Abstract
The diverse coat types of cashmere goats significantly influence cashmere yield, yet the regulatory mechanisms mediated by non-coding RNAs (ncRNAs) remain poorly characterized. In this study, we collected skin tissues from two distinct coat types (CHLC_ESA and CHSC_ESA) of The Jinlan Cashmere Goat for transcriptome sequencing analysis. Our findings revealed 130 differentially expressed (DE) lncRNAs and 341 DE mRNAs. GO functional enrichment analysis uncovered some crucial terms such as keratin filament, intermediate filament, cytokine activity, and cytokine receptor binding, along with two pivotal DE lncRNAs: TCONS_00050130 and TCONS_00050142. Furthermore, KEGG pathway analysis identified several important pathways including the Estrogen signaling pathway, Amino sugar and nucleotide sugar metabolism, Adipocytokine signaling pathway, and cytokine-cytokine receptor interaction. Lastly, by constructing a ceRNA regulatory network among DE lncRNAs and DE mRNAs, we identified multiple ceRNA regulatory axes targeting genes such as EPPK1, IRF4, CCL20, and CXCR7. These mechanistic insights not only advance our understanding of ncRNA-mediated hair follicle biology but also provide actionable targets for precision breeding strategies to enhance cashmere quality and yield across caprine species.
Similar content being viewed by others
Introduction
Cashmere goats are a valuable livestock breed renowned for their production of high-quality cashmere, often referred to as the “fiber gem” or “soft gold”1. Studies have shown that the fleece of cashmere goats consists of two distinct fibers: guard hair (coarse hair) and cashmere undercoat, which are originating from primary and secondary follicles, respectively2,3.
Biologically, the development of a double coat is essential for the goat’s adaptation to harsh environments4. The guard hair offers protection against the elements, while the undercoat cashmere provides insulation during cold winters, safeguarding the goat’ skin5. This adaptation allows cashmere goats to thrive in diverse climates, from the Himalayas to other regions6. The fine, soft undercoat known as cashmere, highly valued for its warmth and luxurious feel, is a sought-after commodity7,8 utilized in the production of pashmina, sweaters, and other cashmere products2,6. In contrast, the coarser guard hair is removed and often discarded or repurposed for low-value items such as brushes or interlinings6.
Goat hair follicle development is a complex, multi-stage process involving intricate cellular interactions and signaling pathways9. This process begins with the formation of the hair follicle, which serves as the structural unit responsible for hair growth10. The hair follicle undergoes a cyclical pattern of growth and regression, known as the hair cycle, encompassing three main phases: anagen (AN), catagen (CA), and telogen (TE)11,12.
While the early stage of the anagen (ESA) phase in hair follicles is also crucial for initiating hair growth and ensuring the proper development of hair fibers. The ESA is a period of intense activity, marked by the activation of hair follicle stem cells, the rapid proliferation of matrix cells, and the crucial involvement of dermal papilla cells3,13. This phase lays the foundation for the subsequent stages of hair growth and development, ultimately leading to the formation of a new hair shaft14,15. However, there is currently limited research specifically focusing on this early stage of the anagen phase in hair follicles.
The Jinlan Cashmere Goat, a domesticated breed originating from Shanxi Province, China, is renowned for its high-quality cashmere production. The predominant coat type in this population is characterized by the coarse hairs exceeding the length of cashmere (CHLC), while a minority of individuals display the opposite trait, where the coarse hairs are shorter than the length of the cashmere (CHSC). We collected skin tissues from the early stage of anagen of Jinlan Cashmere Goat for transcriptome sequencing research and employs functional enrichment, and regulatory network construction to elucidate the mechanisms behind coat type differences, aiming to screen for key non-coding RNAs that affect the type of goat fleece.
Materials and methods
Ethics statement
Institutional Animal Care and Use Committee of Shanxi Agricultural University provided full approval for this research (SXAU-EAW-2023G.UV.005009210). We confirm that all experiments were performed in accordance with the relevant guidelines and regulations of China. We also confirm that this study was performed in accordance with the ARRIVE guidelines.
Animals and tissues
The Jinlan Cashmere Goat is a domestic breed of goat originating from Shanxi Province, China. The anagen of the hair follicles in Jinlan Cashmere Goats lasts from April to November each year, with a rapid growth phase after September. In July, during the early stage of the anagen phase, we collected skin tissue samples from both the coat types (CHLC and CHSC) of Jinlan Cashmere Goat at 2 years old and the detailed sampling procedure is outlined below. Firstly, administer 0.2 ml of Lumianning Injection (Jilin Huamu Animal Health Products Co., Ltd., Changchun, Jilin, China) to the goat for anesthesia. After 7–10 min, once the goat lies down stably, it indicates that the anesthesia has taken effect. Then, a 5 cm x 5 cm patch of hair was carefully trimmed off from the posterior edge of the left scapula, near the midline of the body. Any residual fine hairs on the skin were then meticulously scraped away with a razor. The skin tissue in this area was then wiped with alcohol and disinfected with iodine. Next, a circular skin biopsy punch with a diameter of 1 cm was used to collect the skin sample, with Yunnan Baiyao powder swiftly applied to the wound to stop any bleeding.
