Abstract
Metabolic reprogramming within the tumor microenvironment (TME) plays a central role in cancer progression and immune evasion, with L-arginine metabolism emerging as a key regulatory axis. Arginase overexpression depletes intratumoral L-arginine, thus suppressing T-cell proliferation while fuelling tumor growth through polyamine biosynthesis. OATD-02, a novel dual arginase (ARG1/ARG2) inhibitor, reprograms tumor metabolism by restoring L-arginine availability and reducing the levels of polyamines, thereby shifting the TME toward a more immunostimulatory state. Unlike ARG1-selective inhibitors with limited intracellular uptake, OATD-02 effectively inhibits both extracellular and intracellular arginases, thereby addressing a major limitation of first-generation arginase inhibitors. To visualize the pharmacodynamic effects of OATD-02 dosing in mice with spatial resolution, we employed MALDI mass spectrometry imaging (MALDI-MSI), thus enabling direct mapping of metabolic changes within tumor tissues. In preclinical models, OATD-02 treatment led to widespread accumulation of intratumoral L-arginine with concomitant depletion of polyamines and resulted in metabolic shifts that correlated with increased immune cell infiltration and an improved response to immune checkpoint blockade. These findings underscore the role of dual arginase inhibition in reshaping tumor metabolism and overcoming immune suppression by restoring the metabolic fitness of immune cells to fight cancer. The metabolic changes caused by OATD-02 treatment resulted in significantly enhanced antitumor immune responses, increased T-cell infiltration in tumors, expansion of CD8⁺ T cells in draining lymph nodes, and systemic upregulation of T-cell activation markers. These effects translated into a substantial survival benefit in the CT26 tumor model, particularly when combined with anti-PD-1 therapy, where OATD-02 improved checkpoint blockade efficacy by relieving metabolic constraints affecting tumor-infiltrating lymphocytes. By leveraging the unique capabilities of MALDI-MSI, this study provides high-resolution metabolic insights into the mechanism of action of OATD-02, reinforcing its potential as a next-generation metabolic-immunotherapeutic agent. The observed metabolic reprogramming, coupled with enhanced immune activation and prolonged survival, supports the clinical development of OATD-02 as a promising strategy for enhancing cancer immunotherapy efficacy. OATD-02 is currently undergoing clinical evaluation in a phase I/II trial (NCT05759923), which will further elucidate its safety and therapeutic impact. These findings highlight the potential of arginase-targeted therapies in cancer treatment and underscore the value of MALDI-MSI as a powerful tool for tracking metabolic responses to therapy.
Similar content being viewed by others
Background
Cancer cells undergo profound metabolic reprogramming to sustain proliferation, evade immune surveillance, and adapt to environmental stressors such as nutrient deprivation and hypoxia. Among the key metabolic pathways, L-arginine metabolism plays crucial roles in tumor growth and immune regulation1,2,3. L-arginine is a conditionally essential amino acid involved in multiple biochemical processes, including polyamine biosynthesis4,5, nitric oxide (NO) production6,7, and proline metabolism8,9,10, all of which influence tumor progression8,11,12,13 and immune cell function14,15,16.
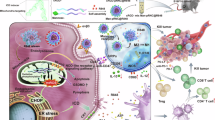
L-arginine degradation is regulated primarily by two enzymes: arginase (ARG1, ARG2) and nitric oxide synthase (NOS). As illustrated in Fig. 1, ARG1, which is predominantly cytosolic, is a key component of the urea cycle in hepatocytes, where it catalyzes the hydrolysis of L-arginine to L-ornithine and urea, facilitating ammonia detoxification. ARG2, which is localized in mitochondria, performs a similar reaction but is more broadly involved in metabolic adaptation. L-ornithine serves as a precursor for polyamine biosynthesis (putrescine, spermidine, and spermine), which promotes tumor proliferation, and for proline synthesis, which plays a key role in extracellular matrix remodeling and redox homeostasis8,17. NOS, on the other hand, converts L-arginine into NO, which can support tumor growth by promoting angiogenesis or inducing tumor cell death through immune-mediated cytotoxicity7. Dysregulated L-arginine metabolism significantly influences the tumor microenvironment (TME), mediating interactions between tumor cells, stromal fibroblasts, and immune cells18,19. Elevated arginase activity is detected in multiple malignancies, including lung, colorectal, breast, and prostate cancers, where it is correlated with poor prognosis, increased tumor invasiveness, and enhanced metastatic potential20,21. Arginase-driven L-arginine depletion promotes tumor cell proliferation by increasing L-ornithine availability, enhancing polyamine synthesis, cell cycle progression, and resistance to apoptosis. Additionally, proline biosynthesis contributes to extracellular matrix remodeling, further facilitating tumor progression8. Notably, L-arginine depletion impairs T-cell activation and proliferation by downregulating the expression of the CD3ζ chain, a key component of the T-cell receptor (TCR) complex, thereby suppressing antitumor immunity22. Myeloid-derived suppressor cells (MDSCs) and tumor-associated macrophages (TAMs) further increase ARG1 expression, exacerbating L-arginine depletion and creating an immune-privileged environment that facilitates tumor immune evasion23. This metabolic alteration reduces the efficacy of immune checkpoint inhibitors, highlighting the need for strategies targeting arginase activity14,15.
In addition to its role in metabolic regulation, ARG2 is now recognized as a critical regulator of both tumor-intrinsic metabolism and immune responses. In contrast to ARG1, which primarily depletes extracellular L-arginine, ARG2 functions within mitochondria, where it directly influences tumor metabolic adaptation, nitrogen balance, and immune regulation (Fig. 1). In tumors, ARG2 expression is linked to glutaminolysis, polyamine biosynthesis, and mitochondrial metabolism, allowing cancer cells to survive in nutrient-deprived environments24,25. The loss of ARG2 in pancreatic ductal adenocarcinoma (PDA) models results in nitrogen accumulation and impaired tumor growth, highlighting its role as a metabolic vulnerability factor in certain cancers24. In addition to its metabolic functions, ARG2 acts as a cell-intrinsic regulator of CD8⁺ T cell activation, persistence, and antitumor efficacy26. Unlike ARG1, which primarily affects T cells by depleting extracellular arginine, mitochondrial ARG2 regulates intracellular arginine metabolism, shaping T-cell fate independently of extracellular arginine availability. Deletion of ARG2 in CD8⁺ T cells enhances their expansion, effector function, and persistence, leading to improved tumor control in preclinical cancer models. Moreover, ARG2-deficient CD8⁺ T cells exhibit strong synergy with PD-1 blockade, suggesting that targeting ARG2 could enhance the efficacy of immune checkpoint therapies26.
Modulation of L-arginine metabolism by the arginase inhibitor OATD-02 in cytosolic and mitochondrial compartments. OATD-02 inhibits arginase 1 (ARG1) in the cytosol and arginase 2 (ARG2) in mitochondria, altering key metabolic pathways. ARG1 inhibition prevents L-arginine catabolism into L-ornithine and urea, thereby reducing polyamine synthesis (putrescine, spermidine, and spermine) catalyzed by L-ornithine decarboxylase (ODC), spermidine synthase (SRM), and spermine synthase (SMS), which may limit tumor cell proliferation. ARG2 inhibition restricts L-ornithine availability in mitochondria, reducing proline biosynthesis via L-ornithine aminotransferase (OAT) and the pyrroline-5-carboxylate (P5C) pathway, which involves P5C synthase (P5CS), P5C dehydrogenase (PDG), and P5C reductase (P5CR). This metabolic shift may impair tumor adaptation to hypoxia and metabolic stress. The dual inhibition of ARG1 and ARG2 by OATD-02 may enhance antitumor effects by modulating polyamine metabolism, amino acid availability, and redox homeostasis.
Arginase inhibitors have been studied for over a century, with early studies identifying α-amino acids as weak inhibitors. A major breakthrough came with N-hydroxy-nor-arginine (nor-NOHA), a micromolar inhibitor that provided structural insights into arginase inhibition27. This led to the development of boronic acid-based inhibitors such as 2(S)-amino-6-boronohexanoic acid (ABH) and (S)-(2-boronoethyl)-L-cysteine (BEC), which are known for their potency and selectivity17. These compounds laid the groundwork for more advanced inhibitors, such as OATD-02 and numidargistat. Preclinical studies have demonstrated that numidargistat (INCB001158) reversed T-cell immunosuppression and reduced tumor growth in multiple syngeneic mouse models, particularly when combined with anti-PD-L1 therapy, highlighting its potential to modulate the tumor immune microenvironment28. However, despite pharmacodynamic evidence of arginase inhibition and increased plasma arginine levels in a phase I/II clinical trial, its antitumor efficacy has remained limited, suggesting the complexity of arginine metabolism in cancer29. A key distinction between numidargistat and OATD-02 lies in their ability to inhibit intracellular ARG2. While numidargistat primarily targets extracellular ARG1, its limited intracellular penetration restricts its effect on ARG2-dependent metabolism in tumor and immune cells28. In contrast, OATD-02 is designed to effectively inhibit both intracellular and extracellular arginases, granting it a broader and potentially stronger pharmacodynamic profile30,31. The ongoing phase I/II clinical trial of OATD-02 aims to evaluate these multidimensional antitumor properties in patients with advanced solid tumors, providing a more comprehensive understanding of its therapeutic potential32.
