Abstract
The aim of this study was to establish a nomogram based on clinical, radiomics, and deep transfer learning (DTL) features to predict meningioma grade. Three hundred forty meningiomas from one hospital composed the training set, and 102 meningiomas from another hospital composed the test set. The enhanced T1 WI images were used for analysis. The clinical, radiomics and DTL features were selected to construct the model. Radiomics and DTL scores were calculated. The deep transfer learning radiomics (DTLR) nomogram was developed on the basis of selected clinical features, radiomics scores and DTL scores. Receiver operating characteristic (ROC) curves and decision curve analysis (DCA) curves were drawn. The clinical features of sex, shape, indistinct margin and peritumoral edema were selected and used to construct the clinical model. Thirty-two radiomics features and 28 DTL features were selected for model construction. The clinical model had an AUC of 0.788. (95% CI: 0.6996–0.8756), with an accuracy of 0.745, a sensitivity of 0.941, and a specificity of 0.549 in the test set. The DTLR nomogram had the highest AUC of 0.866 (95% CI: 0.7984–0.9340), with an accuracy of 0.804, a sensitivity of 0.745, and a specificity of 0.863 in the test set. Compared with the other models, the DTLR nomogram had the greatest net benefit according to the DCA. There was a significant difference between the DTLR nomogram and the clinical model, no significant difference between the rest models in DeLong test.The DTLR nomogram has superior predictive value in DCA and could be a valuable method in clinical decision-making. Given the results of DeLong test, only the radiomics model is sufficient and there is no need to add DTL features. As a new attempt, the DTLR nomogram needs to be improved in the future study.
Similar content being viewed by others
Introduction
Meningiomas are the most prevalent intracranial tumors observed in adults, accounting for approximately 33.8% of all intracranial tumors1,2. They are classified into three grades (1, 2, and 3) according to the 2021 criteria established by the World Health Organization (WHO)3. Low-grade tumors (grade 1) exhibit different biological behaviours than high-grade tumors (grades 2 and 3), which are known for their aggressive nature, tendency to recur, and poor prognosis4,5,6. Hence, the treatment approaches for high- and low-grade tumors vary, with surgical resection being of utmost importance for high-grade tumors that are not amenable to alternative treatments4. Alternative therapies such as cytotoxic chemotherapy, hormone therapy, and targeted therapy can be considered for high-grade tumors that cannot be completely removed by surgery7. Long-term follow-up, stereotactic radiotherapy and surgery are more favorable treatment options in patients with low-grade tumors8. Hence, precise preoperative prediction of meningioma grade may help guide treatment planning for the development of effective treatment strategies and for improving prognosis.
Radiomics and deep learning (DL) have been employed for many diseases9,10,11,12,13. Radiomics is a novel approach that uses high-throughput extraction of texture parameters from medical images to quantitatively and objectively predict results, enabling the prediction of outcomes through modelling. While convolutional neural networks (CNNs) are widely used in deep learning and are based on many images from reality, building a CNN medical model can be challenging due to the limited sample size in medical research. Therefore, many studies have focused on deep transfer learning (DTL). DTL involves training a pretrained model on a vast dataset, such as ImageNet, to recognize diverse image features. These features can serve as a foundation for other image-related tasks. By fine-tuning a pretrained model on a smaller dataset that is specific to a new task, the model can learn to recognize new features and improve its performance on the new task.
Several studies have utilized radiomics or DTL to predict meningioma grade14,15,16,17,18,19,20,21. However, to the best of our knowledge, no investigation has employed a nomogram that integrates clinical, radiomics, and DTL features to predict meningioma grade. In this study, our aim was to establish a valuable evaluation criterion for meningioma grading and assist in formulating clinical treatment strategies by developing a nomogram based on clinical, radiomics, and DTL features to predict meningioma grade for the first time. In the constructed nomogram, clinical, radiomics and DTL features are given point values, and the total points can be calculated. Each total point corresponds to a risk value. The risk of every patient for high-grade meningioma can be determined in this way. The outstanding advantages of nomograms in clinical practice are visual representation and ease of communication. Nomograms provide a clear and intuitive way to represent complex mathematical relationships. Nomograms are useful for conveying information to nonspecialists or for quick decision-making to estimate patient risk.