The four skin tissues collected in each group (CHLC and CHSC) were promptly placed into a 2 ml sterile tube and preserved in liquid nitrogen.
RNA extraction, library construction and sequencing
Total RNA was extracted using TRNzol Universal Reagent (Tiangen, China). The ribosomal RNA was depleted from total RNA samples using the rRNA Removal Kit (Tiangen, China). Sequencing libraries were generated using the Fast RNA-seq Lib Prep Kit V2 (ABclonal Technology, China) following the manufacturer’s recommendations and index codes were added to attribute sequences to each sample. At last, library quality was assessed on the Agilent 5400 system and quantified by QPCR (1.5 nM). The qualified libraries were pooled and sequenced on the Illumina novaseq 6000 platform with PE150 strategy in Novogene Bioinformatics Technology Co., Ltd (Beijing, China).
Bioinformatics analysis
The original fluorescence image files obtained from the Illumina platform are transformed into short reads (Raw data) by base calling and these short reads are recorded in FASTQ format, which contains sequence information and corresponding sequencing quality information. Then quality control was performed and it is an essential step applied to guarantee the meaningful downstream analysis. We used Fastp (version 0.23.1)16 to perform basic statistics on the quality of the raw reads and clean reads were finally obtained.
Clean reads of each sample were then mapped to the reference genome (NCBI_CAPRA_HIRCUS_GCF_001704415.2_ARS1_2) by HISAT2 (v2.0.5)17. We utilized StringTie (v1.3.3b)18 to perform transcript assembly and merging and novel transcripts were then obtained with the software gffcompare (v0.10.6). After predicting the protein-coding capability of novel transcripts using the softwares CNCI (v2.0)19, PFAM (v1.6)20, and CPC2 (v3.2.0)21, transcripts lacking coding potential in all three software were identified as novel lncRNAs, while those exhibiting coding potential in all three were classified as novel mRNAs.
The prediction of target genes for lncRNAs was performed utilizing two strategies: co-location and co-expression. Co-location focuses on the potential regulation of neighboring genes within 100 kb of the lncRNA, based on genomic proximity22. The co-expression targets of lncRNAs are predicted based on the correlation of expression levels between the lncRNA and other genes across multiple samples23. And the Pearson correlation coefficient | r | > 0.75 was used in the study.
Differentially expressed lncRNAs (DE lncRNAs) and genes (DE genes) were identified with P_value < 0.05 using the DESeq2 package (v1.20.0)24. Functional enrichment analysis including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)25 was performed using clusterProfiler (v3.8.1) with a significance threshold of P_value < 0.0526. The targeting relationship of the ceRNA network is predicted using the miRanda (v3.3a), and its visualization is carried out using Cytoscape (3.8.2).
Results
Sequencing data quality control and alignment
Upon completion of the sequencing process, we amassed a comprehensive dataset comprising a total of 692,785,222 raw reads. After rigorous filtering, an impeccable 678,782,742 clean reads were obtained, constituting an impressive 97.98% of the original dataset (Table S1). Notably, each sample boasted a clean read proportion exceeding 97.26% (CHSC_2_ESA). Moreover, the GC content of these clean reads varied gracefully within a narrow range, spanning from 43.53 to 45.21% across all samples (Table S1), further underscoring the uniformity and quality of the data. A substantial proportion of over 96.29% of the clean reads from the CHSC_3_ESA sample, and similarly high percentages for other samples, were successfully mapped onto the reference genome (Table S2). This achievement underscores the precision and efficacy of our alignment strategies.
Collectively, these exhaustive findings paint a compelling picture of the exceptional quality of the sequencing data obtained, attesting to the rigor and meticulousness of our experimental protocols and analyses.
Differential expression analysis
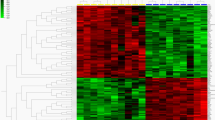
We identified a total of 130 DE lncRNAs across the two comparison groups. Compared to the CHLC_ESA group, the CHSC_ESA group exhibited a significant alteration in the expression profile of these lncRNAs, with 68 lncRNAs being upregulated and 62 downregulated (Fig. 1A). Concurrently, we also identified 341 DE mRNAs, among which 165 were upregulated and 176 were downregulated (Fig. 1B). In addition, we performed heatmap clustering analysis to visualize the expression patterns (Fig. 1C,D). The results showed a clear and distinct pattern of expression clusters and successfully grouped samples from the same experimental condition together.
