4250–4260 (2020). https://doi.org/10.1245/s10434-020-08559-7.
Funding
This work was supported by the Municipal Government of Quzhou (Grant 2023D007, Grant 2023D015, Grant 2023D033, Grant 2023D034, Grant 2023D035).
Author information
Authors and Affiliations
Contributions
Han, S: Conceptualization, Resources, Supervision, Validation, Writing – review editing, Formal analysis, In- vestigation. Zhang, T: Conceptualization, Data Curation, Methodology, Software, Project Administration, Validation, Visualization, Writing – Original Draft. Deng, W: Conceptualization, Data Curation, Methodology, Software, Writing – review editing. Han, S: Resources, Validation, Formal analysis. Wu, H: Visualization, Methodology, Software. Jiang, B: Resources, Validation. Xie, W: Data Curation, Formal analysis, Writing – review editing. Chen, Y: Investigation, Conceptualization, Software. Deng, T: Validation, Investigation. Wen, X: Supervision, Software. Liu, N: Project administration, Funding acquisition, Writing – review editing. Fan, J: Resources, Supervision, Funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Institutional Review Board of The First Affiliated Hospital of Wenzhou Medical University (Ethics approval No. 2024R043) and with the 1964 Helsinki Declaration and its later amendments. All written informed consent to participate in the study was obtained from the patient.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Han, S., Zhang, T., Deng, W. et al. Deep learning progressive distill for predicting clinical response to conversion therapy from preoperative CT images of advanced gastric cancer patients. Sci Rep 15, 17092 (2025). https://doi.org/10.1038/s41598-025-01063-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-01063-6