Materials and methods
Patients
This study was performed in accordance with the Declaration of Helsinki. The institutional review board of the Affiliated Hospital of Qingdao University and Qilu Hospital (Qingdao) of Shandong University, Cheeloo College of Medicine approved this retrospective study. The requirement for informed consent was waived by the institutional review board of the Affiliated Hospital of Qingdao University and Qilu Hospital (Qingdao) of Shandong University, Cheeloo College of Medicine. This study was performed in accordance with relevant guidelines and regulations. We analysed patients who underwent surgical resection for meningioma between January 1, 2012, and June 31, 2022, at the two hospitals. All the tumors were graded according to the 2021 WHO criteria. The inclusion criteria were (1) patients who underwent enhanced T1-weighted imaging prior to surgery, (2) patients who did not undergo any treatment prior to magnetic resonance imaging (MRI) examination or surgery, and (3) patients who received a pathological diagnosis of meningioma with a clear grade after surgery. The exclusion criteria included (1) severe artefacts on MR images that made image analysis unsatisfactory and (2) no pathological diagnosis or grade. We recorded clinical information, including age and sex.
MRI examination and MRI feature analysis
Signa 1.5T and 3.0T MRI from GE and Prisma/Skyra 3.0T MRI machines from Siemens were used. Following the administration of 0.1 mL/kg Gd-DTPA, enhanced T1 WI images were obtained with the following MRI parameters: a TR of 1800 ms, a TE of 10 ms, a slice thickness of 5 mm, and an FOV of 25 cm. The same MRI acquisition parameters were used in the training and test sets.
A consensus was reached by two neuroradiologists with ten and twenty years of experience who analysed the MRI features. The MRI features were analysed via enhanced T1 WI images. The MRI features consisted of location, size, shape, heterogenous enhancement, necrosis or cystic degeneration, dural tail, indistinct margin, peritumoral edema, and surrounding invasion. Skull base meningiomas included meningiomas that occurred in the anterior cranial fossa/olfactory groove, sphenoid orbital region, middle cranial fossa, sellar region/cavernous sinus region, and posterior cranial fossa. Other locations are non-skull base. Round and round-like shape were defined as regular shape, while others were unregular. Surrounding invasion referred to invasion of brain, skull, venous or other surrounding tissues.
Image preprocessing and tumor segmentation
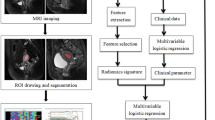
The workflow is shown in Fig. 1.
Image preprocessing and tumor segmentation were performed via 3D slicer software (version 4.11, https://www.slicer.org/). To ensure data standardization, image preprocessing was performed, including the removal of low-frequency intensity nonuniformity via N4ITK MRI bias correction22 and the discretization of image intensities using the same bin width of 25. N4ITK MRI bias correction can enhance overall image quality and improve the accuracy of subsequent analysis tasks. Removing intensity nonuniformity reduces variability in data, which can be especially important in longitudinal studies or when comparing MRI data across different subjects. Discretization reduces the complexity of image data by dividing the continuous range of pixel intensities into a finite number of bins or levels. This can make subsequent image processing and analysis algorithms computationally more efficient. We selected a bin width of 25 based on a balance between computational efficiency and preserving sufficient detail in the intensity histograms. This value has been used in several radiomics studies13,23, where it demonstrated reliable performance in capturing meaningful texture patterns while reducing noise from minor intensity variations. To maintain scale and direction conservation, images were resampled with a voxel size of 1 × 1 × 1 mm324. Resampling allows the voxel size to be standardized across different imaging datasets. Standardizing the voxel size ensures consistency, making it easier to compare and combine data from different sources. Resampling ensures that data are directly comparable among participants or subjects, which is critical for drawing meaningful conclusions.
The region of interest (ROI) was delineated on the processed images along the tumor edge while avoiding adjacent structures. Each tumor slice was included, and a 3D ROI was obtained for each tumor.