GO enrichment analysis
We predicted the target genes of DE lncRNA across the entire genome, yielding 552 co-location (Table S3) and 303 co-expression (Table S4) relationship pairs. Notably, these relationships encompassed 109 and 43 DE lncRNAs, respectively. Finally, we carried out GO functional enrichment analysis (Tables S5 and S6) on the target genes of DE lncRNAs in the study and the top 30 GO terms were shown in (Fig. 2).
Figure 2A shows that the proportion of Cell component (CC) classification is the highest, followed by Molecular function (MF) classification, and finally Biological process (BP) classification. Among them, the keratin filament, intermediate filament and intermediate filament cytoskeleton terms were closely related to the growth of hair follicles and hair. Further analysis revealed that all the three terms were enriched with 24 same genes, which corresponded to two lncRNAs with co-location relationship, namely TCONS_00050130 and TCONS_00050142 (Table 1). We also found some protein-related and nucleic acid-related entries, such as protein heterodimerization activity, protein dimerization activity, DNA binding, nucleic acid binding, DNA packaging complex, protein-DNA complex and so on. Hair follicle development and hair growth are inseparable from the regulation of proteins and nucleic acids.
The top 30 entries of the GO functional enrichment analysis of the co-expression target genes of 43 DE lncRNA are presented in Fig. 2B, and the MF terms are found to be the most abundant, with up to 25 entries. Among these top 30 entries, we found multiple receptor activity and signal transduction-related entries, including cytokine activity, cytokine receptor binding, receptor regulator activity, receptor ligand activity, transmembrane signaling receptor activity, signaling receptor activity, signaling receptor binding, signal transducer activity and G-protein-coupled receptor signaling pathway. Additionally, a significant portion of enzymes related to metabolic activities were also identified, for instance, serine-type endopeptidase activity, serine-type peptidase activity, serine hydrolase activity and pyrophosphatase activity. These findings suggested that cytokine activity, signal transduction regulation and enzymatic activity might play a role in modulating follicle development.
KEGG pathway analysis
KEGG pathway enrichment analysis of target genes for DE lncRNAs was also performed (Tables S7 and S8) and the top 20 KEGG pathways are shown in Fig. 3. The enrichment results of co-location target genes revealed several important pathways (Fig. 3A) such as the Estrogen signaling pathway, Amino sugar and nucleotide sugar metabolism and Adipocytokine signaling pathway. The KEGG pathway analysis of co-expression target genes has identified the Cytokine-cytokine receptor interaction pathway, which echoes the findings of cytokine activity and cytokine receptor binding terms from the GO enrichment results. Furthermore, our analysis has enriched Oxidative phosphorylation and Chemokine signaling pathway, suggesting their potential involvement in the growth and development of hair follicles or hair.
Construction of CeRNA regulatory network
We utilized the DE lncRNAs and mRNAs to construct a ceRNA regulatory network, incorporating all detected miRNAs (Fig. 4 and Table S9). The Pearson correlation coefficients (r) between the expression levels of miRNA and lncRNA, as well as those between miRNA and mRNA, were used to further screen lncRNA-miRNA-mRNA regulatory pairs. We identified a total of 197 lncRNA-miRNA-mRNA regulatory pairs with | r | > 0.75, which include 11 DE lncRNAs and 52 DE mRNAs. We have identified multiple ceRNA regulatory axes with target genes such as EPPK1, IRF4, CCL20, and CXCR7, which are closely related to hair follicle or hair development. This network provides valuable insights into potential regulatory mechanisms and pathways of interest.
Discussion
Cashmere, prized for its softness and warmth, originates from secondary hair follicles and displays a finer, shorter, and less variable diameter compared to guard hair. Conversely, guard hair, produced by primary hair follicles, is characterized by its coarser, longer, and more variable fibers, serving as a protective outer coat7. Studies have explored the impact of external factors, such as nutrition, on hair follicle development. Nutritional restriction, for example, can significantly affect the development and transcriptome of hair follicles in sheep27. Furthermore, studies have delved into the genetic basis of these differences, identifying candidate genes associated with hair traits. For instance, the expression of fibroblast growth factor 21 (FGF21) has been found to differ significantly between cashmere and guard hair follicles, suggesting its potential involvement in regulating hair type-specific development28; Another study has demonstrated that the overexpression of thymosin beta-4 (Tβ4) in the hair follicle tissue of Alpas cashmere goats results in an increase in cashmere yield and promotes hair follicle development.
However, there have been relatively few studies on lncRNA to hair development in goats. This study collected skin tissues from Jinlan Cashmere Goats with two different coat types at the early stage of the anagen (ESA) for transcriptome sequencing analysis. Finally, a total of 130 DE lncRNAs were obtained in the comparison group CHSC_ESA vs. CHLC_ESA.
