Abstract
Bioinvasions represent a major environmental issue, particularly when they take place in biodiversity hotspots, such as mangrove ecosystems that serve as shelter for many marine species and fisheries resources. In this work, we used an integrative approach based on DNA and morphological analyses to identify individuals and the putative presence of cryptic diversity in the invasive tiger prawn (Penaeus monodon) along a mangrove area on the northern coast of Brazil, referred to as “Blue Amazon”. A fragment of the mitochondrial cytochrome c oxidase subunit I (COI) gene was selected for DNA Barcode and associated with distance-based (ABGD—Automatic Barcode Gap Discovery) and probabilistic (GMYC—Generalized Mixed Yule Coalescent and bPTP—Bayesian Poisson tree processes) species delimitation methods. Furthermore, the maternal origin of collected specimens was tracked. The molecular analyses recovered two genetically divergent lineages (7.7%) within the tiger prawn, indicating the occurrence of two species of this bioinvader on the northern coast of Brazil. Even though no differences in external morphology were detected, both lineages could be differentiated by their internal structures. The molecular traceability of the origin of samples showed that lineages I and II shared haplotypes with specimens from 11 and nine countries, respectively, including a shrimp breeding center in Vietnam. This is the first record of two species of tiger prawn along the Brazilian continental shelf. These findings are useful to the development of effective management policies in a region of particular biological relevance.
Similar content being viewed by others
Introduction
Biological invasions or bioinvasions represent a major threat to local biodiversity, considered one of the main causes of population decline in native species1. The tiger prawn (Penaeus monodon, Fabricius, 1798) is a native crustacean from Indo-Pacific region2 that has invaded numerous countries3,4,5,6, including records of established invasive populations in some regions4,7. In general, these invasions are related to improper management in aquaculture systems that allow cultivated specimens escaping into natural aquatic habitats8. Accordingly, the tiger prawn is an important fisheries resource that has been intensively cultivated in many countries, particularly India, Indonesia, Australia, Philippines, Taiwan, Thailand, Vietnam, Sri Lanka, Malaysia, Mozambique, South Africa, Saudi Arabia and Singapore2.
In Brazil, the first report of bioinvasion by P. monodon dates to 1987 in a coastal site of Maranhão, northeastern Brazil9. Ever since, this species has been recorded in several localities along northern and northeastern Brazil, such as the continental shelf of the state of Pará10, at the mouth of the Oiapoque River in the state of Amapá11, and the coastal zones of the states of Maranhão12 and Rio Grande do Norte13. Species invasion can lead to both ecological14 and economic15 losses, including the dissemination of diseases and competition with native species16. In fact, the tiger prawn is recognized as a potential vector of several pathogens17,18. Furthermore, the high fecundity rate19, the aggressive behavior and the non-specialist carnivorous habits of this invasive species would increase the harmful impacts over native crustaceans.
The tiger prawn belongs to the family Penaeidae, which comprises of 27 genera, 226 species and 13 subspecies20. Nonetheless, the taxonomy of penaeids remains poorly understood, with cases of species complexes of species complexes and evidence of cryptic diversity21,22. In this context, DNA-based tools represent an efficient approach to providing a precise identification of crustacean species, such as the sequencing of the first portion of the cytochrome c oxidase subunit I (COI) gene, referred to as DNA Barcode23,24.
The DNA Barcode based on COI allows comparative analyses with public databases such as the BOLD Systems, assuming a minimum threshold of 2% in the genetic divergence to discriminate species25. In addition, these data can be analyzed by distinct algorithms of molecular species delimitation based on distance or coalescence methods26,27,28 to identify accurately distinct lineages of many taxonomic groups, including crustaceans22,29.
Furthermore, when possible, integrative taxonomic methods are highly recommended to be performed by associating, for instance, molecular and traditional morphological data. This approach should provide reliable inferences about the species delimitation, including the unambiguous identification of cryptic diversity and a detailed documentation of biodiversity.
The correct discrimination of species is essential to provide a deep knowledge of regional biodiversity and invasive species, thus being useful to effective management plans focused on the conservation of ecosystems and native biota. Therefore, the goal of this study was to validate the occurrence of the giant tiger prawn in coastal and mangrove areas of the eastern “Blue Amazon” using DNA Barcode tool. This area is of relevant interest because it has a high biodiversity, in addition to presenting pressure from human actions on habitats and species that are threatened with extinction in almost all of their extension30, and the invasion of tiger shrimp has been frequently reported in this region9,10,12,13. Following an integrative approach, we tested the putative occurrence of cryptic diversity in the tiger prawn and the traceability of the maternal origin of invasive specimens along the northern coast of Brazil.
Material and methods
Ethics declarations
The specimens were collected dead from artisanal fishermen who practice subsistence fishing, thus dispensing with the approval of ethics in the use of animals, according to Law n°. 11,794/2008.
