Abstract
Oil palm plantations face serious challenges from Ganoderma boninense, a pathogen that causes basal stem rot (BSR), leading to significant productivity losses, with an estimated economic impact of 68.73%. Ganoderma spreads through direct root contact and airborne spores, affecting plantations across Indonesia, Malaysia, and other countries. Understanding the mechanisms of oil palm resistance to Ganoderma is crucial for developing effective strategies. Metabolomic profiling, ¹H NMR spectroscopy, offers a promising tool for identifying and quantifying metabolic changes associated with Ganoderma resistance. This study, ¹H NMR was employed to analyze root tissues of resistant, susceptible, and control oil palm seedlings exposed to Ganoderma. The results indicated that PCA effectively differentiated resistant palms from susceptible ones, while PLS-DA identified 14 significant metabolites. Further analysis using OPLS-DA and ROC revealed that ascorbic acid, D-gluconic acid, D-fructose, and 2-oxoisovalerate could serve as potential biomarkers for screening resistant palms. The metabolites identified in this study hold considerable promise for supporting breeding programs to develop oil palm varieties with enhanced resistance to BSR.
Similar content being viewed by others
Introduction
Oil palm (Elaeis guineensis) is a vital crop cultivated in tropical regions within ± 10° latitude of the equator, including parts of Africa, Southeast Asia, and South and Central America. These regions offer favorable climatic conditions, such as high rainfall, for oil palm growth1. In Indonesia, oil palm has been commercially cultivated since 1911 under Dutch rule2. To date, Indonesia has been the largest exporter of palm oil in the global market for the past two decades3. The palm oil industry has become an important part of Indonesia’s economy, contributing to employment, rural development, and foreign exchange earnings.
Oil palm plantations face serious challenges from Ganoderma boninense, a pathogen responsible for causing basal stem rot (BSR). Ganoderma infects the base or middle of the oil palm trunk, leading to tissue decay and ultimately plant death4. The disease spreads rapidly through direct contact between infected and healthy roots, as well as via airborne basidiospores, which facilitate long-distance dispersal5. Ganoderma infections are not limited to Indonesia and Malaysia but have also been reported in several African countries and Colombia6. According to research by Kamu et al.7, economic losses due to Ganoderma infections are estimated to reach 68.73%.
No effective method for controlling BSR has been found, as technical cultural approaches, chemical control, and biological control have all been employed without success8. Breeding for oil palm resistance against Ganoderma offers a potential solution, but it remains challenging because disease resistance is controlled by multiple genes9. Developing multigenic resistance is complex, as resistance (R) genes are typically unlinked, making it challenging and labor-intensive to create and maintain R gene combinations in breeding programs10.
Understanding the mechanisms behind oil palm resistance to Ganoderma is essential for effectively managing this disease. Therefore, research exploring these resistance mechanisms, particularly at the metabolomic level, is highly promising, as it is closely linked to the phenotypic level11. Metabolomic profiling facilitates identifying and quantifying metabolic changes within a biological system in response to various stimuli, such as internal factors like genetic alterations and external factors like pathogen interactions12. By gaining insights into the metabolic changes associated with resistance, appropriate strategies can be developed to tackle the Ganoderma problem in oil palm plantations in the future.
Proton nuclear magnetic resonance (¹H NMR) spectroscopy is an advanced technique in metabolomic profiling that has been utilized in previous studies to investigate differences in oil palm resistance to Ganoderma13,14. Unlike other spectrometry techniques, ¹H NMR is nondestructive, unbiased, easily quantifiable, and requires minimal or no chromatographic separation, sample treatment, or chemical derivatization, while also enabling the routine identification of novel compounds15. In this study, we applied untargeted metabolomics to analyze root tissues from resistant, susceptible, and control oil palm seedlings challenged with Ganoderma boninense. Root tissue was selected due to its direct contact with the pathogen, which is critical for early-stage infection. The aim was to identify metabolite signatures associated with resistance, enabling the discovery of potential biomarkers for early screening. These findings are expected to support breeding programs by facilitating the selection of resistant genotypes and enhancing our understanding of the metabolic mechanisms underlying oil palm defense.