The GO enrichment analysis of the co-location target genes of lncRNAs revealed several entries that are directly associated with hair follicle or hair development. Keratin filaments are composed of two type keratin proteins: type I, which number 28, and type II, which number 2629. They are essential for hair follicle development and hair growth, providing structural integrity to the hair shaft, contributing to follicle formation, and regulating hair follicle cycling30,31. For example, Keratin 15 (K15) is a keratin protein mainly expressed in hair follicle stem cells (HFSCs) in humans and mice32,33. Study have shown that K15 is excessively degraded by KLHL24-ΔN28 in KLHL24-mutant mice and patients, leading to a cytoskeleton network disturbance in HFSCs, which then causes premature hair loss, HFSCs decrease, and finally hair follicle degeneration34. The intermediate filament cytoskeleton, comprising intermediate filaments and associated proteins, is essential for maintaining tissue integrity and responding to mechanical stress35. The intermediate filament cytoskeleton refers to the network formed by all intermediate filaments within a cell, including keratin filaments36,37. This network plays a crucial role in maintaining cell shape, resisting mechanical stress, and facilitating cell signaling38.
Additionally, we also found some protein-related and nucleic acid-related GO terms. Proteins and nucleic acids are essential for hair follicle development and hair growth, playing complementary roles in this intricate process. Proteins, particularly keratin, provide the structural integrity of hair shafts, giving them strength and resilience39,40. Nucleic acids, specifically DNA and RNA, are responsible for encoding the genetic information that dictates the synthesis of these proteins, including keratin41. Furthermore, proteins are involved in signaling pathways that regulate the hair follicle cycle, ensuring proper growth, regression, and resting phases42,43. In essence, proteins provide the building blocks and structural components of hair, while nucleic acids act as the blueprints and regulators, ensuring the precise synthesis and function of these proteins. This intricate interplay between proteins and nucleic acids is fundamental for healthy hair growth and development.
The KEGG pathway enrichment analysis of the co-location target genes of DE lncRNA suggests that the Estrogen signaling pathway, Amino sugar and nucleotide sugar metabolism, and Adipocytokine signaling pathway may play important roles. The estrogen signaling pathway includes multiple genes and proteins, including estrogen and its receptors44,45. Two important cell populations in HF, hair follicle stem cells (HFSCs) and dermal papilla (DP) cells, are required for initiating the first step of a new hair cycle46. And estrogen receptors are expressed in both the DP and HFSCs, suggesting a direct influence on hair follicle activity47. Another study showed Estrogen could modulate the hair follicle cycle by influencing the expression of genes involved in hair growth and differentiation48. Amino sugars and nucleotide sugars are essential building blocks for various macromolecules, including glycosaminoglycans (GAGs), which are crucial components of the extracellular matrix (ECM) surrounding hair follicles49. The ECM provides structural support and regulates cell signaling within the hair follicle, influencing its development and growth50. Adipocytokines, secreted by adipocytes, play a crucial role in regulating hair follicle activity. These signaling molecules can influence hair follicle cycling, growth, and even hair loss51. For example, leptin, an adipocytokine, has been shown to promote hair growth by stimulating hair follicle stem cell proliferation52.
Cytokine-related entries were identified in both GO and KEGG enrichment results of co-expression target genes for DE lncRNAs. Cytokines are small, secreted proteins that act as signaling molecules, mediating communication between cells within the immune system and other tissues53. They are essential for regulating a wide range of biological processes, including inflammation, immune responses, and cell growth and differentiation54. Within the hair follicle, cytokines play a crucial role in regulating hair growth, cycling, and even hair loss55. They act as messengers, mediating complex interactions between immune cells and hair follicle cells, ultimately shaping the fate of hair54,55,56.
Oxidative phosphorylation and Chemokine signaling pathway were also found in the TOP 20 pathways. Oxidative phosphorylation (OXPHOS) is a fundamental metabolic process that occurs in mitochondria, the powerhouses of cells. It involves the breakdown of glucose and other nutrients to generate ATP, the primary energy currency of cells57. In hair follicles, OXPHOS is essential for supporting the high energy demands of hair growth and cycling58. Study shows chemokine signaling continues to play a critical role in regulating hair growth and cycling59. Chemokines like CCL2 are involved in attracting immune cells to the hair follicle during the anagen (growth) phase, contributing to the formation of the hair shaft60.