To evaluate the intracellular effects of OATD-02 on tumor metabolism directly in tissue, the advanced imaging technique MALDI-MSI was used to assess spatially resolved metabolic alterations induced by arginase inhibition33,34. This approach enabled the direct visualization of changes in key metabolites, including L-arginine and polyamines, within tumor tissue, complementing bulk quantification methods such as HPLC and LC-MS. By integrating MALDI-MSI with systemic pharmacokinetic and metabolic analyses, this study examines how OATD-02 modulates the tumor microenvironment. The spatial distribution of metabolic changes offers insights into the extent and localization of OATD-02-induced effects, contributing to a broader understanding of its potential as a metabolic modulator in cancer therapy and its role in enhancing immune responses in combination with immune checkpoint inhibitors.
Methods
Chemical compounds
OATD-02 was synthesized at Molecure SA. For in vitro assays, OATD-02 was dissolved in Milli-Q water (Millipore) at a stock concentration of 20 mM and stored at −20 °C until use. For in vivo studies, OATD-02 was dissolved in sterile saline at a final concentration of 10 mg/mL and stored at −20 °C until use. Prior to administration, the solution was prewarmed to RT and used within its validated minimal stability period. Unless otherwise specified, all additional chemical reagents, including analytical standards, were obtained from Merck (Germany).
Cell culture
The CT26.WT (CRL-2638, mouse colon carcinoma) and K562 (CCL-243, human chronic myelogenous leukemia) cell lines were purchased from ATCC®. Both cell lines were maintained in RPMI-1640 medium (Gibco, Life Technologies) supplemented with 10% fetal bovine serum (FBS, Thermo Fisher/Gibco™), 100 U/mL penicillin G and 100 µg/mL streptomycin (Antibiotic-Antimycotic, Gibco®) at 37 °C in a humidified atmosphere containing 5% CO₂. CT26 cells were cultured as adherent monolayers and passaged upon 70–80% confluence using 0.25% trypsin-EDTA (Gibco®). Nonadherent K562 cells were maintained in suspension culture and passaged every 2–3 days by dilution with fresh medium to sustain logarithmic growth. The absence of mycoplasma contamination in both cell lines was confirmed via the MycoAlert™ Mycoplasma Detection Kit (Lonza).
Cellular metabolite detection via HPLC
The detection of amino acids and polyamines in CT26 and K562 cell extracts was performed via high-performance liquid chromatography (HPLC) with dabsyl derivatives, following a modified protocol of Krause et al.35. CT26 adherent cells were seeded at a density of 750,000 cells per 10 mL in 75 cm² flasks and treated with 10 µM or 30 µM OATD-02. K562 suspension cells were seeded at a density of 2 × 10⁶ cells per 10 mL in 75 cm² flasks and treated with OATD-02 at the same concentrations. Treatment was initiated either 2 h postseeding, resulting in a total incubation time of 96 h, or two days postseeding, resulting in a total incubation time of 48 h. At the experimental endpoint, the CT26 monolayers were washed with 5 mL of prewarmed (37 °C) 0.3 M mannitol solution. For K562 nonadherent cells, an initial centrifugation step was performed to pellet the cells, followed by rinsing with warm 0.3 M mannitol. Metabolite extraction was carried out via the addition of 1.5 mL of a cold extraction mixture consisting of HPLC-grade methanol, acetonitrile, and water at a 2:2:1 ratio. The cells were incubated on a laboratory shaker at 4 °C for 5 min and then detached via cell scrapers (for CT26) or resuspended (for K562) in the extraction mixture. The resulting suspensions were centrifuged (10,000 × g, 10 min, 4 °C), and the supernatants were transferred to coded tubes to ensure blinded analysis before being stored at − 80 °C until further HPLC processing. Dabsyl derivatization was performed by mixing 50 µL of NaHCO₃ buffer (0.4 M, pH 9) with 50 µL of a deproteinized sample or standard. Two hundred microliters of freshly prepared dabsyl chloride reagent (4 mg/mL in acetone) was subsequently added. The mixture was incubated at 70 °C for 21 min with intermittent mixing. The reaction was terminated by adding 200 µL of cold dilution buffer (acetonitrile, ethanol, and sodium acetate buffer at a 2:1:1 ratio), followed by incubation on ice for 5 min. The samples were then centrifuged, and the clear supernatants were immediately analyzed by HPLC. HPLC separation was performed via a Dionex 3000 ICS system equipped with an Agilent Zorbax SB-C18 column (4.6 × 250 mm) maintained at 40 °C. The mobile phase consisted of 45 mM sodium acetate buffer (pH 3.5; mobile phase A) and 100% acetonitrile (mobile phase B). The compounds were eluted at a flow rate of 1 mL/min via a gradient starting at 20% phase B, increasing to 100% over 40 min, followed by re-equilibration to 20% over 20 min. The total run time was 60 min. Detection was performed via a UV–VIS detector set at 436 nm. Quantification was achieved by comparing peak areas to standard calibration curves. Standard calibration curves were prepared via serial dilutions of reference standards in the same solvent matrix as the samples to ensure accurate quantification.
LC-MS/MS analysis
The concentrations of OATD-02 and L-arginine in blood serum were quantified via liquid chromatography coupled with tandem mass spectrometry (LC‒MS/MS). Sample preparation involved protein precipitation with acetonitrile, followed by centrifugation. The resulting supernatants were analyzed via hydrophilic interaction liquid chromatography (HILIC) coupled with tandem mass spectrometry, which employs optimized multiple reaction monitoring (MRM) transitions. The quantification of OATD-02 was performed via matrix-matched calibration and quality control samples. Owing to the endogenous nature of L-arginine, a surrogate matrix approach was applied to ensure accurate detection and quantification.
MALDI-MSI imaging
Frozen mouse tumors were sectioned at 12-µm thickness and mounted onto indium-tin oxide (ITO)-coated glass slides (Bruker Daltonics, Bremen, Germany). The slides were stored in slide boxes, vacuum-sealed in foil (CASO, Arnsberg, Germany), and kept at − 80 °C until further processing. Prior to matrix application, the slides were equilibrated to room temperature (RT), removed from vacuum packaging, and dried for 10 min in a vacuum desiccator (SP Bel-Art). A deuterated standard of the OATD-02 derivative (5 mg/mL in 50% methanol) was prepared, and 100 µL was mixed with 5 mL of DHB matrix solution (40 mg/mL in 70% methanol). The matrix was uniformly applied via an M5 pneumatic sprayer (HTX Technologies LLC, Chapel Hill, North Carolina, USA) with the following parameters: 11 layers, flow rate of 0.05 mL/min, velocity of 1200 mm/min, track spacing of 3 mm, and CC pattern at a nozzle temperature of 65 °C. MALDI-MSI measurements were performed via a timsTOF flex mass spectrometer (Bruker Daltonics, Bremen, Germany). External mass calibration was conducted via sodium formate clusters in electrospray ionization (ESI) mode, and online calibration was performed with the deuterated standard. Imaging was carried out with a lateral step size of 40 μm in the mass range of m/z 90–1300 in positive ion mode. Spectra were acquired via 400 shots per pixel. The following MS parameters were applied: Funnel 1 RF: 250 Vpp, Funnel 2 RF: 250 Vpp, Multipole RF: 250 Vpp, MS1 collision energy: 2 eV, Collision cell RF: 650 Vpp, low mass: m/z 90, TOF transfer time: 70 µs, TOF prepulse storage: 4 µs. The MALDI–MS data were processed via DataAnalysis 6.1 and SCiLS Lab 2025 software (Bruker Daltonics). The spectral intensities were normalized to the deuterated standard to ensure quantitative reliability. Statistical and spatial analyses were performed to assess metabolite distribution in the tissue sections.
Animal studies
All in vivo experiments were conducted using 7–9-week-old female BALB/c (BALB/cAnNCrl) mice obtained from Charles River Laboratories (certified SPF status). All procedures complied with the Public Health Service Policy on Humane Care and Use of Laboratory Animals and the Guidelines for the Welfare and Use of Animals in Cancer Research. Ethical approval for the study was granted by the 1 st Local Ethics Committee for Animal Experiments in Warsaw, Poland (approval no. 891/2019). CT26 cells were harvested during the exponential growth phase, with viability exceeding 90%, as confirmed by trypan blue exclusion. The mice were subcutaneously implanted in the right flank with 5 × 10⁵ CT26 cells suspended in 50 µL of PBS. The animals were then randomized into experimental groups. Tumor growth was monitored at least three times per week via caliper measurements and calculated according to the following formula: width × length × depth × π/6, assuming an ellipsoidal shape. Humane endpoints were defined as body weight loss exceeding 20%, a tumor volume surpassing 2000 mm³, or the presence of persistent signs of pain or distress. Animals meeting these criteria were euthanized. For LC–MS and MALDI–MSI analyses, tumor-bearing mice (n = 3) were orally administered OATD-02 at a dose of 100 mg/kg. Blood, tumors and selected organs were collected after either three doses (16 h time point) or four doses (2 h time point). The control animals received an equivalent volume of vehicle (saline). The collected tissues were snap-frozen and stored at − 80 °C until further analysis (see the MALDI-MSI Imaging section for a detailed description). In the efficacy study, the mice (n = 18) were treated orally with OATD-02 at 100 mg/kg twice daily, starting one day after tumor implantation. The anti-mouse CD279 (PD-1) IgG2a rat antibody (clone RMP1-14, cat. 114115, lot B306588, BioXCell) was administered intraperitoneally at 2.5 mg/kg on days 6, 10, 14, and 18 postimplantation. The control animals received saline via oral gavage and an anti-KLH isotype control IgG2b rat antibody (clone LTF-2, cat. BE0090, lot 629816D1, BioXCell) via intraperitoneal injection. On day 15, randomly selected mice (n = 6) from each group (n = 18) were sacrificed, and the tumors, spleens, and tumor-draining lymph nodes were collected for cytometric analysis (see the LC–MS/MS analysis and flow cytometry sections for details on sample processing). Remaining animals (n = 12) were used for survival analysis. To minimize animal discomfort, appropriate analgesic and anesthetic measures were applied, and humane endpoints were consistently enforced. At the end of the study, the animals were deeply anesthetized with an intraperitoneal injection of ketamine (150 mg/kg) and xylazine (15 mg/kg). Euthanasia was performed via cervical dislocation in accordance with ethical guidelines.