Radiomics feature extraction
Two neuroradiologists with ten and twenty years of experience independently drew the ROIs of 20 randomly chosen meningiomas to calculate the intraclass correlation coefficient (ICC) of radiomics features. In cases where discrepancies arose between their segmentations, a consensus process was implemented. Specifically, both neuroradiologists reviewed the discrepant segmentations together and discussed the differences until they reached a mutual agreement on the final segmentation. This collaborative review process ensured that the final ROIs used for analysis were consistent and accurately represented the tumor boundaries. Features demonstrating high stability (ICC ≥ 0.8) were retained for subsequent analysis.
DTL feature extraction
Given the computational complexity and hardware resources, 2D ROI was used for DTL feature extraction instead of 3D ROI. An 2D max ROI was first cropped from the 3D ROI in the axis images for feature extraction and fine-tuning the model. The image size was normalized to 224 × 224 for input. We utilized ResNet50, a well-established pretrained CNN model, to fine-tune the model and extract DTL features for our analysis. Fine-tuning ResNet-50 was crucial for task-specific learning, domain adaptation to unique dataset features, and the prevention of overfitting through rigorous techniques such as data augmentation and dropout layers. Fine-tuning offered the flexibility required for transfer learning, allowing the model to adapt to the nuances of our specific dataset, including the distinct features and patterns of medical images. This adaptation, coupled with regularization, contributed to improved model generalization, making fine-tuning the preferred approach in our study. We fine-tuned the ResNet50 model using the maximum ROIs from both the training and test sets. The fine-tuning process involved setting the batch size and epochs to 32 and 30, respectively. We used a total of 442 images for fine-tuning. The fine-tuning process was conducted using a learning rate of 0.01, with stochastic gradient descent (SGD) as the optimization algorithm. We trained the model for 30 epochs. The fine-tuning process was stopped when the model gained the best performance. To evaluate the model’s effectiveness in grade prediction, we employed multiple metrics, including accuracy, sensitivity, specificity, and AUC, providing a comprehensive assessment of the model’s performance. Following this, we obtained a fine-tuned model from which we extracted DTL features. We fed all the images into the fine-tuned model and extracted DTL features for both the training and test sets. The stability of DTL features was assessed by calculating the ICC across 20 different images. These images were randomly selected from the training dataset and were not part of the testing set to avoid data leakage. The same set of 20 images was used throughout the stability analysis to ensure consistency in feature comparison. The DTL features with high stability (ICC ≥ 0.8) were retained for the next analysis.
Clinical, radiomics, and DTL features selection
Single-factor regression analysis and multivariate regression analysis were used to select clinical features. The selected features were used for clinical model construction.
The normalization of the z score was applied to the radiomics and DTL features before analysis. The Spearman correlation coefficient and least absolute shrinkage and selection operator (LASSO) were used to select features. The correlation coefficient of every two features was calculated, and only the feature with better performance was retained if the correlation coefficient was greater than 0.9. The pairwise feature correlations enhanced interpretability and simplicity. These methods are easily understood and computationally efficient compared with correlations involving multiple features. The identification of highly correlated feature pairs was crucial for selecting features, reducing redundancy, simplifying models, and improving interpretability. LASSO regression was applied to reduce the dimensions of the features following Spearman analysis. The radiomics and DTL scores were calculated from the selected features and the respective coefficients via LASSO regression.
Model construction
Clinical, radiomics and DTL models were built via the selected features with a support vector machine (SVM). The selected clinical features, rad score and DTL score were used to build the DTLR nomogram model. The clinical, radiomics, DTL models and the DTLR nomogram were compared.
The comparison between the different models included performance metrics and visual aids. The performance metrics included accuracy, sensitivity, specificity, and the area under the receiver operating characteristic curve (AUC). The visual aids included ROC curves, calibration curves and decision curve analysis (DCA) curves. The calibration of the model was assessed via a calibration curve. The clinical usefulness of the models was evaluated via DCA curves.
Statistical analysis
The data were analysed via Python. For categorical variables and continuous variables, the chi-square test or t test, respectively, was used to analyse the clinical features of two groups. Single-factor regression analysis and multivariate regression analysis were performed via Cox regression to select clinical features. The DeLong test of AUCs was used to compare the performance of the models. The Hosmer-Lemeshow test was used to evaluate model goodness-of-fit. A significance level of p < 0.05 was established. The one-key AI platform (http://www.medai.icu) was used to process the data, and the code is available at https://gitee.com/wangqingbaidu/OnekeyCompo? from=gitee_search.