Sampling
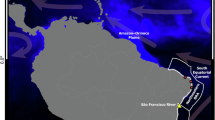
The collection of samples was carried out in distinct sites from the largest continuous mangrove ecosystem in the world along the “Blue Amazon”, northern Brazil. Most of them were located within areas of environmental protection, including the extractive reserves (RESEX) of Araí-Peroba, Gurupi-Piriá, Caeté-Taperaçú, and Canelas Island (Fig. 1).
In the RESEX of Caeté-Taperaçú, the specimens were collected by using fixed traps (fishing corral), while the samples from the remaining localities were obtained by trawl nets (20 mm of mesh size). In all collection expeditions, we were assisted by artisanal fishermen from the local communities. The number of samples per locality and their respective identification codes are described in Table 1.
After collection and preliminary in-field identification, the specimens were transported to the Laboratory of Applied Genetics (LAGA) at the Institute of Coastal Studies (IECOS), UFPA, Bragança—PA. Samples of muscle tissue were obtained from the specimens identified as P. monodon based on the specialized literature31 and stored in 90% ethanol at − 20 °C. Voucher specimens were fixed in 10% formaldehyde, washed and then transferred to 70% ethanol32 to be deposited in the Fisheries Zoological Collection from LAGA.
DNA sequencing and alignment of COI sequences
The total DNA was isolated using the Wizard Genomic DNA (Promega) kit according to the manufacturer’s instructions. Afterwards, the DNA samples were quantified in a NanoDrop™ 2000/2000c spectrophotometer (Thermo Fisher).
The COI fragment was amplified via polymerase chain reaction (PCR) using the primers LCO-1490 and HCO-219833. The PCR conditions were optimized as follows: 2.4 μL of dNTPs (1.25 mM), 1.4 μL of buffer (10x), 0.5 μL of MgCl2 (50 mM), 0.6 μL of each primer (50 ng/μL), 0.1 μL of Taq DNA polymerase (U/ μL), 15 to 30 ng of template DNA and ultrapure water to a final volume of 15 µL. The PCR steps encompassed a first denaturation at 95 °C for 3 min, followed by 40 cycles of denaturation at 95 °C for 45 s, annealing at 45 °C for 45 s and extension at 72 °C for 45 s, plus a final extension at 72 °C for 5 min.
The amplicons were purified in polyethylene glycol (PEG 8000) as reported by34 and then sequenced by the dideoxy method35 using the Big Dye 3.1 (ABI Prism TM Dye Terminator Cycle Sequencing Ready Reaction, Thermo Fisher) kit, according to the manufacturer’s instructions. Then, the precipitated products were sequenced in an ABI 3500 XL automatic sequencer (Thermo Fisher).
The electropherograms were individually inspected using the software BioEdit v. 7.2.536 to evaluate sequence quality and correction when necessary. Subsequently, the sequences were automatically aligned using the ClustalW tool37,38 available in BioEdit.
Molecular identification and species delimitation analyses
The haplotypes in the sequence dataset were identified using the software DNAsp v639. After the first screening, a final dataset was generated based on a single representative from each haplotype to provide a refined search. Each haplotype was compared separately to the following public databases: GenBank (National Center for Biotechnology Information—http://www.ncbi.nlm.nih.gov) using BLAST (Basic Local Alignment Search Tool) in the Nucleotide BLAST (BLAST-N) window, and BOLD (Barcoding of Life Database—http://www.barcodinglife.org ) system40.
Besides the similarity analyses, a neighbor-joining (NJ) tree was built assuming the Kimura-2 parameter (K2P) model41 and 1,000 pseudoreplicates of bootstrap42 in the software MEGA v. X43. The NJ tree was composed of the sequences from the present study as well as 16 public COI sequences available in BOLD systems, represented by two specimens per species from eight genera of Penaeidae.
The species delimitation tests were performed assuming the HKY + G evolutionary model as recommended based on the Bayesian Information Criterion (BIC) by the jModelTest2 available in ACCESS 2.1.644,45, using the CIPRES Science Gateway v3.3 platform46. A Bayesian-inference (BI) tree was generated in the software BEAST v. 1.8.447,48, following a strict clock and Yule speciation prior as defaults. The support values of posterior probabilities were determined based on 100 million generations and a burn-in of 10%. The log files were checked in Tracer v. 1.5, to evaluate the adequacy of burn-in and the chain convergence, assuming as appropriate only those chains with ESS (effective sample size) values above 200. The tree generated in BEAST was summarized in TreeAnnotator, v. 1.10.4.47,48, visualized in FigTree v.1.4.449 and edited in the software Inkscape v. 1.3.2. The edition of the BI tree was based on the putative groups indicated by the delimitation species tests.