Results
1H NMR metabolites profile of oil palm root tissues infected with Ganoderma
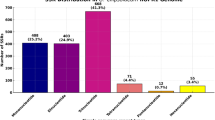
The1H NMR profiling of oil palm root tissues at the nursery stage identified a total of fifty-three (53) metabolites across three sample groups: resistant, susceptible, and control. Specifically, 35 metabolites were detected in resistant root tissues, 39 in susceptible roots, and 44 in control root tissues (Supplementary Table 1).
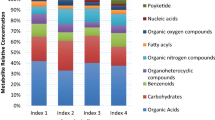
These metabolites were categorized through enrichment analysis into 11 primary classes, including organooxygen compounds (galactitol, glycerol, glyceric acid, myo-inositol, D-sorbitol, D-gluconic acid, D-fructose, glycogen, 2-propanol, threonic acid, acetone, L-arabitol, propylene glycol, threitol, xylitol); carboxylic acids and derivatives (betaine, glycine, guanidoacetic acid, fumaric acid, L-alanine, L-proline, L-isoleucine, L-serine, methylmalonic acid, propionate, sarcosine, 2-aminobutyric acid, aminoadipic acid, N-acetylglycine, L-leucine, malonate, L-valine, trans-aconitic acid); hydroxy acids and derivatives (glycolic acid, lactate, alpha-hydroxyisobutyric acid); sulfonyls (dimethyl sulfone); organic sulfonic acids and derivatives (taurine); dihydrofurans (ascorbic acid); keto acids and derivatives (2-oxobutyrate, 2-oxoisovalerate, acetoacetate); fatty acyls (adipic acid, isovaleric acid, beta-hydroxyisovaleric acid, azelaic acid, sebacic acid, valerate); azoles (allantoin); organonitrogen compounds (ethanolamine, TMAO); and phenols (homovanillic acid, pyrocatechol). The results showed that organooxygen compounds and carboxylic acids and derivatives were the dominant classes across all samples associated with disease resistance (Fig. 1).
This research identified distinct relative metabolites compared to previous studies on1H NMR profiling of oil palm resistance to Ganoderma. In earlier research,1H NMR profiling of oil palm leaf tissues infected with Ganoderma revealed 11 main classes of organic compounds, including benzamides, fatty alcohols, organic dicarboxylic acids, alcohols, polyols, sulfonic acids, cholines, furanones, alkanolamines, fatty acids and conjugates, amino acids and peptides, and monosaccharides14. Meanwhile,1H NMR profiling of stem tissues from oil palms infected with Ganoderma identified 10 main classes of organic compounds: polyketides, nucleic acids, organooxygen compounds, fatty acyls, organonitrogen compounds, organoheterocyclic compounds, benzenoids, carbohydrates, and organic acids13.
Classification and identification of metabolites in oil palm seedling roots
Multivariate data analysis using PCA (2D and 3D) effectively distinguished the metabolite profiles of resistant, susceptible, and control root tissues in oil palm seedlings, as shown in Fig. 2, with PC1, PC2, and PC3 explaining 62.3% of the variation. The analysis clearly separated resistant palms from susceptible ones. Resistant and control roots were relatively grouped together, as neither showed symptoms of Ganoderma infection, in contrast to the susceptible palms, which exhibited disease symptoms, resulting in altered metabolite profiles. Consistent with these findings, PCA analysis of NMR data by Rahmadi et al.14 also successfully differentiated resistant and susceptible palms. Supporting data (Supplementary Fig. 1) from pairwise 2D PCA comparisons between resistant vs. susceptible, resistant vs. control, and susceptible vs. control showed similar patterns. Resistant and control root samples were closely clustered, whereas resistant and susceptible ones formed distinctly separate groups.
PLS-DA analysis was conducted to identify significant metabolites differentiating resistant, susceptible, and control root tissues. Fourteen metabolites with VIP scores > 1 were identified as significant (Fig. 3a). The metabolites included 2-oxoisovalerate, myo-inositol, homovanillic acid, fumaric acid, L-valine, L-alanine, L-arabitol, ascorbic acid, galactitol, glyceric acid, lactate, D-fructose, TMAO, and D-gluconic acid. Notably, three metabolites (2-oxoisovalerate, myo-inositol, and TMAO) were concurrently identified in the PLS-DA findings of Rahmadi et al.14. Furthermore, four metabolites (D-gluconic acid, L-arabitol, ascorbic acid, and D-fructose) were also found to overlap with the significant metabolite profiles reported by Pancoro et al.13 in their investigation of oil palm responses to Ganoderma infection.