Flow cytometry
The collected tumors, spleens, and lymph nodes were cut into small fragments (2–3 mm). Tumors and lymph nodes were enzymatically digested in DMEM containing collagenase (0.8 mg/mL) and DNase I (15 U/mL) for 40 min at 37 °C in a CO₂ incubator. Following digestion, tumor and lymph node suspensions, along with spleen fragments, were passed through cell strainers to obtain single-cell suspensions. The cells were then washed with PBS and treated with ACK erythrocyte lysis buffer for 5 min on ice. Lysis was stopped by the addition of PBS, followed by centrifugation. The resulting cell pellets were resuspended in PBS, and the cell density and viability were assessed. Prior to antibody staining, dead cells were labeled via the Zombie Aqua™ Fixable Viability Kit (BioLegend). The following anti-mouse antibodies were used for flow cytometry: CD8 (PerCP-Cy5.5, clone 53 − 6.7, BioLegend), CD3 (APC, clone 17 A2, eBioscience), CD4 (BV605, clone GK1.5, BioLegend), CD69 (PE, clone H1.2 F3, eBioscience), CD45.2 (V500, clone 104, BioLegend), CD3e (PE-Cy7, clone 145-2 C11, eBioscience), and CD11b (PE, clone M1/70, eBioscience), CD44 (FITC, clone IM7), CD62L (PE-Cy7, clone MEL-14), LAG-3 (BV421, clone C9B7 W), Granzyme B (PE-Cy7, clone NGZB), and IFN-γ (PE, clone XMG1.2). For cytokine detection, ex vivo restimulation of splenocytes was performed prior to intracellular staining. Cells were treated with ionomycin (2 µg/mL) and PMA (5 ng/mL) for 2 h, followed by incubation with brefeldin A (5 µg/mL) for an additional 6 h. The samples were analyzed via a CytoFLEX Analyzer (Beckman Coulter), and the data were processed via FlowJo software (BD Biosciences). The gating strategy and representative flow cytometry plots are provided as Supplementary information (Supplementary Figs. 4 (A–H) and the file named Representative_flow_plots), to illustrate the analysis workflow and reproducibility across biological replicates.
Statistical analysis
Statistical analyses were performed using GraphPad Prism (version 10.0). Data distribution was assessed via D’Agostino-Pearson and Shapiro-Wilk normality tests. The sample size for in vivo studies was determined on the basis of a retrospective analysis of previous optimization experiments, ensuring a statistical power of approximately 80% with an alpha level of 0.05. Given the relatively small sample sizes in the in vivo experiments, which were balanced to maintain statistical sensitivity while adhering to the 3R principles, and the nonnormal distribution observed for some variables, statistical comparisons were performed via nonparametric tests to ensure methodological consistency and robustness. The results are presented as the median, with individual data points shown as scatter dot plots. Multiple group comparisons were conducted via the Kruskal‒Wallis test followed by Dunn’s post hoc test, and nonsignificant differences were reported alongside the observed trends when applicable. All comparisons were performed using unpaired tests, as all measurements were based on independent biological replicates. Metabolite concentrations obtained by HPLC were normalized relative to untreated control values to facilitate cross-condition comparisons. Normalization was performed using the formula “Normalized value = Treated/Mean untreated”. The resulting normalized values were averaged across replicates and visualized as heatmaps, illustrating relative changes in metabolite levels in response to treatment. Survival analysis was conducted using Kaplan-Meier survival curves, with comparisons performed using the log-rank (Mantel‒Cox) test. Additionally, the Gehan-Breslow-Wilcoxon test was applied to assess early differences in survival dynamics. Hazard ratios (HRs) with 95% confidence intervals (CIs) were calculated via the Mantel–Haenszel method to estimate the relative risk of death between treatment groups. Median survival times and statistical comparisons were reported for each treatment condition. Differences were considered statistically significant at p ≤ 0.05.
Results
OATD-02 remodels tumor cell metabolism
To evaluate the metabolic impact of intracellular arginase inhibition by OATD-02, we analyzed the intracellular levels of key metabolites involved in L-arginine metabolism, including L-arginine, L-ornithine, polyamines (spermidine and spermine), glutamine, and proline, in CT26 and K562 cell extracts via HPLC.
We selected CT26 (murine colorectal carcinoma) and K562 (human chronic myeloid leukemia) cells because of their distinct tumor biology and relevance to in vivo models, allowing us to assess the direct anticancer metabolism-related effects of OATD-02. CT26 cells represent a murine model, enabling in vivo studies in an immunocompetent tumor environment, whereas K562 cells are human lymphoblasts that exhibit elevated ARG2 expression, making them an optimal system for assessing the impact of dual ARG1/ARG2 inhibition36. Furthermore, OATD-02 has been tested in vivo in both models30, reinforcing its relevance in evaluating tumor metabolism and therapeutic efficacy.
The selection of OATD-02 concentrations was guided by pharmacokinetic data from in vivo studies, where a 100 mg/kg dose of OATD-02 resulted in plasma and tumor concentrations of approximately 5 µM and 30 nmol/g, respectively, two hours postdosing30. Given the relatively short exposure, 10 and 30 µM were selected for in vitro experiments as physiologically relevant concentrations, reflecting the levels observed in the animal model.
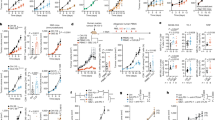
Heatmap visualization (Fig. 2) illustrates the relative metabolite levels normalized to those of untreated controls across different treatment conditions. OATD-02 treatment resulted in a dose- and time-dependent increase in the intracellular L-arginine level. After 48 h, compared with untreated control cells, CT26 (Fig. 2, left panel) and K562 cells (Fig. 2, right panel) treated with 30 µM OATD-02 presented 1.31-fold and 1.43-fold increases in L-arginine levels, respectively. After 96 h, the L-arginine level further increased to 1.62-fold in K562 cells, whereas in CT26 cells the highest increase (1.57-fold) was observed at 10 µM. These findings confirm that OATD-02 effectively enhances L-arginine accumulation within tumor cells, supporting its mechanistic role as an intracellular arginase inhibitor.
Changes in the intracellular metabolism of CT26 and K562 cells upon OATD-02 treatment. The heatmap represents the mean relative levels (n = 3) of L-arginine, L-ornithine, spermidine, spermine, glutamine, and proline in CT26 and K562 tumor cells treated with 10 µM or 30 µM OATD-02 for 48 h and 96 h. Metabolite levels are expressed as a fraction of those in the untreated control.
As expected, OATD-02 treatment significantly suppressed L-ornithine levels, confirming effective arginase inhibition. A marked reduction in intracellular L-ornithine was observed in CT26 cells, where levels decreased to 30% of control values after 48 h of treatment with 30 µM OATD-02 and further declined to 19% at 96 h. A similar pattern was detected in K562 cells, where L-ornithine levels decreased to 9% of the control values under prolonged exposure to OATD-02 (Fig. 2). These results confirm the functional inhibition of arginase, preventing the conversion of L-arginine to L-ornithine and thereby limiting its availability for downstream metabolic pathways.
Consistent with the reduction in L-ornithine, OATD-02 treatment led to a dose- and time-dependent depletion of intracellular polyamines, specifically spermidine and spermine, which rely on L-ornithine as a biosynthetic precursor. The most pronounced effect was observed for spermine in CT26 cells at 96 h, where its level was reduced to 43% of the control value following treatment with 30 µM OATD-02 (Fig. 2). In K562 cells, spermine levels decreased to 55% under the same conditions. Spermidine levels exhibited a modest decline in both tested cell lines (Fig. 2). These findings suggest that OATD-02 limits polyamine biosynthesis by depleting the substrate pool, further reinforcing its role in the metabolic reprogramming of tumor cells.
In contrast to the substantial changes observed in L-arginine metabolism, the levels of glutamine and proline remained largely unaffected by OATD-02 treatment. The fluctuations were minor, with variations remaining within a ± 15% range relative to untreated controls, indicating that OATD-02 specifically modulates the L-arginine metabolic axis without broadly perturbing other amino acid pathways (Fig. 2).
These findings demonstrate that OATD-02 effectively increases intracellular L-arginine while depleting L-ornithine and polyamines in both the CT26 and K562 tumor models. The observed metabolic shifts are consistent with potent arginase inhibition, providing a mechanistic basis for the potential antitumor effects of OATD-02. The inclusion of 10 µM OATD-02, on the basis of in vivo exposure data, further strengthens the clinical relevance of these findings.