Three species delimitation algorithms were used being based on single-locus (COI) trees and genetic distances, as follows: (i) ASAP (Analysis of Species by Automatic Partitioning) using the web ASAP version available at https://bioinfo.mnhn.fr /abi/public/asap/asapweb.html50 based on .FASTA database and assuming the Kimura (K80) TS/TV evolutionary model; (ii) GMYC (Generalized Mixed Yule-Coalescent) using the Splits package available in the (software R v. 3.251) and a summarized ultrametric tree generated by BEAST as input; c) PTP (Poisson Tree Process) carried out on the PTP website (https://species.h-its.org/ptp/;26) following the standard defaults available using a maximum likelihood tree as input in the software IQ-TREE v. 2.1.3. and assuming the GTR + F model and 1000 replications52.
The genetic distance values within and among groups were calculated based on the K2P models41. The verification of polymorphic sites and putative stop codons was performed in the software MEGA v. X43.
Traceability of the maternal origin in tiger prawn samples from the northern coast of Brazil
To trace back the maternal origin of the haplotypes from the present study, we downloaded from BOLD systems the 100 more similar sequences to each haplotype. Therefore, a dataset composed of 517 COI sequences was assembled and inspected using the software DNAsp to verify the shared haplotypes, while the screening of these sequences was performed using WordPad. Some sequences were removed after this step according to the following criteria: (I) repeated sequences, (II) presence of degenerated bases, (III) fragments comprising less the 590 bp, (IV) sequences lacking the origin of samples, and (V) sequences obtained in samples from the same country. A BI tree was built following the abovementioned methodology. The best evolutionary model was HKY + G, while the posterior probabilities were estimated from 100 million generations, assuming a burn-in of 10% and ESS values higher than 200.
Morphological and morphometric identification
The morphological analyses were carried out in the Laboratory of Carcinology at the Institute of Coastal Studies (IECOS), UFPA. The samples were dissected using a scalpel and ultrafine needles (BD Ultra-Fine®, 12.7 × 0.33 mm) onto glass slides, under a binocular stereoscopic microscope (model Q7740SZ). The samples analyzed were the specimens PmoAj07 and PmoLB11, which represent, respectively, lineages I and II. The morphological characteristics measured were the pair of mandibles and the maxilla of each specimen, fundamental structures for the correct identification and discrimination of shrimp. These measurements were performed by a specialist in morphology, with experience in the field of carcinology.
Results
Identification of public databases and species delimitation tests
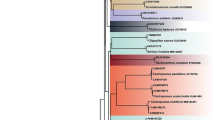
The 17 COI sequences generated in this study encompassed 591 bp, 42 polymorphic sites and five haplotypes, being invariably similar (> 99%) to those referred to as P. monodon. However, these haplotypes were clearly divided into lineage I, comprising three haplotypes (H1, H2, and H3), and lineage II, formed by two haplotypes (H4 and H5). The most frequent haplotypes in the final dataset were H2 (64.7%) and H4 (17.64%) (Table 2). The genetic distance between and within these groups was equal to 7.7% and 0%, respectively. The topology of the NJ tree separated both lineages of the tiger prawn in two highly supported reciprocal monophyletic clusters (Fig. 2), as usually observed for distinct species.
The species delimitation methods recovered a different number of candidate taxa. While the GMYC separated the samples into three clusters, both ASAP and PTP indicated the presence of two species (Fig. 3).
Bayesian inference (BI) tree evidencing the presence of two giant river prawn lineages along the northern coast of Brazil (the number on branches refer to the posterior probabilities). The bars on the right indicate the species delimitation results from each tested algorithm. (PmoAj-Penaeus monodon Ajuruteua; PmoLB Penaeus monodon Lombo Branco; PmoIc-Penaeus monodon Canelas Island; PmoAr- Penaeus monodon Arai; GMYC- Generalized mixed Yule–coalescent; ASAP- Poisson tree processes Assemble; PTP-Species by automatic partitioning. This art was created in the Inkscape software.
Maternal origin of the tiger prawn samples from northern coast of Brazil
The dataset used in the traceability of the maternal origin of collected samples was composed of 35 sequences standardized to 590-bp fragments. Both lineages of the tiger prawn shared haplotypes with specimens from, at least, 15 countries (Australia, Vietnam, Thailand, Philippines, Egypt, Germany, United States of America, China, Indonesia, India, Taiwan, Bangladesh, South Africa, Mozambique, and Brazil). The lineage I is widespread across 11 countries while the lineage II is found in nine countries, including cases of sympatry (Brazil, Germany, Egypt, India, and China) (Fig. 4).
Bayesian inference (BI) tree based on COI sequences of Penaeus monodon indicating the maternal origin of both lineages in the invasive tiger prawn from Brazil and other localities. The map was prepared by Charles Ferreira using the free access QGIS software. The images of the specimens are merely illustrative of the fact that there are two distinct lineages and do not correspond to the actual coloration. This art was created in the Inkscape program.