(a) PLS-DA analysis result of metabolites in resistant, susceptible, and control root tissues from oil palm seedlings. (b) Heatmap visualization of metabolites in resistant, susceptible, and control root tissues from oil palm seedlings. Up-regulated metabolites were represented in red, while down-regulated metabolites were shown in blue.
Fourteen significant metabolites identified from the PLS-DA analysis were further examined using a heatmap to visualize the varying concentrations of each metabolite among resistant, susceptible, and control root tissues in response to Ganoderma. The heatmap results mirrored the PCA findings, with resistant roots clustering with control roots (Fig. 3b). Several metabolites, such as glyceric acid, lactate, ascorbic acid, D-fructose, and D-gluconic acid, were up-regulated in resistant roots, while these metabolites were down-regulated in susceptible roots. In accordance with Pancoro et al.13, these metabolites were up-regulated in healthy oil palm seedlings compared to those severely affected by Ganoderma. In contrast, 2-oxoisovalerate, L-valine, TMAO, homovanillic acid, fumaric acid, and L-alanine were down-regulated in resistant roots compared to susceptible roots.
In this study, OPLS-DA was utilized to compare two resistance categories, complementing the PLS-DA analysis, which identified significant metabolites across all resistance levels. The analysis (Supplementary Fig. 2) identified 14 significant metabolites between resistant and susceptible samples, 18 significant metabolites between resistant and control samples, and 16 significant metabolites between susceptible and control samples, all with a VIP score > 1.0.
Biomarkers and pathway analysis for oil palm resistance to Ganoderma
Biomarker identification plays a crucial role in the early detection of diseases22. In this study, OPLS-DA and ROC curve analysis were utilized to identify oil palm metabolites capable of distinguishing between resistance and susceptibility to Ganoderma. The OPLS-DA results, along with a ROC curve (AUC) = 1, indicate that these metabolites have the potential to serve as reliable biomarkers. An AUC score 1 signifies that the classifier can accurately differentiate between positive and negative classes, with no false positives13,14. Several metabolites were identified as significant biomarkers (Table 1). Ascorbic acid, D-gluconic acid, D-fructose, and 2-oxoisovalerate exhibited significant differences in resistant roots compared to susceptible roots, suggesting their potential as biomarkers for screening Ganoderma-resistant oil palms. These metabolites are crucial in distinguishing resistance levels, providing valuable insights for breeding programs. To further illustrate these differences, box plots were generated, highlighting the significant variation in ascorbic acid concentrations among resistant, susceptible, and control root tissues (Fig. 4).
Pathway analysis was performed to determine the metabolic pathways associated with oil palm resistance to Ganoderma. Utilizing the KEGG database for pathway enrichment analysis (p-value < 0.05 and pathway impact > 0.1), the results revealed that basal stem rot (BSR) disease affects distinct metabolic pathways in resistant, susceptible, and control root tissues. Notably, the glycine, serine, and threonine metabolism pathways were consistently present across all three root conditions (Fig. 5). However, ascorbate and aldarate metabolism, as well as glycerolipid metabolism, were detected only in resistant and susceptible roots.
Pathway analysis of metabolites in (a) resistant, (b) susceptible, and (c) control root tissues from oil palm seedlings (p-value < 0.05 and pathway impact > 1). The horizontal dotted line represents the threshold for statistical significance (p-value), while the vertical line denotes the cutoff for pathway impact scores.
Discussion
The analysis of 1 H NMR spectra of oil palm root tissues revealed key differences in the metabolic profiles among resistant, susceptible, and control oil palm seedlings. Enrichment analysis highlighted that sulfonyl compounds were exclusively detected in resistant roots. These compounds are known as plant immune-priming agents that enhance disease resistance23. Sulfonyl compounds trigger the biosynthesis of salicylic acid (SA), which strengthens the plant’s defense against pathogens, including fungi, bacteria, and viruses24. Among the sulfonyl compounds identified, dimethyl sulfone was notably up-regulated in resistant roots (Supplementary Fig. 3). Supporting this finding, dimethyl sulfone has demonstrated antifungal activity against Botrytis cinerea in a separate study25.