OATD-02 increases systemic L-arginine levels and reshapes tumor metabolism in vivo
To assess the biological consequences of the metabolic changes observed in vitro, we evaluated the pharmacodynamic effects of OATD-02 in vivo in CT26 tumor-bearing mice. The selected dose of 100 mg/kg (PO, BID) was confirmed in prior toxicokinetic studies to be safe and within the therapeutic window for long-term administration in BALB/c mice30.
Mice were treated with OATD-02 via oral gavage twice daily, and blood and tumor samples were collected at two pharmacokinetically relevant time points: 2 h after the last dose (peak exposure) and 16 h after the last dose (trough levels before the next administration) (Fig. 3A). In contrast to the extended exposure in vitro (Fig. 2), the 2 h and 16 h time points in vivo reflect early pharmacodynamic responses following multiple doses of OATD-02, during which a tissue-level equilibrium was likely established. enabling detection of metabolic shifts shortly after the final dose. LC–MS/MS analysis of the plasma samples confirmed that OATD-02 remained detectable at both time points, with mean plasma concentrations of 2.97 µg/mL at 2 h and 1.05 µg/mL at 16 h (Fig. 3B, left panel). These pharmacokinetic data indicate that OATD-02 achieves substantial systemic exposure, supporting its bioavailability and stability in circulation.
Consistent with its role as an intracellular arginase inhibitor, OATD-02 treatment led to a marked increase in systemic L-arginine levels (Fig. 3B, right panel). At 2 h posttreatment, the serum L-arginine concentration was approximately 10-fold greater than that in untreated control mice, with the mean concentration increasing from ~ 140 µM in controls to ~ 1.20 mM in treated mice. Although L-arginine levels decreased by 16 h, they remained significantly elevated (~ 930 µM) relative to those of the untreated controls, indicating a sustained pharmacodynamic effect of OATD-02 on systemic L-arginine homeostasis (Fig. 3B, right panel). Further correlation analysis revealed a strong positive correlation between the OATD-02 plasma concentration and the serum L-arginine level (Spearman r = 0.797, p = 0.0153), confirming the dose-dependent modulation of L-arginine metabolism.
Pharmacodynamic effects of OATD-02 in CT26 tumor-bearing mice. (A) Experimental design: Mice (n = 3) were treated with OATD-02 (100 mg/kg PO, BID), and blood and tumor samples were collected at 2 h and 16 h after the last dose. (B) LC-MS/MS analysis of the serum concentrations of OATD-02 (left) and L-arginine (right) at the indicated time points. (C) Representative MALDI-MSI ion images of L-arginine, proline, spermine, and spermidine levels in tumors of untreated and OATD-02-treated mice (n = 2) at 2 h and 16 h after the last dose.
To investigate the spatial distribution of metabolic alterations in tumors, MALDI-MSI was employed to visualize L-arginine and related metabolites, including proline, spermine, and spermidine, in tumor sections at 2 h and 16 h posttreatment (Fig. 3C, Suppl. Figure 1). Consistent with the serum data, intratumoral L-arginine levels were substantially increased in the OATD-02-treated mice compared with the untreated controls. This effect was evident as early as 2 h posttreatment and persisted at 16 h, with widespread but heterogeneous L-arginine enrichment across tumor tissue (Fig. 3C, Suppl. Figure 1). Notably, regions with the highest L-arginine accumulation coincided with the tumor parenchyma rather than the stromal compartments, suggesting a preferential metabolic impact on tumor cells. However, spatial analysis also revealed localized regions with lower L-arginine enrichment, which may correspond to hypoxic or necrotic areas within the tumor core.
In parallel with L-arginine accumulation, MALDI-MSI analysis revealed a time-dependent reduction trend in the levels of the polyamines spermine and spermidine in tumor tissue. At 2 h posttreatment, a moderate decrease in polyamine levels was observed, particularly in the central regions of larger tumors. By 16 h, spermine and spermidine depletion became more pronounced and spatially widespread, maintaining this trend over time, suggesting that OATD-02-driven inhibition of arginase limits L-ornithine availability for polyamine biosynthesis over time (Fig. 3C, Suppl. Figure 1). Interestingly, the strongest reduction in polyamines was observed in tumor regions where L-arginine accumulation was highest, suggesting a metabolic shift favoring L-arginine retention over its downstream utilization.
Unlike the substantial changes observed in L-arginine and polyamines, tumor proline levels exhibited only minor fluctuations in response to OATD-02 treatment (Fig. 3C, Suppl. Figure 1). Proline distribution appeared relatively uniform across tumor sections, suggesting that its biosynthesis from L-arginine-derived intermediates remained largely unaffected. This further supports the selective metabolic action of OATD-02 on the L-arginine-polyamine axis.
Collectively, these findings demonstrate that OATD-02 effectively increases systemic and intratumoral L-arginine levels while simultaneously reducing polyamine metabolites in tumors. Spatial metabolic analysis suggests that these changes are not uniform across the tumor microenvironment, with larger tumors displaying distinct metabolic heterogeneity, potentially influenced by variations in vascularization and hypoxia. The temporal dynamics of these changes suggest that OATD-02 exerts a sustained metabolic effect, reshaping the tumor microenvironment in a manner that may contribute to its antitumor efficacy. Notably, the moderate impact observed for some metabolites, such as proline and early-stage polyamine depletion, may reflect the relatively short duration of OATD-02 administration in this study (only 3–4 doses before sample collection at 2 h and 16 h, respectively). Longer treatment regimens or combination therapies targeting hypoxic tumor regions may further amplify these metabolic shifts, potentially leading to a more pronounced reprogramming of tumor metabolism over time.
OATD-02 improves survival and enhances antitumor immune response in vivo
Having established the strong in vivo metabolic effects of OATD-02, we next evaluated its therapeutic potential in combination with immune checkpoint blockade. CT26 tumor-bearing mice were treated with OATD-02 (100 mg/kg, PO, BID) alone or in combination with anti-PD-1 antibodies (2.5 mg/kg, IP) according to the schedule outlined in Fig. 4A. Tumor growth and survival were monitored, and immune profiling of tumors, spleens, and lymph nodes was conducted on day 15 postimplantation.
Kaplan-Meier survival analysis revealed that both the OATD-02 and anti-PD-1 monotherapy significantly extended median survival compared with the vehicle-treated controls (Fig. 4B). Compared with control mice, animals receiving OATD-02 alone had a median survival of 29.5 days, whereas anti-PD-1 monotherapy extended survival to 32 days. The log-rank test confirmed significant differences between the vehicle group and the anti-PD-1 group (p = 0.0097) and between the vehicle group and the OATD-02 group (p = 0.0470). However, the greatest survival benefit was observed in the combination therapy group (OATD-02 + anti-PD-1), with a median survival of 41.5 days, which was significantly longer than that in the monotherapy group (p = 0.0479 vs. anti-PD-1 alone). Hazard ratio (Mantel–Haenszel) analysis indicated that, compared with anti-PD-1 therapy alone, combination therapy reduced mortality risk by approximately 62% (HR = 0.382, 95% CI: 0.147–0.991), supporting a synergistic effect.
To assess the immune mechanisms underlying this survival benefit, we analyzed tumor-infiltrating immune cells via flow cytometry. OATD-02 treatment led to a significant increase in the proportion of CD45⁺ T cells within tumors (Fig. 4C, top left panel). This effect was further enhanced in the combination therapy group (p = 0.0429, Dunn’s test), suggesting more robust immune infiltration. Given that L-arginine availability is crucial for T-cell activation and proliferation, particularly within metabolically restricted tumor microenvironments, these findings indicate that OATD-02-mediated arginase inhibition relieves metabolic constraints on intratumoral T cells, facilitating their recruitment and expansion. In contrast, CD11b⁺ myeloid cell populations, including myeloid-derived suppressor cells (MDSCs) and tumor-associated macrophages (TAMs), did not significantly change following treatment (p > 0.05 for all comparisons, Dunn’s test; Fig. 4C, bottom left panel). However, a visible trend toward reduced myeloid cell infiltration was observed in the combination therapy group, suggesting a potential shift in the tumor immune landscape. These results imply that while OATD-02 effectively promotes T-cell activation, its effects on immunosuppressive myeloid populations may require longer treatment durations or additional combinatorial approaches for full efficacy.
To determine whether these effects extend beyond the tumor microenvironment, we analyzed tumor-draining lymph nodes (TDLNs), which are critical sites for priming antitumor immune responses. Compared with those in the vehicle group, the numbers of CD4⁺ and CD8⁺ T cells were significantly greater in the mice that received OATD-02 + anti-PD-1 (p = 0.0058 for CD4⁺ and p = 0.0099 for CD8⁺; Dunn’s test; Fig. 4C, middle panels). Neither monotherapy alone induced significant changes relative to the controls, suggesting that increased systemic L-arginine availability in the combination treatment group supports the expansion of tumor-reactive T cells in lymphoid organs, potentially enhancing long-term antitumor immunity.