Morphological analyses
Description of samples from lineage I
Mandibles: asymmetrical with a distinct gap between the incisive and molar processes. Left mandible presents incisive process with four slightly prominent teeth. Molar process with protuberances, being more robust than the incisive. Right mandible presents incisive process with tree few prominent teeth. Molar process with protuberances, being more robust than the incisive (Fig. 5, @1A). Maxilla: Scaphognatite with numerous marginal plumose setae; distal and proximal lobes of the basal and coxal endites with numerous distal plumose setae, with the proximal lobe of the coxal endite being smaller with a few distal setae (Fig. 5 1B). Non-segmented endopod with two distal spines (Fig. 5 1C).
Internal structures of lineages I (1A-1C) and II (2A-2C) of P. monodon from the northern coast of Brazil. (1A- mandible pair, 1B- maxilla, 1C- non-segmented endopod with two distal spines; 2A- mandible pair, 2B- maxilla, 2C- non-segmented endopod with five distal spines). This art was created in the Inkscape program.
Description of samples from lineage II
Mandibles: Palpi present. Asymmetrical mandibles with a distinct gap between the incisor and molar processes. Left mandible presents incisive process with four well-developed teeth. Molar process with protuberances, being more robust than the incisive. Right mandible presents incisive process with two wide teeth. Molar process with protuberances, being more robust than the incisive (Fig. 5 2A). Maxilla: Scaphognatite with numerous marginal plumose setae; distal and proximal lobes of the basal and coxal endites with numerous distal plumose setae, with the proximal lobe of the coxal endite being smaller with a few distal setae (Fig. 5 2B). Non-segmented endopod with five distal spines (Fig. 5 2C).
Discussion
Cryptic diversity in the invasive tiger prawn P. monodon along the largest mangrove area worldwide
Two well-defined, reciprocally monophyletic and highly divergent lineages (genetic distance of 7.7% in COI) of the tiger prawn P. monodon were identified along protected areas of the “Blue Amazon” in northern Brazil. In fact, previous reports have already suggested the P. monodon should encompass cryptic species. For example,53 indicated the presence of two lineages of the tiger prawn based on analyzes of COI data in samples collected in Southeastern Pacific, Southwestern Atlantic and Southeastern Indian oceans. Recently,22 recovered three Molecular Operational Taxonomic Units (MOTUs) in P. monodon after screening 1680 COI sequences available in the public database from BOLD systems. Additionally, these authors established a minimum value of 1.8% in genetic distances as a threshold to distinguish between intra and interspecific variation within the tribe Peneaini. Therefore, samples exhibiting values above this threshold would refer to unique MOTUs22. However, none of these reports have reported sympatric lineages or have analyzed the invasive populations of P. monodon in Brazil.
The species delimitation tests provided contrasting results, since both ASAP and PTP methods identified two candidate species while the GMYC indicated the existence of three clusters. Even though the GMYC and PTP algorithms are based on coalescent approaches54, the latter usually recovers smaller numbers of putative groups than the GMYC53. Accordingly, some reports have demonstrated that GMYC tends to determine an overestimated division of lineages55,56. Therefore, we consider that the presence of two cryptic species in the giant river prawn from northern Brazilian coast would be more likely, as suggested by ASAP and PTP methods and corroborated by the phylogenetic inferences and values of genetic distance as well.
In addition to the genetic differences identified, the lineages exhibit clear differences in their internal morphologies, in the structures of the pair of mandibles and maxillae. This was the first study to report morphological differences between the lineages, which is very important, as it reinforces the idea that they may represent distinct species beyond genetic evidence22.
In fact, molecular species delimitation methods are highly effective in detecting hidden diversity of several groups22,55,56. In the case of Penaeidae, most of their diversity remains largely overlooked and associated with evidence of cryptic forms, thus hindering a reliable species discrimination53. On the other hand, the DNA barcode has proved to be a reliable tool for the identification of cryptic species, as recently reported in taxa of Penaeini22, one of the largest tribes in Penaeidae that includes the giant river prawn57. In addition,22proposed that genetic distance values equal or above 1.8% would be compatible with the presence of distinct molecular operational taxonomic units or MOTUs in this group of crustaceans.
The two invasive lineages/species of tiger prawn identified in this study are sympatrically widespread throughout the largest continuous mangrove zone in the world58. Moreover, a recent study in the same area also recorded the bioinvasion by the rainbow shrimp Mierspenaeopsis sculptilis, a native species from the Indo-Pacific region59. Considering the importance of mangroves as nursery habitats for the marine fauna60, the presence of bioinvaders in these ecosystems are potentially harmful, as already reported in mangroves of Bengal, India61.
As a matter of fact, the mangrove zones along the Amazon coast have been recognized as priority areas for conservation by the Brazilian legislation that defined six Extractive Marine Reserves (RESEX) along this region. Two of them (RESEX Filhos do Mangue and RESEX Viriandeua were implemented recently62,63. Thus, genetic information about the diversity of bioinvaders is useful to design proper monitoring policies for the management and conservation of these ecosystems.