In contrast, phenolic compounds were exclusively detected in susceptible roots infected with Ganoderma. Plants under stress or infection often increase the production of phenolic compounds as a response to cellular damage, aiding in defense against pathogens26. The elevated phenol levels in susceptible plants are believed to indicate that the plants are attempting to respond to infection or stress, but their defense mechanisms are not yet strong enough to effectively combat the pathogens. The phenolic compounds up-regulated in susceptible plants were identified as homovanillic acid and pyrocatechol (Supplementary Fig. 3). The role of homovanillic acid in plants is not well understood, though it acts as a potential antioxidant in animal cells27. Pyrocatechol, on the other hand, plays a role in protecting plants against pathogens and contributes to nitrogen detoxification28. In tomatoes, pyrocatechol has been shown to prevent disease symptoms after infection by Fusarium oxysporum29.
The grouping of resistant and control root tissues in the PCA analysis suggests that resistant palms maintain a metabolite profile closer to that of healthy, uninfected plants. This implies that resistant palms possess innate or early-induced defenses that minimize metabolic disruption during Ganoderma infection, thereby preventing the drastic biochemical changes observed in susceptible plants. According to Zhu et al.30, plant immunity inducers can prime plant defense mechanisms and significantly reduce the occurrence and severity of plant diseases. Moreover, resistant plants often maintain homeostasis by producing specific defense-related metabolites that inhibit pathogen growth. Similarly, Jian et al.31 reported that plants produce defense molecules that directly suppress pathogen growth and development through both constitutive and inducible mechanisms.
PLS-DA analysis revealed key metabolites that are significantly different across the resistance categories. Moreover, the heatmap results further support these findings, showing a pattern where metabolites up-regulated in resistant roots are consistent with known defense-related compounds. This up-regulation suggests that these metabolites play a crucial role in strengthening the plants defense against Ganoderma infection, while their down-regulation in susceptible roots implies a weakened or compromised defense mechanism. Furthermore, according to Anjali et al.32, up-regulated metabolites in plants due to pathogen attacks contribute to the formation of a robust defense system, which boost plant survival. In addition, several studies have shown that these defense-related metabolites not only act as direct antimicrobial agents but also serve as signaling molecules that activate further immune responses33,34,35,36,37.
Biomarker analysis identified ascorbic acid, D-gluconic acid, D-fructose, and 2-oxoisovalerate as significant biomarkers with potential for screening resistant palms, as they play essential roles in plant defense mechanisms. Ascorbic acid, particularly, is well known for protecting plant cells from oxidative stress induced by pathogen attacks38. Furthermore, in vitro studies have demonstrated that the growth of the soil-borne hemibiotrophic fungus Macrophomina is significantly inhibited in media containing ascorbic acid compared to those without it, suggesting its direct antifungal properties35. Similarly, D-gluconic acid exhibits antifungal activity and has potential as a biocontrol agent, as it disrupts fungal growth and enhances plant resistance39,40. In addition, cultivating Rhodosporidium paludigenum with gluconic acid improved its ability to control green mold infections in citrus fruit34. Moreover, D-fructose is recognized as a key regulatory molecule in plant defense responses to fungal pathogens33. Notably, elevated sugar levels in plant tissues enhance the plant’s immune response against fungal pathogens41. Unfortunately, the specific role of 2-oxoisovalerate in plant defense against pathogens remains poorly understood. Therefore, further research is needed to elucidate its potential function in plant immunity and stress responses.
Pathway analysis revealed the involvement of glycine, serine, and threonine metabolism pathways in all three root conditions (resistant, susceptible, and control root tissues). This finding aligns with previous studies by Pancoro et al.13 and Rahmadi et al.14. These pathways are fundamental to protein synthesis and play a crucial role in plant responses to environmental stress42. Their significance has also been demonstrated in other studies, such as mandarin fruit responses to cyclic lipopeptides from Bacillus subtilis43. Furthermore, several proteins within the glycine-rich protein (GRP) superfamily have been identified as key players in cellular signaling and stress responses in plants44.