Combination therapy with OATD-02 and anti-PD-1 enhances antitumor immunity and prolongs survival in CT26 tumor-bearing mice. (A) Experimental design: CT26 tumor-bearing mice (n = 18) were treated with OATD-02 (100 mg/kg, PO, BID) from day 1 until the end of the study. Anti-PD-1 antibodies (2.5 mg/kg, IP) were administered on days 6, 10, 14, and 18. On day 15, a subset of animals (n = 6) was sacrificed for flow cytometry analysis, while the remaining mice (n = 12) were monitored for survival. (B) Kaplan–Meier survival curves comparing treatment groups (Vehicle, Anti-PD-1, OATD-02, and combination therapy). Log-rank test significance: *p < 0.05. (C) Flow cytometry analysis of immune cell populations in tumors, lymph nodes, and spleens on day 15. Effector CD4⁺ and CD8⁺ T cells were identified as CD44⁺CD62L⁻ (within CD4⁺ or CD8⁺ populations, respectively). In panel C, “Combined” refers to animals treated with a combination of anti-PD-1 and OATD-02. Statistical analysis: Kruskal–Wallis test followed by Dunn’s post hoc test (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
To further assess systemic immune activation, we measured the expression of CD69, a marker of early T-cell activation, on splenic CD4⁺ and CD8⁺ T cells (Fig. 4C, right panels). While both monotherapies induced a modest increase in CD69 expression, the combination treatment led to highly significant upregulation compared with the vehicle (p < 0.0001 for CD4⁺, p = 0.0005 for CD8⁺, Dunn’s test). This finding suggests that systemic T-cell activation extends beyond the tumor site, potentially priming circulating T cells for enhanced antitumor responses. To explore the functional polarization of T cells, we assessed the abundance of effector T cell subsets (CD44⁺CD62L⁻) in the spleen (Fig. 4C, right panels). Both CD4⁺ and CD8⁺ effector T cells were significantly increased in the combination therapy group compared with the vehicle-treated controls (p = 0.0223 for CD4⁺, p = 0.0165 for CD8⁺; Dunn’s test), suggesting that OATD-02 augments systemic T-cell effector differentiation when combined with anti-PD-1 antibody. To further characterize the functional status of T cells, we analyzed the expression of key effector molecules in splenic T cells following ex vivo restimulation. Although not statistically significant, combination treatment with OATD-02 and anti–PD-1 suggested a tendency toward increased frequencies of IFN-γ⁺ and GzmB⁺ CD8⁺ T cells (p = 0.0762 and p = 0.0825, respectively; Dunn’s test; Supplementary Fig.2), indicating enhanced cytotoxic potential. These findings suggest that OATD-02 promotes not only T cell expansion, but also their functional polarization toward an effector phenotype. Finally, to assess potential exhaustion-associated changes, we evaluated the expression of the inhibitory receptor LAG-3 (Supplementary Fig. 3). A trend toward reduced LAG-3 expression was observed in splenic CD4⁺ T cells following OATD-02 treatment, whereas LAG-3 levels on CD8⁺ T cells remained largely unchanged. While these observations suggest potential modulation of exhaustion-associated signaling, further studies will be needed to confirm their functional relevance.
These findings demonstrate that OATD-02 enhances antitumor immunity by increasing intratumoral T-cell infiltration, expanding tumor-reactive T cells in the lymph nodes, and increasing systemic T-cell activation. When combined with anti-PD-1 therapy, these effects translate into a significant survival benefit, with a 62% reduction in mortality risk compared with anti-PD-1 therapy alone (Fig. 4B). Notably, the most pronounced immunological effects, including enhanced T-cell activation in the spleen and lymph nodes, were exclusive to the combination therapy group, underscoring the potential of OATD-02 as a metabolic adjuvant for immune checkpoint blockade.
Discussion
Metabolic reprogramming in the tumor microenvironment (TME) plays a pivotal role in cancer progression and immune evasion. Our study demonstrated that the dual ARG1/ARG2 arginase inhibitor OATD-02 effectively modulates the L-arginine metabolic axis, enhancing antitumor immune responses. Using a combination of in vitro, in vivo, and spatial metabolomics approaches, we demonstrated that OATD-02 increases L-arginine availability within tumors, depletes immunosuppressive polyamines, and enhances the efficacy of immune checkpoint inhibitors (ICIs). Notably, OATD-02 significantly elevated both intracellular and systemic L-arginine levels while concurrently reducing L-ornithine and polyamine concentrations. This effect was observed in CT26 colon carcinoma and K562 leukemia cell models, as well as in CT26 tumor-bearing mice. Our findings are consistent with those of previous studies demonstrating that arginase-mediated L-arginine depletion suppresses T-cell activation and facilitates tumor immune evasion37,38. Unlike numidargistat (CB-1158), an arginase inhibitor with predominantly extracellular activity and limited efficacy against ARG2-driven pathways28,29, OATD-02 effectively inhibits both extracellular and intracellular arginase isoforms. This distinction is critical, as ARG2 plays a tumor-intrinsic role in metabolic adaptation, regulating nitrogen balance and polyamine biosynthesis39,40,41. The ability of OATD-02 to penetrate tumor cells and inhibit intracellular ARG2 represents a unique mechanistic advantage over other arginase-targeting therapies.
While systemic metabolic alterations are critical for therapeutic efficacy, spatial heterogeneity within the tumor microenvironment may further shape treatment outcomes. To characterize the metabolic landscape of tumors, we employed MALDI mass spectrometry imaging (MALDI-MSI) to map metabolite distributions with high spatial resolution. Our results revealed heterogeneous L-arginine accumulation across tumor sections. Given that larger tumors often develop hypoxic cores with impaired circulation and tissue necrosis42, these spatial differences may reflect microenvironmental constraints on metabolic fluxes. Although the inclusion of tumors with differing volumes might have introduced variability, we deliberately included such samples to capture potential differences in intratumoral metabolic architecture. In line with this approach, Andersen et al.43 demonstrated that MALDI-MSI enables the spatially resolved detection of metabolic alterations in prostate cancer tissues, revealing distinct metabolic differences between tumor and normal regions. MALDI-MSI has also demonstrated that anti-cancer drugs frequently fail to penetrate tumors in both patients and mouse models44,45. These findings underscore the power of MALDI-MSI in identifying spatial metabolic heterogeneity and highlight its value in assessing the metabolic reprogramming induced by OATD-02 in our study. Beyond the heterogeneous distribution of L-arginine, we observed a trend towards preferential depletion of polyamines, particularly in central tumor regions. This suggests that metabolic adaptation to OATD-02 treatment is influenced by tumor perfusion and oxygen availability. Hypoxic tumor regions are known to favor metabolic pathways that sustain proliferation under stress, including increased glutaminolysis and alternative nitrogen metabolism46. The differential impact of OATD-02 across spatially distinct tumor compartments suggests that L-arginine-polyamine axis modulation may be influenced by oxygen and nutrient gradients within the tumor microenvironment. Since hypoxic regions often exhibit increased polyamine turnover to support tumor survival13,40,42, this adaptation may shape the response to OATD-02.
Expanding on these insights, our results show that metabolic modulation by OATD-02 can significantly enhance the efficacy of ICIs. By increasing L-arginine availability and reducing polyamines, OATD-02 creates a microenvironment more permissive for T-cell activation, thereby potentiating the effects of PD-1 blockade. OATD-02 combined with anti-PD-1 therapy led to increased T-cell infiltration in tumors, enhanced T-cell activation in tumor-draining lymph nodes, and improved systemic immune responses. In addition to promoting T-cell infiltration and expansion, OATD-02 also increased the abundance of effector CD4⁺ and CD8⁺ T cells in the spleen and enhanced systemic T-cell activation, as evidenced by increased CD69 expression. These systemic immune changes likely contribute to the observed survival benefit when OATD-02 is combined with PD-1 blockade. Observed trends toward increased IFN-γ and granzyme B production in CD8⁺ T cells support the notion that OATD-02 promotes functional cytotoxic polarization. These findings are consistent with those of Sosnowska et al.37, who reported that arginase inhibition relieves metabolic constraints on T cells, thereby enhancing PD-1 blockade. Similarly, Grzywa et al.47 and our previous studies31 demonstrated that OATD-02 boosts antitumor immunity in multiple preclinical models, whereas Pilanc et al.48 reported increased immune cell infiltration and potentiation of anti-PD-1 therapy by OATD-02 in glioblastoma models, underscoring the broad applicability of this cancer-agnostic mechanism across multiple tumor types. Furthermore, whereas CB-1158, an ARG1-restricted inhibitor, showed only modest increases in plasma L-arginine in clinical trials – likely owing to its limited intracellular activity29 – our findings indicate that the dual targeting of both the intracellular and the extracellular arginases by OATD-02 results in superior metabolic and immunomodulatory effects. Indeed, our results align with those of previous experiments30, in which we directly compared OATD-02 to CB-1158 and found that OATD-02 produced greater tumor growth inhibition in an ARG2-dependent model.
The above observations underscore the crucial role of L-arginine metabolism in shaping the tumor microenvironment (TME) and highlight the importance of considering spatial metabolic heterogeneity when evaluating arginase-targeting therapies. The uneven metabolic reprogramming observed in OATD-02-treated tumors suggests the need to address regional metabolic constraints, particularly in hypoxic tumor cores. Arginases, particularly ARG2, regulate tumor metabolism and immune suppression1,17. Their inhibition restores L-arginine availability, crucial for effective T-cell activation and antitumor immunity. Given its role in mitochondrial metabolism and nitrogen balance, intracellular ARG2 is a particularly relevant therapeutic target1. By inhibiting both isoforms, OATD-02 may provide broader therapeutic benefits than agents that selectively target ARG129,49. Tumor cells adapt metabolically to sustain growth and evade immune surveillance. L-arginine, polyamine, and proline metabolism play key roles in these processes, and OATD-02 disrupts their balance, reshaping the TME.