Maternal origin of invasive populations in the tiger prawn from northern coast of Brazil and the effects of aquaculture activities
Both lineages or species of the tiger prawn identified along the northern coast of Brazil are also present in, at least 16 countries, revealing multiple origins and invasion routes. A similar finding was reported for alien populations of P. monodon in Colombia64. As a matter of fact, the tiger prawn is one of the main crustaceans used in aquaculture, ranking the fourth most cultivated species worldwide in 2020. Furthermore, crustaceans represent the second most produced resource in aquaculture, particularly in Asia, followed by Americas, Europe, Oceania and Africa2. Following this trend, Brazil has imported two major stocks of P. monodon for local shrimp farm from the Philippines, in the late 1970s65, and Taiwan, in 198466. It should be highlighted that the Philippines remained as the largest producer of giant river prawn in the 1990s67.
Accordingly, the identification of shared haplotypes between the invasive Brazilian populations and specimens from the Philippines and Taiwan suggests that the imported stocks for shrimp farming activities contributed to the bioinvasion of P. monodon in natural Brazilian habitats, as previously suggested by other authors9,68. Furthermore, one of the haplotypes observed in lineage I from this study has already been recovered in a shrimp breeding center from Vietnam69, reinforcing the present hypothesis of invasion routes.
This is the first report about the presence of two invasive lineages of the tiger prawn along the “Blue Amazon” in Brazil. We provided robust evidence that these lineages refer to two species of P. monodon distributed along the northern coast of Brazil. We also showed that both species have multiple mitochondrial origins and have already spread to 19 countries. This information is essential to design proper management and conservation policies of these areas. Based on the molecular data, we confirmed that aquaculture activities, particularly the importation of specimens for shrimp farming, have been the main vector of bioinvasion.
Data availability
The sequences obtained were deposited in GenBank, with the following access codes: lineage_I: PQ433754-PQ433766 and lineage_II: PQ433767-PQ433770.
References
Ehrenfeld, J. G. Ecosystem consequences of biological invasions. Annu. Rev. Ecol. Evol. Syst. 41, 59–80. https://doi.org/10.1146/annurev-ecolsys-102209-144650 (2010).
FAO. Penaeus monodon. Cultured Aquatic Species Information Programme. In: Fisheries and Aquaculture. Roma. Updated on 07/29/2005. https://www.fao.org/fishery/en/culturedspecies/penaeus_monodon/en. Acessed 21 May 2024. (2024)
Wakida-Kusunoki, A. T., De Anda-Fuentes, D. & López-Téllez, N. A. Presence of giant tiger shrimp Penaeus monodon (Fabricius, 1798) in eastern Peninsula of Yucatan coast, Mexico. Latin Am. J. Aquat. Res. 44(1), 155–158. https://doi.org/10.3856/vol44-issue1-fulltext-16 (2016).
Aguirre-Pabón, J. C., Orozco Berdugo, G. & Narváez Barandica, J. C. Genetic status, source and establishment risk of the giant tiger shrimp (Penaeidae: Penaeus monodon), an invasive species in Colombian Caribbean waters. Acta Biol. Colomb. 20(1), 117–127. https://doi.org/10.15446/abc.v20n1.41946 (2015).
Alfaro-Montoya, J., Monge-Ortiz, A. M., Martínez-Fernández, D. & Herrera-Quesada, E. First record of the nonindigenous Penaeus monodon Fabricius, 1798 (Penaeidae) in the Caribbean Sea of Costa Rica, Central America, with observations on selected aspects of its reproductive biology. Bioinvasions Rec. 4, 217–222 (2015).
Khafage, A. R., Taha, S. M. & Attallah, M. A. Presence of tiger shrimp Penaeus monodon Fabricius, 1798 (Penaeidae) in the Egyptian commercial shrimp catch, Alexandria, Egypt. Egypt. J. Aquat. Res. 45, 183–187. https://doi.org/10.1016/j.ejar.2019.05.002 (2019).
Hill, J. M., Caretti, O. N. & Heck, K. L. Jr. Recently established Asian tiger shrimp Penaeus monodon Fabricius, 1798 consume juvenile blue crabs Callinectes sapidus Rathbun, 1896 and polychaetes in a laboratory diet-choice experiment. Bioinvasions Rec. 6, 233–238. https://doi.org/10.3391/bir.2017.6.3.08 (2017).
May-kú, M. A., Rubio-Piña, J., Ek-Huchim, J. P. & Ardisson, P. L. Genetic identification of Penaeus monodon from incipient invasions in the Yucatán Peninsula, south-southeast Gulf of Mexico. Bull. Mar. Sci. 100, 373–382. https://doi.org/10.5343/bms.2023.0059 (2024).
Fausto-Filho, J. Registro da captura de Penaeus monodon Fabricius, no litoral do Estado do Maranhão, Brasil (Crustacea: Penaeidae). Arq. Ciên. Mar. 26, 81–82. https://doi.org/10.32360/acmar.v26i1-2.31444 (1987).