Results
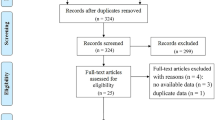
We collected 170 and 51 cases of high-grade meningioma from the two hospitals, respectively. To minimize selection bias, an equal number of low-grade meningioma cases were randomly selected to match the high-grade group, considering the considerably larger number of low-grade cases. A total of 442 patients were included in the study. A total of 47 and 28 patients were excluded according to exclusion criteria 1 and 2, respectively. A total of 340 patients from one hospital composed the training set, and 102 patients from another hospital composed the test set.
Table 1 shows the results of the clinical features analysis of the two groups in the training and test sets, which revealed significant differences in sex, size, shape, heterogenous enhancement, necrosis or cystic degeneration, indistinct margin, peritumoral edema and surrounding invasion in the training set. There was a significant difference in age, sex, size, shape, heterogenous enhancement, necrosis or cystic degeneration, indistinct margin, peritumoral edema in the test set. There was no significant difference between training and test set in low grade meningioma (Supplemental Data Table 1). There was a significant difference in age, size and necrosis or cystic degeneration between training and test set in high grade meningioma (Supplemental Data Table 1). Table 2 presents the results of the single-factor and multivariate regression analyses. The features of sex, shape, indistinct margin and peritumoral edema were ultimately selected and used to construct the clinical model.
There were 1143 radiomics features extracted from each ROI. After conducting ICC analysis, Spearman analysis and LASSO regression, 32 features were selected, and LASSO regression indicated a λ value of 0.0193. The LASSO regression procedure is presented in Fig. 2A, B, and the correlation coefficients of the selected features are shown in Fig. 3A.
Using the selected features, the Rad score was calculated via Formula 1(Supplemental Data). The accuracy, sensitivity, specificity, and AUC (95% CI) of the fine-tuned model was 0.725, 0.627, 0.824, 0.793(0.7066–0.8786). For the DTL features, 2048 features were initially extracted, with 28 features being selected by ICC analysis, Spearman analysis and LASSO regression. The λ value determined via LASSO regression was 0.0687. The LASSO regression procedure is presented in Fig. 2C, D, and the correlation coefficients of the selected features are shown in Fig. 3B. The DTL score was calculated via Formula 2 (Supplemental Data).
The DTL model is visualized by Grad CAM (Fig. 4, Supplemental Data Fig.1–10).
The DTLR nomogram is presented in Fig. 5A.
The DTL nomogram included the clinical features of sex, shape, indistinct margin, peritumoral edema, Rad score and DTL score (A). The ROC curves (B) and calibration curves (C) of the clinical model, radiomics model, DTL model and DTLR nomogram. DTL Deep transfer learning, DTLR Deep transfer learning radiomics.
Table 3 shows the results of the clinical, radiomics, DTL and DTLR nomogram models. The DTL model had the highest AUC of 0.942 (95% CI: 0.9179–0.9660), with an accuracy of 0.874, a sensitivity of 0.824 and a specificity of 0.924 in the training set. The DTLR nomogram had the highest AUC of 0.866 (95% CI: 0.7984–0.9340), with an accuracy of 0.804, a sensitivity of 0.745 and a specificity of 0.863 in the test set.
The ROC curves and calibration curves of the four models are shown in Fig. 5B, C. The AUCs of the clinical, radiomics, DTL, and DTLR nomogram models were compared with those of the DeLong test (Table 4). There was a significant difference between the DTLR nomogram and the clinical model.
The Hosmer–Lemeshow test results for the clinical, radiomics, DTL and DTLR nomogram were 0.098, 0.414, 0.387 and 0.414, respectively. The DCA curve in Fig. 6 indicates that the DTLR nomogram has the largest net benefit across all threshold probabilities compared with the other models.