Interestingly, both resistant and susceptible roots infected with Ganoderma showed activation of the ascorbate and aldarate metabolism and glycerolipid metabolism pathways. Meanwhile, in the control treatment, these metabolic pathways were not detected. This indicates that the ascorbate and aldarate metabolism and glycerolipid metabolism pathways are only active when Ganoderma infection occurs. Based on Fick et al.45, some genes involved in plant immune response activation are triggered during pathogen infection. The activation of these metabolic pathways suggests a potential role in the plant’s defense mechanisms, possibly linked to oxidative stress regulation and lipid signaling.
Ascorbate and aldarate metabolism is crucial for protecting cells from oxidative damage, with ascorbic acid acting as a potent antioxidant that neutralizes reactive oxygen species (ROS) generated during pathogen invasion. Studies have shown that plants with lower ascorbate levels are more susceptible to fungal infections46,47. Glycerolipid metabolism, which regulates lipid signaling and ROS metabolism, plays a vital role in plant defense. Lipids and lipid-derived compounds can exhibit antimicrobial activity and contribute to plant defense against pathogens48,49,50. Furthermore, detecting fatty acids in root tissues suggests a potential activation of glycerolipid metabolism, which is critically involved in membrane remodeling during pathogen response51.
These findings imply that Ganoderma infection triggers specific metabolic adjustments that could be crucial for the plant’s adaptive response to pathogen stress. The activation of these pathways highlights the dynamic nature of the plant’s defense mechanisms. Overall, this study highlights several key metabolites and pathways differentially regulated in resistant versus susceptible oil palm seedlings, offering insights into the underlying mechanisms of Ganoderma resistance and potential biomarkers for breeding and early screening.
Conclusion
¹H NMR successfully identified metabolites that distinguish between oil palm seedlings resistant and susceptible to basal stem rot (BSR) caused by Ganoderma. Multivariate data analysis effectively differentiated resistant and susceptible palms, revealing that ascorbic acid, D-gluconic acid, D-fructose, and 2-oxoisovalerate could serve as potential biomarkers for screening resistant palms. Pathway analysis indicated that both resistant and susceptible roots infected with Ganoderma exhibited similar metabolic pathways, particularly ascorbate and aldarate metabolism, as well as glycerolipid metabolism, which play crucial roles in oil palm defense mechanisms against Ganoderma infection. Furthermore, these findings enhance our understanding of disease resistance mechanisms and can facilitate the breeding of new oil palm varieties with improved resistance to Ganoderma.
Methods
Sample collection
The oil palm root tissues used in this study were collected from an oil palm nursery at the Indonesian Oil Palm Research Institute (IOPRI) in Marihat, Simalungun Regency, North Sumatra, Indonesia. Three categories of samples were used in this study: resistant, susceptible, and control roots. Six-month-old oil palm seedlings, both resistant and susceptible that had been previously inoculated with Ganoderma were used. Seedling infection was conducted using the rubber wood block (RWB) method, with inoculum prepared by growing isolates on blocks of sterilized rubber wood. These blocks were then placed beneath germinating seedlings growing in soil-filled polybags (sitting germinated seed technique) (Fig. 6). The resistant seedlings, or healthy plants, were identified based on the absence of mycelium on the roots or necrosis on the basal stems and leaves. Susceptible seedlings were identified by the presence of mycelium on the roots, necrosis on more than 25% of the basal stem, and the appearance of Ganoderma fruiting bodies. Additionally, six-month-old control seedlings that had not been treated with Ganoderma infection were included. Each treatment had five individuals as biological replicates, totaling 15 oil palm seedlings tested using ¹H NMR.
Metabolite extraction
The roots of oil palm seedlings were thoroughly washed and then oven-dried at 60 °C for 5 days or until a constant weight was achieved. One gram of the dried root sample was ground using a mortar and liquid nitrogen, then dissolved in 30 mL of methanol16. The mixture was subjected to maceration on a shaker at 115 rpm overnight. Ultrasonic extraction was subsequently carried out at 65 °C for 30 min, followed by centrifugation at 3500 rpm for 20 min. The supernatant was collected and evaporated with a rotary evaporator to yield the crude extract.