Cintra, I. H. A., Paiva, K. S., Botelho, M. N. & Silva, K. C. A. Presence of Penaeus monodon in the continental shelf of the State of Para, Northern Brazil (Crustacea, Decapoda, Penaeidae). Revista de Ciências Agrárias Amazon. J. Agric. Environ. Sci. 54, 314–317 (2011).
Vieira, I. M. et al. Novo registro do camarão-tigre gigante Penaeus monodon Fabricius, 1798 na foz do rio Oiapoque, Amazônia Oriental, Amapá. Brasil. Rev. Contemp. 4, 2510. https://doi.org/10.56083/RCV4N3-040 (2024).
Andrade, K. S. P., Filgueira, C. H. M. S. & Nunes, J. L. S. Extension of occurrence of the non-native shrimp Penaeus monodon Fabricius, 1798 (Decapoda: Penaeidae) in Brazilian Amazon coast. Bol. Lab. Hidrobiol. 31, 1–4. https://doi.org/10.18764/1981-6421e2021.7 (2021).
Souza, E. M. Jr., Garcia, J. Jr., Araújo, P. V. N., Alencar, C. E. R. D. & Freire, F. A. M. Second report of the occurrence of giant tiger prawn, Penaeus monodon Fabricius, 1798 (Crustacea: decapoda), in Rio Grande do Norte state. Northeast Brazil. Arq. Ciên. Mar. 48, 116–120 (2015).
Yadav, S. P. S. et al. Invasive alien plant species (Banmara): Investigating its invasive potential, ecological consequences on biodiversity, and management strategies. J. Agr. Food Res. 15, 101031. https://doi.org/10.1016/j.jafr.2024.10103 (2024).
Cuthbert, R. N. et al. Economic impact disharmony in global biological invasions. Sci. Total Environ. 913, 169622. https://doi.org/10.1016/j.scitotenv.2023.169622 (2024).
Crowl, T. A., Crist, T. O., Parmenter, R. R., Belovsky, G. & Lugo, A. E. The spread of invasive species and infectious disease as drivers of ecosystem change. Front. Ecol. Environ. 6, 238–246. https://doi.org/10.1890/070151 (2008).
Jiravanichpaisal, P., Miyazaki, T. & Limsuwan, C. Histopathology, biochemistry, and pathogenicity of Vibrio harveyi infecting black tiger prawn Penaeus monodon. J. Aquat. Anim. Health. 6, 27–35. (1994).
Flegel, T. W. Major viral diseases of the black tiger prawn (Penaeus monodon) in Thailand. World J. Microb. Biot. 13, 433–442. https://doi.org/10.1023/A:1018580301578 (1997).
Uddin Sk, N., Ghosh, S., & Maity, J. Reproductive biology, maturation size and sex ratio of black tiger shrimp (Penaeus monodon Fabricius, 1798) from fishing grounds of Digha coast, West Bengal, India. Int. J. Aquat. Biol. 3, 372–378. https://doi.org/10.22034/ijab.v3i6.2 (2015).
ITIS from the Integrated Taxonomic Information System Retrieved Accessed 21 May 2024, https://doi.org/10.5066/F7KH0KBK
Hurzaid, A., Chan, T. Y., Nor, S. A. M., Muchlisin, Z. A. & Chen, W. J. Molecular phylogeny and diversity of penaeid shrimps (Crustacea: Decapoda) from South-East Asian waters. Zool. Scr. 49, 596–613. https://doi.org/10.1111/zsc.12428 (2020).
Farias, M. C. L., Coelho, P. F., Pontes, A. I. & Jacobina, U. P. DNA barcode reveals high cryptic diversity in the commercially important Penaeini shrimps (Decapoda, Penaeidae). Org. Divers. Evol. 23, 857–869. https://doi.org/10.1007/s13127-023-00616-9 (2023).
Hebert, P. D., Cywinska, A., Ball, S. L. & DeWaard, J. R. Biological identifications through DNA barcodes. Proc. Roy. Soc. London, Ser. B, Biol. Sci. 270, 313–321. https://doi.org/10.1098/rspb.2002.2218 (2003).
Ratnasingham, S. & Hebert, P. D. A DNA-based registry for all animal species: the Barcode Index Number (BIN) system. PLoS ONE 8, e66213. https://doi.org/10.1371/journal.pone.0066213 (2013).
Carvalho-Batista, A., Terossi, M., Zara, F. J., Mantelatto, F. L. & Costa, R. C. A multigene and morphological analysis expands the diversity of the seabod shrimp Xiphopenaeus Smith, 1869 (Decapoda: Penaeidae), with descriptions of two new species. Sci. Rep. 9, 15281. https://doi.org/10.1038/s41598-019-51484-3 (2019).
Zhang, J., Kapli, P., Pavlidis, P. & Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29, 2869–2876. https://doi.org/10.1093/bioinformatics/btt499 (2013).