Discussion
In the present study, we developed a nomogram that integrates clinical, radiomics, and DTL features to predict meningioma grade by using enhanced T1WI images. Our findings suggest that the DTLR nomogram demonstrated superior predictive performance, with an AUC of 0.866 (95% CI: 0.7984–0.9340), an accuracy of 0.804, a sensitivity of 0.745 and a specificity of 0.863 in the test set.
Numerous studies have utilized radiomics to predict meningioma grade. For example, Chu H et al.25 analysed 98 patients, including 16 patients with high-grade meningioma, via logistic regression. They reported areas under the curve of 0.958 and 0.948 in the training and test groups, respectively. In Chen C et al.‘s study26, linear discriminant analysis (LDA) and an SVM were used to construct a radiomic model for predicting meningioma grade. The study sample included 150 cases, including 89 high-grade meningiomas, and the combination of LASSO + LDA yielded the highest accuracy of 75.6% among the LDA-based models.
Several studies have utilized deep learning techniques for meningioma grade prediction. Zhu Y et al.20 developed a deep learning radiomics model for preoperative grading and reported an AUC of 0.811 (95% CI, 0.635–0.986), a sensitivity of 0.769, and a specificity of 0.898 in the validation cohort. Yang L et al.21 investigated the clinical usefulness of an enhanced-T1WI-based deep learning radiomics model (DLRM) in differentiating low- and high-grade meningiomas. Their results revealed that the DLRM outperformed conventional radiomics and deep learning models in predicting meningioma grade, with training AUCs of 0.988, 0.980, and 0.892 for DLRM, radiomics, and deep learning, respectively, and test AUCs of 0.935, 0.918, and 0.718, respectively.
Compared with previous studies, our study had several improvements. First, in this study, we included a relatively larger sample size of 221 cases of high-grade meningioma, surpassing the sample sizes of most other radiomic studies focused on predicting meningioma grade. Radiomics and deep learning, which are rooted in artificial intelligence and big data, benefit from the use of larger sample sizes, as they contribute to more dependable and robust outcomes. Second, to the best of our knowledge, no previous study has employed a DTLR nomogram for the prediction of meningioma grade. Although some studies have used radiomics nomograms for meningioma grade prediction, our study revealed that DTL performed better than radiomics did. Radiomics is concerned with extracting quantitative characteristics from medical images, whereas deep transfer learning focuses on automatically learning features from vast datasets of images27,28,29. Radiomics requires proficiency in image processing and statistical analysis to extract meaningful features, whereas deep transfer learning requires expertise in deep learning and computer vision to fine-tune pretrained networks for a specific task27,28,29.
We selected the 2D maximum ROI from the 3D ROI along the image axis. The rationale for this choice was to focus on the slice that best captured the most significant and representative features of the tumor, reducing potential noise and variability that could arise from including non-representative slices. This approach has been used in previous studies to effectively extract key features with minimal computational complexity23,30,31. We chose ResNet50 due to its balance between depth and computational efficiency. ResNet50 provided a deeper architecture which enabled it to capture more complex and hierarchical features. In the context of our study, where subtle differences in imaging features were crucial for accurate prediction of meningioma grade, we considered ResNet50’s depth to be advantageous. Additionally, ResNet50 has been widely adopted and proven effective in transfer learning tasks for medical imaging32,33, which gave us confidence in its suitability. Even so, the comparison of different pretrained models was essential. We will consider compare deep transfer learning results across different pretrained models in the future. In the Grad CAM of ResNet50, warm colors (red, yellow, orange) represented areas of the image that had the highest positive influence on the model’s prediction. Red typically indicated the most important regions in the image for making a decision. The red regions related to enhanced part of the tumor were identified by the model, which had an important impact on meningioma grade, due to different grade meningioma had different enhanced pattern.
In our study, we constructed the first DTLR nomogram to predict meningioma grade, which combines clinical, radiomics, and DTL features. The DTLR nomogram had an AUC of 0.866 (95% CI: 0.7984–0.9340), with an accuracy of 0.804, a sensitivity of 0.745 and a specificity of 0.863. Furthermore, the DTLR nomogram exhibited a greater net benefit across the entire range of threshold probabilities, as demonstrated by the decision curve analysis (DCA) curve, surpassing the performance of the other models. Moreover, the DTLR nomogram is visual and quantifiable for doctors, which allows a patient’s risk of high-grade meningioma to be calculated and an individualized treatment plan to be formulated.