LncRNA can function as competing endogenous RNAs (ceRNAs) by competitively binding to microRNAs (miRNAs), thereby regulating the expression of target messenger RNAs (mRNAs)61. Emerging evidence suggests that lncRNAs, acting as ceRNAs, are vital for proper hair follicle development and regeneration62,63. Epiplakin (EPPK1), a member of the plakin protein family that is exclusively expressed in epithelial tissues where it binds to keratins64. Previous studies have found that the expression of EPPK1 was highest in catagen of Yak Hair Follicle Cycle, which was consistent with hair keratin expression65. The above research suggested that EPPK1 might be one of the proteins involved in hair growth. IRF4 has been linked to hair graying, as it is involved in the production and storage of melanin, the pigment that determines hair, skin, and eye color66. IRF4 could cause excessive cell death in hair follicles when not regulated properly, which could lead to hair loss67,68. The ceRNA regulatory network reveals that chi-let-7a-3p directly acts on IRF4, and there are up to 5 DE lncRNAs interacting with the miRNA. CCL20 is a chemokine that plays a role in diverse physiological and pathological processes69. Scharschmidt et al.70 identified CCL20 as a HF-derived, microbiota-dependent chemokine and found its receptor, Ccr6, to be preferentially expressed by Tregs in neonatal skin. CeRNA regulatory network indicates that CCL20 is regulated by chi-let-7a-3p, which corresponds to six DE lncRNAs. ACKR3, also known as CXCR7, represents an atypical chemokine receptor that control the migration of normal human epidermal melanocytes71,72,73. And the melanocyte stem cell located in the hair follicle bulge contribute to follicular structures during each anagen phase of the hair cycle, synchronizing periodic activities to impact color to the hair74. The above DE genes, as well as upstream DE lncRNAs and miRNAs in the ceRNA network, all have the potential to regulate the development of different goat coat types.
Conclusions
In this study, we collected skin tissues from goats with different coat types for transcriptome sequencing analysis. GO functional enrichment analysis revealed keratin filament, intermediate filament, cytokine activity, and cytokine receptor binding terms. And KEGG pathway analysis found Estrogen signaling pathway, Amino sugar and nucleotide sugar metabolism and Adipocytokine signaling pathway. Finally, the analysis of the ceRNA regulatory network between DE lncRNAs and DE mRNAs also revealed several ceRNA regulatory axes, including those with EPPK1, IRF4, CCL20, and CXCR7 as target genes. This study identifies critical DE lncRNAs and regulatory pathways, which can be explored in future functional studies to elucidate their roles in coat type determination.
Data availability
Sequence data that support the findings of this study have been deposited in National Genomics Data Center (https://ngdc.cncb.ac.cn/) with the primary accession code CRA018956. Sequencing data is available via GSA accession code CRA018956.
References
Jin, M. et al. Genetic signatures of selection for cashmere traits in Chinese goats. Animals (Basel) 10 https://doi.org/10.3390/ani10101905 (2020).
Zhang, Y. et al. Transcriptome profiling reveals transcriptional and alternative splicing regulation in the early embryonic development of hair follicles in the cashmere goat. Sci. Rep. 9 https://doi.org/10.1038/s41598-019-54315-7 (2019).
Diao, X. et al. Hair follicle development and cashmere traits in Albas goat kids. Animals (Basel) 13 https://doi.org/10.3390/ani13040617 (2023).
Coelho, L. S., Fonseca, C. E. M. D., Melo, A. L. P. D., Neves, M. A. & Rodrigues, M. T. First assessment of the double coated produced by goats Raised in Brazil. Small Ruminant Res. 187, 106094 (2020).
Parsad, R. et al. Climate resilience in goats: a comprehensive review of the genetic basis for adaptation to varied Climatic conditions. Mamm. Genome. https://doi.org/10.1007/s00335-024-10101-z (2024).
Vasu, M., Ahlawat, S., Arora, R. & Sharma, R. Deciphering the molecular drivers for cashmere/pashmina fiber production in goats: a comprehensive review. Mamm. Genome. https://doi.org/10.1007/s00335-025-10109-z (2025).
Saleh, A. A., Rashad, A. M. A., Hassanine, N. N. A. M., Sharaby, M. A. & Zhao, Y. Assessment of hair and cashmere properties and their genetic background of several goat breeds in Southwest China. Sci. Rep. 12, 11135. https://doi.org/10.1038/s41598-022-14441-1 (2022).
Kerkhoff, K., Cescutti, G., Kruse, L. & Mussig, J. Development of a DNA-analytical method for the identification of animal hair fibers in textiles. Text. Res. J. 79, 69–75 (2009).
Gao, Y. et al. Developmental mapping of hair follicles in the embryonic stages of cashmere goats using proteomic and metabolomic construction. Animals (Basel) 13 https://doi.org/10.3390/ani13193076 (2023).
Gong, G. et al. Identification of genes related to hair follicle cycle development in inner mongolia cashmere goat by WGCNA. Front. Vet. Sci. 9, 894380. https://doi.org/10.3389/fvets.2022.894380 (2022).
Su, R. et al. Transcriptomic analysis reveals critical genes for the hair follicle of inner Mongolia cashmere goat from catagen to Telogen. PLoS One. 13, e0204404. https://doi.org/10.1371/journal.pone.0204404 (2018).