1H NMR profiling
This research employed an untargeted metabolomics approach based on ¹H NMR17. A total of 50 ± 0.1 mg of the crude extract was accurately weighed, and 1.0 mL of methanol-d4 (Merck-Sigma, USA) containing 0.001% trimethylsilyl propionic acid-d4 sodium salt (TMSP) was added as an internal standard. The mixture was vortexed for 60 s, sonicated for 20 min, and then centrifuged at 10,000 rpm for 5 min. The supernatant was placed into an NMR tube for ¹H NMR profiling. The analysis was conducted using a JEOL JNM-ECZR NMR spectrometer (Tokyo, Japan) operating at a proton frequency of 500 MHz, with water signal suppression applied at a chemical shift of 4.89 ppm.
Preprocessing data
The NMR spectra were processed using Mestrenova software version 14.3, which included Fourier transformation, phase correction, and baseline adjustment to ensure data accuracy18. The spectra were calibrated using the TMSP-2,2,3,3-d4 signal at δ 0.0 ppm as the reference standard. Metabolite identification and quantification from the ¹H NMR spectra were performed using the ASICS package in the R programming language, which enables automated spectral deconvolution for precise metabolite profiling19.
Multivariate statistical analysis
The normalized NMR data matrix was analyzed using MetaboAnalyst 5.0 20, with Pareto scaling applied prior to statistical and multivariate analyses. The analyses included principal component analysis (PCA), partial least squares discriminant analysis (PLS-DA), and orthogonal projections to latent structures discriminant analysis (OPLS-DA) to classify and differentiate samples based on multidimensional data. Additionally, heatmap visualization was used to highlight metabolite variations across different sample groups. Variable importance in projection (VIP) scores and receiver operating characteristic (ROC) curves were employed to identify potential biomarkers associated with Ganoderma resistance. Pathway analysis, incorporating pathway enrichment techniques, was performed to identify critical pathways linked to resistance mechanisms. Significant metabolic pathways were determined based on the p-value (< 0.05) and impact value score (> 0.1)13,21. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used as a metabolic pathway identification reference.
Data availability
The datasets generated and analyzed during this study are available from the corresponding author upon reasonable request.
References
Sapey, E., Adusei-Fosu, K., Agyei-Dwarko, D. & Okyere-Boateng, G. Collection of oil palm (Elaeis guineensis Jacq.) germplasm in the Northern regions of Ghana. Asian J. Agric. Sci. 4, 325–328 (2012).
Corley, R. H. V. & Tinker, P. B. The Oil Palm (Wiley-Blackwell, 2016).
Tandra, H. & Suroso, A. I. The determinant, efficiency, and potential of Indonesian palm oil downstream export to the global market. Cogent Econ. Financ. 11, 2189671 (2023).
Khoo, Y. W. & Chong, K. P. Ganoderma boninense: general characteristics of pathogenicity and methods of control. Front. Plant. Sci. 14, 1156869 (2023).
Merciere, M. et al. About Ganoderma boninense in oil palm plantations of Sumatra and Peninsular Malaysia: ancient population expansion, extensive gene flow and large-scale dispersion ability. Fungal Biol. 121, 529–540 (2017).
Castillo, S. Y. et al. Ganoderma zonatum is the causal agent of basal stem rot in oil palm in Colombia. J. Fungi. 8, 230 (2022).
Kamu, A., Khim, P. C., Abu, S. I., Gabda, D. & Chong, M. Ho. Estimating the yield loss of oil palm due to Ganoderma basal stem rot disease by using bayesian model averaging. J. Oil Palm. Res. (2020).
Rupaedah, B. et al. Evaluation of microbial biocontrol agents for Ganoderma boninense management in oil palm nurseries. J. Saudi Soc. Agric. Sci. 23, 236–244 (2024).
Tuzun, S. The relationship between pathogen-induced systemic resistance (ISR) and Multigenic (horizontal) resistance in plants. Eur. J. Plant. Pathol. 107, 85–93 (2001).
Jost, M. et al. Plant and pathogen genomics: essential approaches for stem rust resistance gene stacks in wheat. Front. Plant. Sci. 14, 1223504 (2023).