The nomogram presented in Fig. 5A allows clinicians to calculate the individualized risk of high-grade meningioma based on patient characteristics. The score components include sex, shape, indistinct margin, peritumoral edema, Rad score and DTL score. For example, a male patient with an irregular, indistinct margin, peritumoral edema meningioma and high Rad score and DTL score will score higher on the nomogram, corresponding to a predicted higher risk of high-grade meningioma. In addition to providing individual risk predictions, we performed DCA to evaluate the clinical utility of the model. DCA compares the net benefit of using the model at different probability thresholds against the strategies of treating all or no patients. For instance, at a threshold of 40%, the model provides a net benefit over treating all patients, indicating that it is a valuable tool for identifying those at risk for high-grade meningioma. Clinicians can use this information to guide treatment decisions: for patients whose predicted risk exceeds the chosen threshold, the model suggests they may benefit from treatment. Conversely, patients below the threshold may not require immediate intervention. We recommend using the model to inform decisions when the risk threshold is between 20% and 80%, as this range shows the greatest net benefit.
It should be pointed out that there was a significant difference between the DTLR nomogram and the clinical model, no significant difference between the rest models in DeLong test. If only considered DeLong test, it did not suggest significant benefit of adding DTL features to radiomics features. It seemed that only the radiomics model was sufficient and there was no need to add DTL features which would add increased computational requirements and potentially more technical expertise. But it was undeniable that DTLR nomogram was a new attempt and the AUC value did improve in the test set.
Although our study yielded promising results, we must acknowledge several limitations. First, the retrospective nature of the study may have introduced selection bias and overestimated the accuracy of the diagnosis. Second, skull stripping was not performed as part of the preprocessing step in our current study. However, the presence of non-brain structures, particularly the bright, dense skull, may affect image intensity discretization and potentially introduce unwanted artifacts into the feature extraction process. This could influence the model’s performance and the accuracy of the radiomics features. we planned to incorporate skull stripping into the preprocessing pipeline in future studies and reanalyse the data to determine its impact on feature extraction and model performance. Third, the ROI was based solely on enhanced T1WI images instead of incorporating multiparametric MR images. We only used enhanced T1WI images mainly for three reasons. First, the boundary between the tumor and the surrounding tissue was most distinct on enhanced T1WI images, which facilitated the delineation of the ROI and made it more accurate. Second, the two hospitals from which the patients in this study were sourced were the largest in the region. Most patients were referred from local hospitals, where they had already undergone related examinations. To save medical resources and costs, the surgeons only ordered enhanced T1WI scans before surgery. Although some patients had more comprehensive MRI examinations, the number of such cases was limited. Therefore, to ensure a sufficient sample size, this study only selected enhanced T1WI images. Third, we opted for enhanced T1WI images in this study because it represented a widely used sequence in routine clinical practice and was often readily available across different institutions. Our aim was to create a model that could be easily applicable and generalizable to a broad range of clinical settings. Future studies should explore the use of multiparametric MRI to improve the accuracy of the ROI and the predictive power of the nomogram. Fourth, there was no integration of all the features and DTL into one neural network or self-supervised learning with medical images. We will attempt these methods in future research. Fifth, larger sample size from multicenter is needed to improve predictive efficiency and avoid overfitting in the future study.
Conclusion
In our study, we built a DTLR nomogram by using enhanced T1WI images to predict meningioma grade. Although additional validation is needed before its clinical application, the nomogram has superior predictive value in DCA and could be a valuable tool in clinical decision-making. But the DeLong test indicates that only the radiomics model is sufficient and there is no need to add DTL features which would add increased computational requirements and potentially more technical expertise. It is undeniable that DTLR nomogram is a new attempt and the AUC value did improve in the test set. Further research is necessary to improve the predictive value of the DTLR nomogram.
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).
References
Holleczek, B. et al. Incidence, mortality and outcome of meningiomas: A population-based study from Germany. Cancer Epidemiol. 62, 101562. https://doi.org/10.1016/j.canep.2019.07.001 (2019).