Pazzaglia, I. et al. PDGFA in cashmere goat: A motivation for the hair follicle stem cells to activate. Anim. (Basel). https://doi.org/10.3390/ani9020038 (2019).
Liu, B. et al. A Microarray-based analysis reveals that a short photoperiod promotes hair growth in the Arbas cashmere goat. PLoS One. 11, e0147124. https://doi.org/10.1371/journal.pone.0147124 (2016).
Stenn, K. S. & Paus, R. Controls of hair follicle cycling. Physiol. Rev. 81, 449–494. https://doi.org/10.1152/physrev.2001.81.1.449 (2001).
Paus, R. & Cotsarelis, G. The biology of hair follicles. N Engl. J. Med. 341, 491–497. https://doi.org/10.1056/nejm199908123410706 (1999).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. https://doi.org/10.1093/bioinformatics/bty560 (2018).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 12, 357–360 (2015).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. https://doi.org/10.1038/nbt.3122 (2015).
Sun, L. et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 41, e166. https://doi.org/10.1093/nar/gkt646 (2013).
Mistry, J., Bateman, A. & Finn, R. D. Predicting active site residue annotations in the Pfam database. BMC Bioinform. 8, 298. https://doi.org/10.1186/1471-2105-8-298 (2007).
Kang, Y. J. et al. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 45, W12–w16. https://doi.org/10.1093/nar/gkx428 (2017).
Bao, Z. et al. LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 47, D1034–d1037. https://doi.org/10.1093/nar/gky905 (2019).
Kopp, F. & Mendell, J. T. Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407. https://doi.org/10.1016/j.cell.2018.01.011 (2018).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 https://doi.org/10.1186/s13059-014-0550-8 (2014).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. https://doi.org/10.1089/omi.2011.0118 (2012).
Lv, X. et al. Effect of nutritional restriction on the hair follicles development and skin transcriptome of Chinese Merino sheep. Anim. (Basel). https://doi.org/10.3390/ani10061058 (2020).
Gong, G. et al. The regulation mechanism of different hair types in inner Mongolia cashmere goat based on PI3K-AKT pathway and FGF21. J. Anim. Sci. https://doi.org/10.1093/jas/skac292 (2022).
Jacob, J. T., Coulombe, P. A., Kwan, R. & Omary, M. B. Types I and II keratin intermediate filaments. Cold Spring Harb Perspect. Biol. https://doi.org/10.1101/cshperspect.a018275 (2018).
Bragulla, H. H. & Homberger, D. G. Structure and functions of keratin proteins in simple, stratified, keratinized and cornified epithelia. J. Anat. 214, 516–559. https://doi.org/10.1111/j.1469-7580.2009.01066.x (2009).
Moll, R., Divo, M. & Langbein, L. The human keratins: biology and pathology. Histochem. Cell. Biol. 129, 705–733. https://doi.org/10.1007/s00418-008-0435-6 (2008).
Inoue, K. et al. Differential expression of stem-cell-associated markers in human hair follicle epithelial cells. Lab. Invest. 89, 844–856. https://doi.org/10.1038/labinvest.2009.48 (2009).
Liu, Y., Lyle, S., Yang, Z. & Cotsarelis, G. Keratin 15 promoter targets putative epithelial stem cells in the hair follicle Bulge. J. Invest. Dermatol. 121, 963–968. https://doi.org/10.1046/j.1523-1747.2003.12600.x (2003).
Cui, J. et al. KLHL24-mediated hair follicle stem cells structural disruption causes alopecia. J. Invest. Dermatol. 142 2079–2087, e2078. https://doi.org/10.1016/j.jid.2022.01.007 (2022).
Laly, A. C. et al. The keratin network of intermediate filaments regulates keratinocyte rigidity sensing and nuclear mechanotransduction. Sci. Adv. 7 https://doi.org/10.1126/sciadv.abd6187 (2021).
Coulombe, P. A. Capturing intermediate filament networks. Elife 11 https://doi.org/10.7554/eLife.78248 (2022).
Gruenbaum, Y. & Aebi, U. Intermediate filaments: a dynamic network that controls cell mechanics. F1000Prime Rep. 6 https://doi.org/10.12703/p6-54 (2014).
Mariani, R. A., Paranjpe, S., Dobrowolski, R. & Weber, G. F. 14-3-3 targets keratin intermediate filaments to mechanically sensitive cell-cell contacts. Mol. Biol. Cell. 31, 930–943. https://doi.org/10.1091/mbc.E18-06-0373 (2020).
Trüeb, R. M. Let food be Thy medicine: value of nutritional treatment for hair loss. Int. J. Trichology. 13, 1–3. https://doi.org/10.4103/ijt.ijt_124_20 (2021).