Puillandre, N. et al. Species delimitation and exploration of species partitions with ASAP and LIMES. DNA Barcoding: Methods and Protocols, Springer, New York. https://doi.org/10.1007/978-1-0716-3581-0_20 (2024).
Pons, J. et al. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 55, 595–609. https://doi.org/10.1080/10635150600852011 (2006).
Hurzaid, A., Chan, T. Y., Mohd Nor, S. A., Muchlisin, Z. A. & Chen, W. J. Molecular phylogeny and diversity of penaeid shrimps (Crustacea: Decapoda) from South-East Asian waters. Zool. Scr. 49, 596–613. https://doi.org/10.1111/zsc.12428 (2020).
Magris, R. A. et al. A blueprint for securing Brazil’s marine biodiversity and supporting the achievement of global conservation goals. Divers. Distrib. 27, 198–215. https://doi.org/10.1111/ddi.13183 (2021).
Perez Farfante, I. P. & Kensley, B. F. Penaeoid and sergestoid shrimps and prawns of the world: Keys and diagnoses for the families and genera (Editions du Muséum, 1997).
Ueida, V. S. & Corrêa, R. M. Coleta e fixação de peixes de riachos. In Ecologia de Peixes de Riachos (eds Caramaschi, E. P. et al.) 1–22 (Série Oecologia Brasiliensis, PPGR-UFRJ, 1999).
Folmer, O., Black, M. B., Hoch, W., Lutz, R. A. & Vrijehock, R. C. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–9 (1994).
Paithankar, K. R. & Prasad, K. S. Precipitation of DNA by polyethylene glycol and ethanol. Nucleic. Acids. Res. 19, 1346. https://doi.org/10.1093/nar/19.6.1346 (1991).
Sanger, F., Nicklen, S. & Coulson, A. R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74, 5463–5467. https://doi.org/10.1073/pnas.74.12.5463 (1977).
Hall, T. A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic. Acids. Symp. Ser. 41, 95–8 (1999).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic. Acids. Res. 22, 4673–80. https://doi.org/10.1093/nar/22.22.4673 (1994).
Larkin, M. A. et al. Clustal W and clustal X version 20. Bioinformatics 23, 2947–8. https://doi.org/10.1093/bioinformatics/btm404 (2007).
Rozas, J. et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–302. https://doi.org/10.1093/molbev/msx248 (2017).
Ratnasingham, S. & Hebert, P. D. BOLD: The barcode of life data system. Mol. Ecol. Notes. 7, 355–364 (2007).
Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. https://doi.org/10.1007/BF01731581 (1980).
Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x (1985).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. M. Biol. Evol. 35, 1547–1549. https://doi.org/10.1093/molbev/msy096 (2018).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. Jmodeltest 2: More models, new heuristics and parallel computing. Nat. Methods 9, 772 (2012).
Guindon, S. & Gascuel, O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst. Biol. 52, 696–704. https://doi.org/10.1080/10635150390235520 (2003).
Miller, M. A., Pfeiffer, W. & Schwartz, T. Creating the CIPRES science gateway for inference of large phylogenetic trees gateway computing environments workshop (GCE). New Orleans https://doi.org/10.1109/GCE.2010.5676129 (2010).
Drummond, A. J. & Rambaut, A. Beast: bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. https://doi.org/10.1186/1471-2148-7-214 (2007).
Suchard, M. A. & Rambaut, A. Many-core algorithms for statistical phylogenetics. Bioinformatics 25, 1370–1376. https://doi.org/10.1093/bioinformatics/btp244 (2009).
Rambaut, A. Figtree: A graphical viewer of phylogenetic trees. Versão 1.4.4. Edinburgh, Uk: Institute of Evolutionary Biology, University of Edinburgh (2018).
Puillandre, N., Brouillet, S. & Achaz, G. ASAP: Assemble species by automatic partitioning. Mol. Ecol. Resour. 21, 609–620. https://doi.org/10.1111/1755-0998.13281 (2021).
R Core Team. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org (2012).
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A. & Minh, B. Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. M. Biol. Evol. 32, 268–274. https://doi.org/10.1093/molbev/msu300 (2015).
Ramirez, J. L. et al. DNA barcoding of penaeidae (Decapoda; Crustacea): Non-distance-based species delimitation of the most economically important shrimp family. Diversity 13, 460. https://doi.org/10.3390/d13100460 (2021).
Hubert, N. & Hanner, R. DNA barcoding, species delineation and taxonomy: a historical perspective. DNA Barcodes 3, 44–58. https://doi.org/10.1515/dna-2015-0006 (2015).
Correa, C. et al. Species delimitation in frogs from South American temperate forests: The case of Eupsophus, a taxonomically complex genus with high phenotypic variation. PLoS ONE 12, e0181026. https://doi.org/10.1371/journal.pone.0181026 (2017).
Da Silva, R. et al. Comparative analyzes of species delimitation methods with molecular data in snappers (Perciformes: Lutjaninae). DNA Mitocondrial Parte A 29, 1108–1114. https://doi.org/10.1080/24701394.2017.1413364 (2018).