Nugroho, R. A. P. et al. Metabolomics-assisted breeding in oil palm: potential and current perspectives. Int. J. Mol. Sci. 25, 9833 (2024).
Johnson, C. H., Ivanisevic, J. & Siuzdak, G. Metabolomics: beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell. Biol. 17, 451–459 (2016).
Pancoro, A., Karima, E., Apriyanto, A. & Effendi, Y. ¹H NMR metabolomics analysis of oil palm stem tissue infected by Ganoderma boninense based on field severity indices. Sci. Rep. 12, 21087 (2022).
Rahmadi, H. Y. et al. ¹H NMR analysis of metabolites from leaf tissue of resistant and susceptible oil palm breeding materials against Ganoderma boninense. Metabolomics 20, 89 (2024).
Emwas, A. H. et al. NMR spectroscopy for metabolomics research. Metabolites 9, 123 (2019).
Hernández Bort, J. A. et al. Reduced quenching and extraction time for mammalian cells using filtration and syringe extraction. J. Biotechnol. 182–183, 97–103 (2014).
Zhang, H. et al. ¹H NMR metabolic profiling of gastric cancer patients with lymph node metastasis. Metabolomics 14, 47 (2018).
Zanatta, A. C., Vilegas, W. & Edrada-Ebel, R. UHPLC-(ESI)-HRMS and NMR-based metabolomics approach to assess the seasonality of Byrsonima intermedia and Serjania marginata from Brazilian Cerrado flora diversity. Front. Chem. 9, 710025 (2021).
Lefort, G. et al. ASICS: an R package for a whole analysis workflow of 1D ¹H NMR spectra. Bioinformatics 35, 4356–4363 (2019).
Chong, J. et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46, W486–W494 (2018).
Manjunath, J. et al. Plasma metabolomic profiling reveals a novel Circulating biomarker signature in chronic pruritus of unknown origin. Sci. Rep. 14, 17472 (2024).
Wang, L. & Huang, Y. Evaluating classification performance of biomarkers in two-phase case‐control studies. Stat. Med. 38, 100–114 (2019).
Noutoshi, Y., Ikeda, M., Saito, T., Osada, H. & Shirasu, K. Sulfonamides identified as plant immune-priming compounds in high-throughput chemical screening increase disease resistance in Arabidopsis thaliana. Front. Plant. Sci. 3, 245 (2012).
Vlot, A. C., Dempsey, D. A. & Klessig, D. F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206 (2009).
Petruccelli, V., Brasili, E., Varone, L., Valletta, A. & Pasqua, G. Antifungal activity of dimethyl sulfoxide against Botrytis cinerea and phytotoxicity on tomato and lettuce plants. Plant. Biosyst. 154, 455–462 (2020).
Bhattacharya, A., Sood, P. & Citovsky, V. The roles of plant phenolics in defence and communication during Agrobacterium and Rhizobium infection. Mol. Plant. Pathol. 11, 705–719 (2010).
Marín-Valencia, I. et al. Biochemical diagnosis of dopaminergic disturbances in paediatric patients: analysis of cerebrospinal fluid homovanillic acid and other biogenic amines. Clin. Biochem. 41, 1306–1315 (2008).
Kulma, A. & Szopa, J. Catecholamines are active compounds in plants. Plant. Sci. 172, 433–440 (2007).
Retig, N. & Chet, I. Catechol-induced resistance of tomato plants to Fusarium wilt. Physiol. Plant. Pathol. 4, 469–475 (1974).
Zhu, F. et al. Join the green team: inducers of plant immunity in the plant disease sustainable control toolbox. J. Adv. Res. 57, 15–42 (2024).
Jian, Y. et al. How plants manage pathogen infection. EMBO Rep. 25, 31–44 (2023).
Anjali et al. Role of plant secondary metabolites in defence and transcriptional regulation in response to biotic stress. Plant. Stress. 8, 100154 (2023).
Formela-Luboińska, M. et al. The role of sugars in the regulation of the level of endogenous signaling molecules during defense response of yellow lupine to Fusarium oxysporum. Int. J. Mol. Sci. 21, 4133 (2020).
Zhang, Z. et al. Cultivation of Rhodosporidium paludigenum in gluconic acid enhances effectiveness against Penicillium digitatum in citrus fruit. Postharvest Biol. Technol. 172, 111374 (2021).