Yang, M. et al. The roles of non-coding RNA in the development and regeneration of hair follicles: current status and further perspectives. Front. Cell. Dev. Biol. 9 https://doi.org/10.3389/fcell.2021.720879 (2021).
Wang, X. et al. Regulation of signaling pathways in hair follicle stem cells. Burns Trauma. 10, tkac022. https://doi.org/10.1093/burnst/tkac022 (2022).
Wang, L. et al. Differential expression of proteins associated with the hair follicle cycle—Proteomics and bioinformatics analyses. PLoS One. 11, e0146791. https://doi.org/10.1371/journal.pone.0146791 (2016).
Millar, S. E. Molecular mechanisms regulating hair follicle development. J. Invest. Dermatol. 118, 216–225. https://doi.org/10.1046/j.0022-202x.2001.01670.x (2002).
Vrtačnik, P., Ostanek, B., Mencej-Bedrač, S. & Marc, J. The many faces of Estrogen signaling. Biochem. Med. (Zagreb). 24, 329–342. https://doi.org/10.11613/bm.2014.035 (2014).
Hwang, W. J., Lee, T. Y., Kim, N. S. & Kwon, J. S. The role of Estrogen receptors and their signaling across psychiatric disorders. Int. J. Mol. Sci. https://doi.org/10.3390/ijms22010373 (2020).
Hu, H. M. et al. Estrogen leads to reversible hair cycle retardation through inducing premature catagen and maintaining telogen. PLoS ONE e40124 https://doi.org/10.1371/journal.pone.0040124 (2012).
Oh, H. S. & Smart, R. C. An Estrogen receptor pathway regulates the telogen-anagen hair follicle transition and influences epidermal cell proliferation. Proc. Natl. Acad. Sci. U S A. 93, 12525–12530. https://doi.org/10.1073/pnas.93.22.12525 (1996).
Grymowicz, M. et al. Hormonal effects on hair follicles. Int. J. Mol. Sci. https://doi.org/10.3390/ijms21155342 (2020).
Liang, A. et al. Signaling pathways in hair aging. Front. Cell. Dev. Biol. 11, 1278278. https://doi.org/10.3389/fcell.2023.1278278 (2023).
Park, S. Hair follicle morphogenesis during embryogenesis, neogenesis, and organogenesis. Front. Cell. Dev. Biol. 10, 933370. https://doi.org/10.3389/fcell.2022.933370 (2022).
Chen, J. et al. Emerging role of dermal white adipose tissue in modulating hair follicle development during aging. Front. Cell. Dev. Biol. 9, 728188. https://doi.org/10.3389/fcell.2021.728188 (2021).
Watabe, R. et al. Leptin controls hair follicle cycling. Exp. Dermatol. 23, 228–229. https://doi.org/10.1111/exd.12335 (2014).
Hoffmann, R., Eicheler, W., Huth, A., Wenzel, E. & Happle, R. Cytokines and growth factors influence hair growth in vitro. Possible implications for the pathogenesis and treatment of alopecia areata. Arch. Dermatol. Res. 288, 153–156. https://doi.org/10.1007/bf02505825 (1996).
Gregoriou, S. et al. Cytokines and other mediators in alopecia areata. Mediators Inflamm. 2010 (928030). https://doi.org/10.1155/2010/928030 (2010).
Rahmani, W., Sinha, S. & Biernaskie, J. Immune modulation of hair follicle regeneration. NPJ Regen. Med. 5 https://doi.org/10.1038/s41536-020-0095-2 (2020).
Danilenko, D. M., Ring, B. D. & Pierce, G. F. Growth factors and cytokines in hair follicle development and cycling: recent insights from animal models and the potentials for clinical therapy. Mol. Med. Today. 2, 460–467. https://doi.org/10.1016/1357-4310(96)10045-9 (1996).
Tang, Y. et al. Mitochondrial aerobic respiration is activated during hair follicle stem cell differentiation, and its dysfunction retards hair regeneration. PeerJ 4, e1821. https://doi.org/10.7717/peerj.1821 (2016).
Liu, M. et al. Intrinsic ROS drive hair follicle cycle progression by modulating DNA damage and repair and subsequently hair follicle apoptosis and macrophage polarization. Oxid. Med. Cell. Longev. 2022 8279269, https://doi.org/10.1155/2022/8279269 (2022).
Lefebvre, S., Fliniaux, I., Schneider, P. & Mikkola, M. L. Identification of Ectodysplasin target genes reveals the involvement of chemokines in hair development. J. Invest. Dermatol. 132, 1094–1102. https://doi.org/10.1038/jid.2011.453 (2012).
Wu, W. et al. Mechanical stimuli-induced CCL2 restores adult mouse cells to regenerate hair follicles. Mol. Ther. Nucleic Acids. 32, 94–110. https://doi.org/10.1016/j.omtn.2023.03.002 (2023).