Wiemels, J., Wrensch, M. & Claus, E. B. Epidemiology and etiology of meningioma. J. Neurooncol. 99 (3), 307–314. https://doi.org/10.1007/s11060-010-0386-3 (2010).
WHO classification of tumours editorial board. World Health Organization Classification of Tumours of the Central Nervous System 5th edn (International Agency for Research on Cancer, 2021).
Rogers, L. et al. Meningiomas: knowledge base, treatment outcomes, and uncertainties. A Rano review. J. Neurosurg. 122 (1), 4–23. https://doi.org/10.3171/2014.7.JNS131644 (2015).
Goldbrunner, R. et al. EANO guideline on the diagnosis and management of meningiomas. Neuro Oncol. 23 (11), 1821–1834. https://doi.org/10.1093/neuonc/noab150 (2021).
Whittle, I. R., Smith, C., Navoo, P., Collie, D. & Meningiomas Lancet ;363(9420):1535–1543. doi:https://doi.org/10.1016/S0140-6736(04)16153-9 (2004).
Li, D. et al. Survival impacts of extent of resection and adjuvant radiotherapy for the modern management of high-grade meningiomas. J. Neurooncol. 145 (1), 125–134. https://doi.org/10.1007/s11060-019-03278-w (2019).
Black, P. M., Villavicencio, A. T., Rhouddou, C. & Loeffler, J. S. Aggressive surgery and focal radiation in the management of meningiomas of the skull base: preservation of function with maintenance of local control. Acta Neurochir. (Wien). 143 (6), 555–562. https://doi.org/10.1007/s007010170060 (2001).
Gillies, R. J., Kinahan, P. E. & Hricak, H. Radiomics: images are more than pictures, they are data. Radiology 278 (2), 563–577. https://doi.org/10.1148/radiol.2015151169 (2016).
Kumar, V. et al. Radiomics: the process and the challenges. Magn. Reson. Imaging. 30 (9), 1234–1248. https://doi.org/10.1016/j.mri.2012.06.010 (2012).
Lambin, P. et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur. J. Cancer. 48 (4), 441–446. https://doi.org/10.1016/j.ejca.2011.11.036 (2012).
Liu, S. et al. Deep learning radiomic nomogram to predict recurrence in soft tissue sarcoma: a multi-institutional study. Eur. Radiol. 32 (2), 793–805 (2022).
Zheng, Y. M. et al. A CT-Based deep learning radiomics nomogram to predict histological grades of head and neck squamous cell carcinoma [published online ahead of print, 2022 Nov 30]. Acad. Radiol. S1076-6332 (22), 00612–00612. https://doi.org/10.1016/j.acra.2022.11.007 (2022).
Duan, C. et al. A radiomics nomogram for predicting the meningioma grade based on enhanced T1WI images. Br. J. Radiol. 95 (1137), 20220141. https://doi.org/10.1259/bjr.20220141 (2022).
Duan, C. F. et al. Comparison of different radiomic models based on enhanced T1-weighted images to predict the meningioma grade. Clin. Radiol. 77 (4), e302–e307. https://doi.org/10.1016/j.crad.2022.01.039 (2022).
Park, Y. W. et al. Radiomics and machine learning May accurately predict the grade and histological subtype in meningiomas using conventional and diffusion tensor imaging. Eur. Radiol. 29 (8), 4068–4076. https://doi.org/10.1007/s00330-018-5830-3 (2019).
Lu, Y. et al. The diagnostic value of texture analysis in predicting WHO grades of meningiomas based on ADC maps: an attempt using decision tree and decision forest. Eur. Radiol. 29 (3), 1318–1328. https://doi.org/10.1007/s00330-018-5632-7 (2019).
Ke, C. et al. Differentiation between benign and nonbenign meningiomas by using texture analysis from multiparametric MRI. J. Magn. Reson. Imaging. 51 (6), 1810–1820. https://doi.org/10.1002/jmri.26976 (2020).
Han, Y. et al. Preoperative predictive histopathological grading based on radiomics of MRI. Magn. Reson. Imaging. 77, 36–43. https://doi.org/10.1016/j.mri.2020.11.009 (2021).