L-arginine fuels tumor cell proliferation and supports processes such as extracellular matrix production, but it is also a critical resource for immune cell function (e.g., as a precursor for nitric oxide and polyamines). Tumors frequently exploit L-arginine metabolism by upregulating arginases, which deplete local L-arginine and redirect its metabolic flux toward polyamine and proline biosynthesis to support tumor growth and adaptation17. Conversely, depriving immune cells of L-arginine has profound immunosuppressive effects. L-arginine deficiency impairs T-cell proliferation and activation by downregulating the CD3ζ chain of the T-cell receptor complex, thereby weakening TCR signaling23,50. It also skews T-cell differentiation, leading to diminished IFN-γ and IL-2 production and a shift away from effective Th1 immune responses1. Collectively, these effects create an immunosuppressive environment, highlighting why L-arginine availability is critical for antitumor immunity. L-arginine depletion promotes tumor growth and immune evasion. OATD-02 restores L-arginine levels, reinvigorating T-cell activity and enhancing antitumor immunity37,51. Concurrent targeting of ARG1 and ARG2 may further enhance these effects.
In addition to L-arginine, polyamines play a significant role in tumor progression and immune evasion through complementary metabolic mechanisms. Polyamines, such as spermidine and spermine, are essential for tumor cell proliferation and survival. They stabilize DNA, modify chromatin, and promote ribosome biogenesis, thereby accelerating tumor growth52. These oncometabolites also contribute to immunosuppression. The spermidine secreted by tumor cells can directly inhibit CD8⁺ T-cell activation by preventing proper TCR clustering, effectively acting as a metabolic immune checkpoint53,54 and highlighting how tumors exploit polyamine pathways to evade immune surveillance. Consistent with this notion, our study confirms the critical role of polyamine metabolism in the TME. OATD-02 treatment led to a notable reduction in intracellular spermidine and spermine levels in both CT26 and K562 cells. The depletion of polyamines observed in vitro was mirrored in tumor tissues – MALDI-MSI analysis of tumors from OATD-02-treated mice revealed a progressive decline in polyamine levels over time. Arginase inhibition by OATD-02 alters the metabolic balance in tumors, limiting the production of immunosuppressive polyamines and disrupting a key mechanism of tumor-mediated immune evasion. Given the dual role of polyamines in promoting tumor growth and suppressing immune responses, targeting polyamine synthesis has been explored as a therapeutic strategy to increase the efficacy of immunotherapy52. In this context, the polyamine-lowering effect of OATD-02 may contribute to creating a tumor microenvironment that is less conducive to immune escape, thereby improving the effectiveness of anti-PD-1 therapy. Notably, the sustained reduction in tumor polyamine levels observed in our study further supports the combination of OATD-02 with immune checkpoint inhibitors, particularly in tumors characterized by high polyamine levels and an immunosuppressive microenvironment53.
Proline metabolism represents another aspect of tumor metabolic adaptation, influencing extracellular matrix (ECM) remodeling, redox homeostasis, and survival under stress. Proline biosynthesis contributes to oxidative stress resistance by maintaining the NADP⁺/NADPH balance, buffering redox status, and supporting tumor cell survival, particularly under hypoxic conditions9,55. Additionally, proline plays a crucial role in collagen production, as its derivative, hydroxyproline, is a major structural component of collagen. Increased proline utilization promotes collagen-rich ECM deposition and stiffness, facilitating tumor invasion and metastasis. Notably, high collagen content in tumors often correlates with poor prognosis56,57. Interestingly, despite the metabolic significance of proline, OATD-02 treatment did not significantly alter the intracellular proline levels in our models. In both CT26 and K562 cells, proline concentrations remained relatively stable across all conditions, even as L-arginine and L-ornithine underwent substantial changes. These findings suggest that tumor cells may preserve proline homeostasis through alternative pathways that are unaffected by arginase inhibition. For instance, cancer cells can recycle proline from collagen-derived hydroxyproline or upregulate glutamine metabolism to sustain proline synthesis10,58. Indeed, the high glutamine content in our cell culture medium likely provided an alternative substrate for proline production, potentially masking any secondary effects of arginase inhibition on the L-ornithine–proline axis.
The immunomodulatory and protumorigenic impact of arginases has been well documented in solid tumors, including lung, breast, colorectal, pancreatic, prostate, melanoma, renal, ovarian, and esophageal cancers, where elevated arginase activity frequently correlates with enhanced tumor growth, metastasis, or immune evasion24,51,59,60,61,62,63. These findings underscore the broad relevance of L-arginine metabolism as a therapeutic target in oncology. However, the dependency on arginase activity varies among cancer types. For example, thymic epithelial tumors lack ARG1 expression, whereas in some lung cancers, ARG2 activity does not induce immunosuppression unless it is accompanied by additional factors such as nitric oxide production, highlighting the context-dependent role of arginases64,65. Nevertheless, accumulating evidence suggests that dysregulated L-arginine metabolism also plays a crucial role in hematologic malignancies. Notably, acute myeloid leukemia (AML) cells secrete arginase to deplete L-arginine, leading to T-cell suppression; in AML models, arginase inhibition restores T-cell proliferation and induces leukemia cell apoptosis36,66. Furthermore, multiple myeloma exhibits increased arginase activity in myeloid cells, contributing to systemic L-arginine depletion and T-cell dysfunction67. Chronic myeloid leukemia (CML) provides another example, as CML cells rely on ARG2 to adapt to hypoxic conditions and resist therapy. Inhibiting arginase in CML has been shown to counteract these resistance mechanisms68. Collectively, these findings extend the therapeutic rationale for arginase inhibition to a broad spectrum of malignancies.
Given the broad immunosuppressive role of arginase in the TME, its inhibition represents a promising strategy to enhance cancer immunotherapy. Owing to its dual ARG1/ARG2 targeting ability and potent immunomodulatory effects, OATD-02 is a strong candidate for combination therapies designed to overcome resistance to ICIs. Interestingly, a modest decrease in the expression of the exhaustion marker LAG-3 was observed in splenic CD4⁺ T cells, suggesting a potential alleviation of exhaustion-associated signaling, although the functional implications of this observation require further investigation. Multiple studies have demonstrated that increased arginase activity in tumors suppresses antitumor immunity by impairing CD8⁺ T-cell function and promoting immunosuppressive myeloid populations37. Early trials with arginase inhibitors, such as CB-1158 (numidargistat), validated arginase as a therapeutic target by showing that its inhibition restores T-cell proliferation and improves checkpoint blockade efficacy in preclinical models28. However, CB-1158 primarily targets extracellular ARG1 and has limited cell permeability, which may explain its modest clinical efficacy29. In contrast, the ability of OATD-02 to effectively inhibit both intracellular and extracellular arginases provides broader immunomodulatory effects31, potentially translating into enhanced therapeutic benefits when it is combined with ICIs.
On the basis of preclinical data, combining arginase blockade with ICIs appears to be one of the most promising combinatorial cancer therapies. AZD0011 significantly increased intratumoral L-arginine levels while reducing ornithine, and its combination with anti–PD-L1 therapy resulted in markedly greater tumor growth inhibition than either treatment alone69,70. In addition to checkpoint inhibitors, OATD-02–mediated metabolic reprogramming may enhance other immunotherapies. For example, Ye et al.71 demonstrated that cotargeting arginase and L-arginine-depleting enzymes potentiated T-cell activation and tumor regression in a melanoma model, suggesting that increasing L-arginine availability could improve the outcomes of adoptive T-cell transfer or cancer vaccines. A promising avenue for enhancing antitumor immunity is combining arginase inhibition with other metabolic interventions. In our previous studies30, we reported that adding OATD-02 to an IDO1 (indoleamine-2,3-dioxygenase) inhibitor (epacadostat) alongside anti–PD-L1 therapy resulted in significantly improved tumor control in a murine CT26 colorectal carcinoma model compared with dual or single agent treatments. This finding aligns with earlier studies indicating that simultaneous blockade of L-arginine and tryptophan catabolism can synergistically enhance antitumor immunity28. Given the capacity of OATD-02 to modulate both L-arginine and polyamine pathways, pairing it with IDO1 inhibitors may be particularly effective in counteracting metabolic immunosuppression. Another promising approach for combination therapy involves the integration of arginase inhibitors with targeted anticancer agents. Preclinical studies suggest that arginase blockade can increase the efficacy of certain tyrosine kinase inhibitors. For example, combining an arginase inhibitor with cabozantinib or lenvatinib has been shown to remodel the immune microenvironment and improve tumor control72. These findings suggest that OATD-02 may similarly enhance the effects of targeted therapies by mitigating tumor metabolic resistance mechanisms. In addition to targeted therapies, arginase inhibition may also act synergistically with other treatment modalities, including radiotherapy and NK cell–based immunotherapies. Notably, the combination of arginase inhibition with blockade of the inhibitory NK cell receptor NKG2 A has been reported to improve tumor control and enhance immune activation70, highlighting additional therapeutic opportunities for OATD-02 in multimodal treatment regimens.