Gao, L., Zhao, Y., Ma, X. & Zhang, L. Integrated analysis of lncRNA-miRNA-mRNA CeRNA network and the potential prognosis indicators in sarcomas. BMC Med. Genomics. 14, 67. https://doi.org/10.1186/s12920-021-00918-x (2021).
Zhao, R. et al. Transcriptomic analysis reveals the involvement of lncRNA-miRNA-mRNA networks in hair follicle induction in Aohan fine wool sheep skin. Front. Genet. 11, 590. https://doi.org/10.3389/fgene.2020.00590 (2020).
Tian, D. et al. Comprehensive analysis of the expression profiles of mRNA, LncRNA, circrna, and MiRNA in primary hair follicles of coarse sheep fetal skin. BMC Genom. 25, 574. https://doi.org/10.1186/s12864-024-10427-7 (2024).
Szabo, S. et al. Epiplakin attenuates experimental mouse liver injury by chaperoning keratin reorganization. J. Hepatol. 62, 1357–1366. https://doi.org/10.1016/j.jhep.2015.01.007 (2015).
Li, S. et al. Insights from tandem mass Tag (TMT) proteomic analysis on protein network modification in control of Yak hair follicle cycle. Int. J. Mol. Sci. https://doi.org/10.3390/ijms26041532 (2025).
Paus, R., Sevilla, A. & Grichnik, J. M. Human hair graying revisited: principles, misconceptions, and key research frontiers. J. Invest. Dermatol. 144, 474–491. https://doi.org/10.1016/j.jid.2023.09.276 (2024).
Heilmann-Heimbach, S. et al. Meta-analysis identifies novel risk loci and yields systematic insights into the biology of male-pattern baldness. Nat. Commun. 8, 14694. https://doi.org/10.1038/ncomms14694 (2017).
Ong, J. S. et al. Uncovering the complex relationship between balding, testosterone and skin cancers in men. Nat. Commun. 14 https://doi.org/10.1038/s41467-023-41231-8 (2023).
Buri, C., Gutersohn, A., Hauser, C., Kappeler, A. & Mueller, C. The chemokines CCL11, CCL20, CCL21, and CCL24 are preferentially expressed in polarized human secondary lymphoid follicles. J. Pathol. 204, 208–216. https://doi.org/10.1002/path.1622 (2004).
Scharschmidt, T. C. et al. Commensal microbes and hair follicle morphogenesis coordinately drive Treg migration into neonatal skin. Cell. Host Microbe. 21, 467–477e465. https://doi.org/10.1016/j.chom.2017.03.001 (2017).
Duval, V., Alayrac, P., Silvestre, J. S. & Levoye, A. Emerging roles of the atypical chemokine receptor 3 (ACKR3) in cardiovascular diseases. Front. Endocrinol. (Lausanne). 13, 906586. https://doi.org/10.3389/fendo.2022.906586 (2022).
Meyrath, M. et al. The atypical chemokine receptor ACKR3/CXCR7 is a broad-spectrum scavenger for opioid peptides. Nat. Commun. 11, 3033. https://doi.org/10.1038/s41467-020-16664-0 (2020).
Lee, E. et al. CXCR7 mediates SDF1-induced melanocyte migration. Pigment Cell. Melanoma Res. 26, 58–66. https://doi.org/10.1111/pcmr.12024 (2013).
Zhou, Y., Zhang, Y. X., Xiong, Y. Y. & Li, Y. M. Pathogenesis and regenerative therapy in vitiligo and alopecia areata: focus on hair follicle. Front. Med. (Lausanne). 11, 1510363. https://doi.org/10.3389/fmed.2024.1510363 (2024).
Funding
This work was supported by the Basic Research Project of Shanxi Province (202203021222165), the China Agriculture Research System (CARS-39-24), the Shanxi Province’s Earmarked Fund for Modern Agro-industry Technology Research System (2024CYJSTX14-12), the Biological Breeding-National Science and Technology Major Project (2023ZD0405102-04) and the Shanxi Agricultural University Team Cultivation Project (dkykyzx202207).
Author information
Authors and Affiliations
Contributions
P.Z. and L.Z. wrote the main manuscript text; L.Z. contributing to the formal analysis; Experiments were conducted by P.Z., H.G., J.H., Z.W., Y.X., and L.Z. L.Z. also reviewed and edited the manuscript. Visualization of data was handled by H.G., J.H., Z.W., and Y.X. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhao, P., Guo, H., Han, J. et al. Transcriptome sequencing reveals the expression profiles of lncRNAs and mRNAs in goat skin tissues with different types of wool coats. Sci Rep 15, 18977 (2025). https://doi.org/10.1038/s41598-025-01187-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-01187-9