Burkenroad, M. A natural classification of the Dendrobranchiata, with a key to recent genera. Crustacean Phylogeny Crustacean Issues 1, 279–290 (1983).
Nascimento, J. R., Souza-Filho, P. W. M., Proisy, C., Lucas, R. M. & Rosenqvist, A. Mapeamento de mudanças no maior cinturão contínuo de mangue amazônico usando classificação baseada em objetos de imagens de satélite multissensor. Ciência Estuarina, Costeira e de Plataforma. 117, 83–93 (2013).
Ferreira, C. S. M. et al. First record of rainbow shrimp, exotic species Mierspenaeopsis sculptilis (Heller, 1862), in the Brazilian coastal amazon, validated by DNA barcode. BMC Zool. 8, 11. https://doi.org/10.1186/s40850-023-00176-7 (2023).
Glaser, M. Interrelations between mangrove ecosystem, local economy and social sustainability in Caeté estuary, north Brazil. Wetl. Ecol. Manag. 11, 265–272. https://doi.org/10.1023/A:1025015600125 (2003).
Chakraborty, S. Bioinvasion and environmental perturbation: Synergistic impact on coastal–mangrove ecosystems of West Bengal, India. Impacts of invasive species on coastal environments: coasts in crisis, 171–245. https://doi.org/10.1007/978-3-319-91382-7_6 (2019).
BRASIL. DECRETO Nº 11.959, DE 21 DE MARÇO DE 2024 Cria a Reserva Extrativista Filhos do Mangue, localizada nos Municípios de Primavera e Quatipuru, Estado do Pará. Diário Oficial da República Federativa do Brasil. Brasília, DF, v. 57, p. 6. 21 Mar. 2024. Seção 1. https://www.in.gov.br/en/web/dou/-/decreto-n-11.959-de-21-de-marco-de-2024-549581246 (2024).
BRASIL. DECRETO Nº 11.958, DE 21 DE MARÇO DE 2024 Cria a Reserva Extrativista Viriandeua, localizada nos Municípios de Salinópolis e São João de Pirabas, Estado do Pará. Diário Oficial da República Federativa do Brasil. Brasília, DF, v. 57, p. 4. 21 Mar. 2024. Seção 1. https://www.in.gov.br/en/web/dou/-/decreto-n-11.958-de-21-de-marco-de-2024-549605953 (2024).
Aguirre-Pabón, J., Chasquib, L., Muñoza, E. & Narváez-Barandica, J. Multiple origins define the genetic structure of tiger shrimp Penaeus monodon in the colombian Caribbean Sea. Heliyon 9, e17727. https://doi.org/10.1016/j.heliyon.2023.e17727 (2023).
Leão, T. C. C., Almeida, W. R., Dechoum, M. S. & Ziller, S. R. Espécies Exóticas Invasoras no Nordeste do Brasil: Contextualização, Manejo e Políticas Públicas. Cepan, Recife (2011).
Medeiros, G. F. et al. Current distribution of the exotic copepod Pseudodiaptomus trihamatus Wright, 1937 along the northeastern coast of Brazil. Braz. J. Oceanogr. 54, 241–245 (2006).
Cruz, P. S., Andalecio, M. N., Bolivar, R. B. & Fitzsimmons, K. Tilapia–shrimp polyculture in Negros Island, Philippines: A review. J. World Aquac. Soc. 39, 713–725. https://doi.org/10.1111/j.1749-7345.2008.00207.x (2008).
Silva, G. D. V., Silva, G. A. V. & Maia, R. C. First documented record of the giant-tiger-shrimp, Penaeus monodon (Fabricius, 1798), in the coast of Ceará, Northeastern Brazil. Arq. Ciên. Mar. 55, 154–159. https://doi.org/10.32360/acmar.v55i2.71034 (2022).
Van Quyen, D. et al. Improved genomic resources for the black tiger prawn (Penaeus monodon). Mar. Genom. 52, 100751. https://doi.org/10.1016/j.margen.2020.100751 (2020).
Acknowledgements
Charles Ferreira, thanks the Higher Education Personnel Improvement Coordination (CAPES) for the award Ing of PhD scholarship. The authors thank artisanal fishermen for their support in obtaining samples and National Council for Scientific and Technological Development (CNPq) Universal Call, Process: 407536/2021-3.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by CF, TM, LM, IV and GEG. The first draft of the manuscript was written by JM, ÍL, JS, BC, AM, JC, SM, CH, MV, IS and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interest
The authors have no relevant financial or non-financial interests to disclose.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ferreira, C., Martins, T., Melo, L. et al. DNA reveal new invasive species of tiger shrimp Penaeus monodon (Penaeidae) along the world’s largest mangrove region in the Brazilian Blue Amazon. Sci Rep 15, 18058 (2025). https://doi.org/10.1038/s41598-025-03022-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-03022-7