Our findings provide strong justification for the continued clinical development of OATD-02 as a metabolic-immunotherapeutic agent. While our studies demonstrated that dual ARG1/ARG2 inhibition enhances immune responses by increasing intratumoral L-arginine30 and depleting polyamines, the full extent of tumor adaptation to prolonged L-arginine repletion remains to be explored. Future studies using patient-derived models will be essential to determine how metabolic compensation, such as glutamine or tryptophan dependence, may influence therapeutic outcomes28. Moreover, integrating MSI with functional immune profiling may provide deeper insights into how spatial metabolic heterogeneity impacts therapeutic responses, helping to refine treatment strategies and optimize combination approaches. Encouragingly, the ongoing phase I clinical trial (NCT05759923) will provide critical insights into the safety, pharmacokinetics, and pharmacodynamics of OATD-02 in patients with advanced solid tumors. These results will guide the design of future combination strategies, reinforcing the potential of OATD-02 as a novel metabolic-immunotherapeutic approach.
Conclusions
Over the past years, studies have demonstrated the therapeutic potential of ARG1 inhibition in restoring L-arginine availability and enhancing antitumor immunity28,29,69,70. However, recent studies have revealed a previously underappreciated role of ARG2, the mitochondrial isoform, in tumor metabolism and immune evasion, revealing its critical function in nitrogen balance, mitochondrial adaptation, and metabolic reprogramming39,40,41,73. To our knowledge, this is the first study providing direct metabolic evidence of pharmacological ARG2 inhibition, demonstrating that dual ARG1/ARG2 blockade reshaped the tumor microenvironment by increasing L-arginine availability, depleting polyamines, and enhancing immune responses. This mechanistic advantage over ARG1-restricted inhibitors such as numidargistat29 underscores the importance of targeting both arginase isoforms in metabolic therapy. Using MALDI-MSI spatial metabolomics, we mapped the metabolic consequences of OATD-02 at high spatial resolution, revealing widespread L-arginine accumulation in tumor tissues, a shift that aligns with enhanced T-cell activation. While metabolic adaptations to sustain L-arginine repletion remain a consideration, our findings reinforce the therapeutic relevance of ARG2 inhibition as a metabolic-immunotherapeutic strategy. Future research should focus on identifying tumor types that would benefit the most from dual ARG1/ARG2 inhibition. Dual arginase blockade represents a promising approach for overcoming immune suppression in tumors, offering a novel avenue for metabolic intervention in cancer treatment.
Data availability
The datasets generated and analyzed during the current study are not publicly available due to the data policy of Molecure SA but can be obtained from the corresponding author upon reasonable request.
Abbreviations
- 3R:
-
Replacement, reduction, refinement (ethical principles for animal research)
- ABH:
-
2(S)-amino-6-boronohexanoic acid
- ARG1:
-
Arginase 1
- ARG2:
-
Arginase 2
- BEC:
-
(S)-(2-boronoethyl)-L-cysteine
- BID:
-
twice a day
- CAFs:
-
Cancer-Associated Fibroblasts
- CD:
-
Cluster of Differentiation (e.g., CD8, CD4; surface markers on immune cells)
- CI:
-
confidence interval
- CT26:
-
Murine Colon Carcinoma Cell Line
- FACS:
-
Fluorescence-Activated Cell Sorting
- FBS:
-
Fetal bovine serum
- HPLC:
-
high-performance liquid chromatography
- HR:
-
hazard ratio
- ICIs:
-
Immune checkpoint inhibitors
- IgG2a:
-
Immunoglobulin G, subclass 2a
- IP:
-
intraperitoneal administration
- K562:
-
Human Chronic Myelogenous Leukemia Cell Line
- L-Arg :
-
L-arginine
- LC‒MS/MS:
-
Liquid chromatography‒tandem mass spectrometry
- MALDI-MSI:
-
Matrix-assisted Laser Desorption/Ionization Mass Spectrometry Imaging
- MDSCs:
-
Myeloid-Derived Suppressor Cells
- NO:
-
Nitric oxide
- NOS:
-
nitric oxide synthase
- PBS:
-
Phosphate-buffered saline
- PD1:
-
Programmed Cell Death Protein 1
- PO:
-
per os (oral administration)
- RPMI-1640:
-
Roswell Park Memorial Institute Medium
- RT:
-
room temperature
- SPF:
-
specific pathogen-free
- TAMs:
-
Tumor-associated macrophages
- TCR:
-
T-cell receptor
- TME:
-
Tumor microenvironment
References
Canè, S., Geiger, R. & Bronte, V. The Roles of Arginases and Arginine in Immunity (Nature Reviews Immunology. Nature Research, 2024).
Chen, C., Han, P. & Qing, Y. Metabolic heterogeneity in tumor microenvironment – A novel landmark for immunotherapy. Autoimmun. Rev. 23 (6), 103579 (2024).
Liu, X. et al. The significant role of amino acid metabolic reprogramming in cancer. Cell Commun Signal [Internet]. 2024 Jul 29 [cited 2025 Feb 24];22(1):380. Available from: https://pmc.ncbi.nlm.nih.gov/articles/PMC11285422/
Damiani, E. & Wallace, H. M. Polyamines and cancer. In: Methods in Molecular Biology. Humana Press Inc.; 469–488. (2018).
Wu, J. Y. et al. Polyamine metabolism and anti-tumor immunity. Front Immunol [Internet]. Feb 18 [cited 2025 Feb 24];16:1529337. Available from: https://www.frontiersin.org/articles/ (2025). https://doi.org/10.3389/fimmu.2025.1529337/full
Avtandilyan, N., Javrushyan, H., Petrosyan, G. & Trchounian, A. The Involvement of Arginase and Nitric Oxide Synthase in Breast Cancer Development: Arginase and NO Synthase as Therapeutic Targets in Cancer. Biomed Res Int. ;2018. (2018).
Mintz, J. et al. Current Advances of Nitric Oxide in Cancer and Anticancer Therapeutics. Vaccines (Basel) [Internet]. 2021 Feb 1 [cited 2025 Feb 23];9(2):94. Available from: https://pmc.ncbi.nlm.nih.gov/articles/PMC7912608/
Byers, S. et al. Proline metabolism in tumor growth and metastatic progression. Front. Oncol. | Www Frontiersin Org. [Internet]. 1, 776 (2020). Available from: www.frontiersin.org.
Geng, P., Qin, W. & Xu, G. Proline Metabolism in Cancer. Vol. 53, Amino Acidsp. 1769–1777 (Springer, 2021).
Phang, J. M., Liu, W., Hancock, C. N. & Fischer, J. W. Proline metabolism and cancer: emerging links to glutamine and collagen. Vol. 18, Current Opinion in Clinical Nutrition and Metabolic Care. Lippincott Williams and Wilkins; 71–77. (2015).
Akinjiyan, F. A. et al. DDR2-regulated arginase activity in ovarian cancer-associated fibroblasts promotes collagen production and tumor progression. Oncogene [Internet]. Jan 12 [cited 2025 Feb 24];43(3):189. (2023). Available from: https://pmc.ncbi.nlm.nih.gov/articles/PMC10786713/
Matos, A., Carvalho, M., Bicho, M. & Ribeiro, R. Arginine and Arginases Modulate Metabolism, Tumor Microenvironment and Prostate cancer ProgressionVol. 13 (Nutrients. MDPI, 2021).
Novita Sari, I. et al. Metabolism and Function of Polyamines in cancer ProgressionVol. 519p. 91–104 (Elsevier Ireland Ltd, 2021). Cancer Letters.
Fatima, Z., Abonofal, A. & Stephen, B. Targeting Cancer Metabolism to Improve Outcomes with Immune Checkpoint Inhibitors. J Immunother Precis Oncol [Internet]. 2023 May 1 [cited 2025 Feb 24];6(2):91. Available from: https://pmc.ncbi.nlm.nih.gov/articles/PMC10195018/
Li, H. et al. Targeting T-cell metabolism to boost immune checkpoint inhibitor therapy. Front Immunol [Internet]. 2022 Dec 7 [cited 2025 Feb 24];13:1046755. Available from: https://pmc.ncbi.nlm.nih.gov/articles/PMC9768337/
Wang, J. et al. Engineered microparticles modulate arginine metabolism to repolarize tumor-associated macrophages for refractory colorectal cancer treatment. J. Transl Med. 22 (1), 908 (2024).
Failla, M. et al. Opportunities and Challenges of Arginase Inhibitors in Cancer: A Medicinal Chemistry Perspective. J Med Chem [Internet]. ; (2024). Available from: https://doi.org/10.1021/acs.jmedchem.4c01429
Chen, J., Cui, L., Lu, S. & Xu, S. Amino acid metabolism in tumor biology and therapy. Cell Death & Disease 2024 15:1 [Internet]. 2024 Jan 13 [cited 2025 Feb 24];15(1):1–18. Available from: https://www.nature.com/articles/s41419-024-06435-w
Ricci, J. E. Tumor-induced metabolic immunosuppression: mechanisms and therapeutic targets. Cell. Rep. 44 (1), 115206 (2025).
García-Navas, R., Gajate, C. & Mollinedo, F. Neutrophils drive Endoplasmic reticulum stress-mediated apoptosis in cancer cells through arginase-1 release. Sci. Rep. ;11(1). (2021).
Niu, F. et al. Arginase: an Emerging and Promising Therapeutic Target for cancer TreatmentVol. 149 (Elsevier Masson s.r.l., 2022). Biomedicine and Pharmacotherapy.