Zhu, Y. et al. A deep learning radiomics model for preoperative grading in meningioma. Eur. J. Radiol. 116, 128–134. https://doi.org/10.1016/j.ejrad.2019.04.022 (2019).
Yang, L. et al. A deep learning radiomics model May help to improve the prediction performance of preoperative grading in meningioma. Neuroradiology 64 (7), 1373–1382. https://doi.org/10.1007/s00234-022-02894-0 (2022).
Tustison, N. J. et al. N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging. 29, 1310–1320 (2010).
Duan, C. et al. Performance comparison of 2D and 3D MRI radiomics features in meningioma grade prediction: A preliminary study. Front. Oncol. 13, 1157379. https://doi.org/10.3389/fonc.2023.1157379 (2023). Published 2023 Mar 23.
Depeursinge, A., Foncubierta-Rodriguez, A., Van De Ville, D. & Muller, H. Three-dimensional solid texture analysis in biomedical imaging: review and opportunities. Med. Image Anal. 18, 176–196 (2014).
Chu, H. et al. Value of MRI radiomics based on enhanced T1WI images in prediction of meningiomas grade. Acad. Radiol. 28 (5), 687–693. https://doi.org/10.1016/j.acra.2020.03.034 (2021).
Chen, C. et al. The diagnostic value of Radiomics-Based machine learning in predicting the grade of meningiomas using conventional magnetic resonance imaging: A preliminary study. Front. Oncol. 9, 1338. https://doi.org/10.3389/fonc.2019.01338 (2019). Published 2019 Dec 6.
Lambin, P. et al. Radiomics: the Bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 14 (12), 749–762. https://doi.org/10.1038/nrclinonc.2017.141 (2017).
Aerts, H. J. et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach [published correction appears in Nat Commun 5 4644. Cavalho, Sara [corrected to Carvalho, Sara]]. Nat Commun. 2014 5 4006. (Published, 2014). https://doi.org/10.1038/ncomms5006
Litjens, G. et al. A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88. https://doi.org/10.1016/j.media.2017.07.005 (2017).
Arefan, D., Chai, R., Sun, M., Zuley, M. L. & Wu, S. Machine learning prediction of axillary lymph node metastasis in breast cancer: 2D versus 3D radiomic features. Med. Phys. 47 (12), 6334–6342. https://doi.org/10.1002/mp.14538 (2020).
Meng, L. et al. 2D and 3D CT radiomic features performance comparison in characterization of gastric cancer: A multi-center study. IEEE J. Biomed. Health Inf. 25 (3), 755–763. https://doi.org/10.1109/JBHI.2020.3002805 (2021).
Zhang, Y. et al. Deep Learning-based automatic diagnosis of breast Cancer on MRI using mask R-CNN for detection followed by ResNet50 for classification. Acad. Radiol. 30 (Suppl 2), S161–S171. https://doi.org/10.1016/j.acra.2022.12.038 (2023).
Wang, H., Ying, J., Liu, J., Yu, T. & Huang, D. Harnessing ResNet50 and senet for enhanced ankle fracture identification. BMC Musculoskelet. Disord. 25 (1), 250. https://doi.org/10.1186/s12891-024-07355-8 (2024). Published 2024 Apr 1.
Author information
Authors and Affiliations
Contributions
Nan Li and Chongfeng Duan designed the study. Nan Li and Xuejun Liu develop the method. Xiaona Xia and Gang Wang acquired the data. Xushun Liu and Chongfeng Duan analyzed and interpreted the data. Nan Li wrote and revised the manuscript. Chongfeng Duan supervised the study. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, N., Liu, X., Xia, X. et al. An MRI-based deep transfer learning radiomics nomogram for predicting meningioma grade. Sci Rep 15, 16614 (2025). https://doi.org/10.1038/s41598-025-01665-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-01665-0
Keywords
This article is cited by
-
Multi-task Nomogram Model for Predicting Rebleeding and Survival Outcomes in Cerebral Contusion
Journal of Imaging Informatics in Medicine (2026)
-
Advancements in the application of MRI radiomics in meningioma
Radiation Oncology (2025)