Noor, A. & Little, C. R. Evaluating the role of exogenously applied ascorbic acid in rescuing soybean plant health in the presence of pathogen-induced oxidative stress. Pathogens 11, 1117 (2022).
Hönig, M., Roeber, V. M., Schmülling, T. & Cortleven, A. Chemical priming of plant defense responses to pathogen attacks. Front. Plant. Sci. 14, 1146577 (2023).
Yan, F. et al. Lactic acid induced defense responses in tobacco against Phytophthora nicotianae. Sci. Rep. 14, 9338 (2024).
Boubakri, H. The role of ascorbic acid in plant–pathogen interactions. in Ascorbic Acid in Plant Growth, Development and Stress Tolerance (eds Hossain, M. A. et al.) 255–271 (Springer, Cham, (2017).
Kaur, R., Macleod, J., Foley, W. & Nayudu, M. Gluconic acid: an antifungal agent produced by Pseudomonas species in biological control of take-all. Phytochemistry 67, 595–604 (2006).
De Werra, P., Péchy-Tarr, M., Keel, C. & Maurhofer, M. Role of gluconic acid production in the regulation of biocontrol traits of Pseudomonas fluorescens CHA0. Appl. Environ. Microbiol. 75, 4162–4174 (2009).
Jeandet, P., Formela-Luboińska, M., Labudda, M. & Morkunas, I. The role of sugars in plant responses to stress and their regulatory function during development. Int. J. Mol. Sci. 23, 5161 (2022).
Trovato, M. et al. Editorial: amino acids in plants: regulation and functions in development and stress defense. Front. Plant. Sci. 12, 772810 (2021).
Tunsagool, P. et al. Metabolomic study of stress responses leading to plant resistance in Mandarin fruit mediated by preventive applications of Bacillus subtilis Cyclic lipopeptides. Postharvest Biol. Technol. 156, 110946 (2019).
Czolpinska, M. & Rurek, M. Plant glycine-rich proteins in stress response: an emerging, still prospective story. Front. Plant. Sci. 9, 302 (2018).
Fick, A., Swart, V. & Van Den Berg, N. The ups and downs of plant NLR expression during pathogen infection. Front. Plant. Sci. 13, 921148 (2022).
Smirnoff, N. Ascorbic acid metabolism and functions: A comparison of plants and mammals. Free Radic Biol. Med. 122, 116–129 (2018).
Botanga, C. J. et al. Metabolite profiling of Arabidopsis inoculated with Alternaria brassicicola reveals that ascorbate reduces disease severity. Mol. Plant-Microbe Interact. 25, 1628–1638 (2012).
Seth, T., Asija, S., Umar, S. & Gupta, R. The intricate role of lipids in orchestrating plant defense responses. Plant. Sci. 338, 111904 (2024).
Chanda, B. et al. Glycerol-3-phosphate levels are associated with basal resistance to the hemibiotrophic fungus Colletotrichum Higginsianum in Arabidopsis. Plant. Physiol. 147, 2017–2029 (2008).
Kuźniak, E. & Gajewska, E. Lipids and lipid-mediated signaling in plant–pathogen interactions. Int. J. Mol. Sci. 25, 7255 (2024).
Yu, L., Zhou, C., Fan, J., Shanklin, J. & Xu, C. Mechanisms and functions of membrane lipid remodeling in plants. Plant. J. 107, 37–53 (2021).
Author information
Authors and Affiliations
Contributions
S.N. Contributed to research design, conceptualization, experiment execution, and manuscript writing. H.Y.R. Contributed to research design and experiment execution. A.N.S. Contributed to manuscript writing. A.R.P. Contributed to conceptualization and supervision. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics declarations
We confirm that all methods in this study were conducted in accordance with relevant guidelines and legislation for plant research. This research does not involve any studies with human or animal participants conducted by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nugroho, S., Rahmadi, H.Y., Simamora, A.N. et al. 1H NMR metabolomic profiling of resistant and susceptible oil palm root tissues in response to Ganoderma boninense at the nursery stage. Sci Rep 15, 16784 (2025). https://doi.org/10.1038/s41598-025-01691-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-01691-y