Zea, A. H. et al. l-Arginine modulates CD3ζ expression and T cell function in activated human T lymphocytes. Cell. Immunol. 232 (1–2), 21–31 (2004).
Lu, J. et al. Myeloid-derived suppressor cells in cancer: therapeutic targets to overcome tumor immune evasion. Experimental Hematology & Oncology 2024 13:1 [Internet]. 2024 Apr 12 [cited 2025 Feb 24];13(1):1–24. Available from: https://ehoonline.biomedcentral.com/articles/https://doi.org/10.1186/s40164-024-00505-7
Zaytouni, T. et al. Critical role for arginase 2 in obesity-associated pancreatic cancer. Nature Communications 2017 8:1 [Internet]. 2017 Aug 14 [cited 2025 Feb 24];8(1):1–12. Available from: https://www.nature.com/articles/s41467-017-00331-y
Zhang, H. et al. Elevated expression of HIGD1A drives hepatocellular carcinoma progression by regulating polyamine metabolism through c-Myc–ODC1 nexus. Cancer Metab. ;12(1). (2024).
Martí i Líndez, A. A. et al. Mitochondrial arginase-2 is a cell-autonomous regulator of CD8 + T cell function and antitumor efficacy. JCI Insight ;4(24). (2019).
Pudlo, M., Demougeot, C., Girard-Thernier, C. & Arginase Inhibitors A Rational Approach Over One Century. Med Res Rev [Internet]. 2017 May 1 [cited 2025 Feb 24];37(3):475–513. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/med.21419
Steggerda, S. M. et al. Inhibition of arginase by CB-1158 blocks myeloid cell-mediated immune suppression in the tumor microenvironment. J. Immunother Cancer ;5(1). (2017).
Naing, A. et al. First-in-human phase 1 study of the arginase inhibitor INCB001158 alone or combined with pembrolizumab in patients with advanced or metastatic solid tumours. BMJ Oncol. ;3(1). (2024).
Grzybowski, M. M. et al. OATD-02 validates the benefits of Pharmacological Inhibition of arginase 1 and 2 in Cancer. Cancers (Basel) ;14(16). (2022).
Borek, B. et al. Arginase 1/2 inhibitor OATD-02: from discovery to first-in-man setup in cancer immunotherapy. Mol. Cancer Ther. 22 (7), 807–817 (2023).
Dudek, M. A. et al. 717TiP An open-label, multicentre, dose-escalation, first-in-human phase I study to evaluate safety, tolerability and antineoplastic activity of OATD-02 (dual arginase 1 and arginase 2 inhibitor) in patients with selected advanced and/or metastatic solid tumors. Annals of Oncology [Internet]. 2023 Oct 1 [cited 2025 Mar 10];34:S495. Available from: https://www.annalsofoncology.org/action/showFullText?pii=S0923753423027400
Schulz, S., Becker, M., Groseclose, M. R., Schadt, S. & Hopf, C. Advanced MALDI mass spectrometry imaging in pharmaceutical research and drug development. Curr Opin Biotechnol [Internet]. Feb 1 [cited 2025 Mar 10];55:51–9. (2019). Available from: https://pubmed.ncbi.nlm.nih.gov/30153614/
Spruill, M. L., Maletic-Savatic, M., Martin, H., Li, F. & Liu, X. Spatial analysis of drug absorption, distribution, metabolism, and toxicology using mass spectrometry imaging. Biochem Pharmacol [Internet]. 2022 Jul 1 [cited 2025 Mar 10];201. Available from: https://pubmed.ncbi.nlm.nih.gov/35561842/
Krause, I., Bockhardt, A., Neckermann, H., Henle, T. & Klostermeyer, H. Simultaneous determination of amino acids and biogenic amines by reversed-phase high-performance liquid chromatography of the Dabsyl derivatives. J. Chromatogr. A ;715(1). (1995).
Ng, K. P. et al. The arginase inhibitor Nω-hydroxy-nor-arginine (nor-NOHA) induces apoptosis in leukemic cells specifically under hypoxic conditions but CRISPR/Cas9 excludes arginase 2 (ARG2) as the functional target. PLoS One ;13(10). (2018).
Sosnowska, A. et al. Inhibition of arginase modulates T-cell response in the tumor microenvironment of lung carcinoma. Oncoimmunology ;10(1). (2021).
Borek, B., Gajda, T., Golebiowski, A. & Blaszczyk, R. Boronic acid-based Arginase Inhibitors in cancer ImmunotherapyVol. 28 (Elsevier Ltd, 2020). Bioorganic and Medicinal Chemistry.
Ochocki, J. D. et al. Arginase 2 suppresses renal carcinoma progression via biosynthetic cofactor pyridoxal phosphate depletion and increased polyamine toxicity. Cell. Metab. 27 (6), 1263–1280e6 (2018).
Setty, B. A. et al. Hypoxic proliferation of osteosarcoma cells depends on arginase II. Cell. Physiol. Biochem. 39 (2), 802–813 (2016).
Ino, Y. et al. Arginase II expressed in Cancer-Associated fibroblasts indicates tissue hypoxia and predicts poor outcome in patients with pancreatic Cancer. PLoS One ;8(2). (2013).
Emami Nejad, A. et al. The role of hypoxia in the tumor microenvironment and development of cancer stem cell: a novel approach to developing treatment. Cancer Cell International 2021 21:1 [Internet]. 2021 Jan 20 [cited 2025 Mar 10];21(1):1–26. Available from: https://cancerci.biomedcentral.com/articles/https://doi.org/10.1186/s12935-020-01719-5
Andersen, M. K. et al. Spatial differentiation of metabolism in prostate cancer tissue by MALDI-TOF MSI. Cancer Metab. ;9(1). (2021).
Abu Sammour, D. et al. Quantitative Mass Spectrometry Imaging Reveals Mutation Status-independent Lack of Imatinib in Liver Metastases of Gastrointestinal Stromal Tumors. Sci Rep [Internet]. 2019 Dec 1 [cited 2025 Mar 11];9(1). Available from: https://pubmed.ncbi.nlm.nih.gov/31337874/
Hinsenkamp, I. et al. Inhibition of Rho-Associated kinase 1/2 attenuates tumor growth in murine gastric Cancer. Neoplasia 18 (8), 500–511 (2016).
Tufail, M., Jiang, C. H. & Li, N. Altered metabolism in cancer: insights into energy pathways and therapeutic targets. Molecular Cancer 2024 23:1 [Internet]. 2024 Sep 18 [cited 2025 Mar 11];23(1):1–40. Available from: https://molecular-cancer.biomedcentral.com/articles/https://doi.org/10.1186/s12943-024-02119-3
Grzywa, T. M. et al. Potent but transient immunosuppression of T-cells is a general feature of CD71 + erythroid cells. Commun. Biol. ;4(1). (2021).
Pilanc, P. et al. A novel oral arginase 1/2 inhibitor enhances the antitumor effect of PD-1 Inhibition in murine experimental gliomas by altering the immunosuppressive environment. Front. Oncol. ;11. (2021).
Chen, C. L., Hsu, S. C., Ann, D. K., Yen, Y. & Kung, H. J. Arginine Signaling and cancer MetabolismVol. 13 (Cancers. MDPI, 2021).
Rodriguez, P. C. et al. Arginase I Production in the Tumor Microenvironment by Mature Myeloid Cells Inhibits T-Cell Receptor Expression and Antigen-Specific T-Cell Responses. Cancer Res [Internet]. Aug 15 [cited 2025 Feb 26];64(16):5839–49. (2004). Available from: /cancerres/article/64/16/5839/511544/Arginase-I-Production-in-the-Tumor
Czystowska-Kuzmicz, M. et al. Small extracellular vesicles containing arginase-1 suppress T-cell responses and promote tumor growth in ovarian carcinoma. Nat. Commun. ;10(1). (2019).
Casero, R. A., Murray Stewart, T. & Pegg, A. E. Polyamine Metabolism and Cancer: Treatments, Challenges and OpportunitiesVol. 18p. 681–695 (Nature Publishing Group, 2018). Nature Reviews Cancer.
Hibino, S. et al. Tumor cell derived spermidine is an oncometabolite that suppresses TCR clustering for intratumoral CD8 + T cell activation. Proc. Natl. Acad. Sci. U S A. 120, 24 (2023).
Kay, K. E. et al. Tumor cell-derived spermidine promotes a pro-tumorigenic immune microenvironment in glioblastoma via CD8 + T cell inhibition [Internet]. (2023). Available from: https://doi.org/10.1101/2023.11.14.567048
Linder, S. J. et al. Inhibition of the proline metabolism rate-limiting enzyme P5CS allows proliferation of glutamine-restricted cancer cells. Nat. Metab. 5 (12), 2131–2147 (2023).
D’Aniello, C., Patriarca, E. J., Phang, J. M. & Minchiotti, G. Proline Metabolism in Tumor Growth and Metastatic ProgressionVol. 10 (Frontiers Media S.A., 2020). Frontiers in Oncology.
Wang, D. et al. Targeting the glutamine-arginine-proline Metabolism axis in cancerVol. 39 (Taylor and Francis Ltd., 2024). Journal of Enzyme Inhibition and Medicinal Chemistry.
Phang, J. M. Proline Metabolism in Cell Regulation and cancer Biology: Recent Advances and HypothesesVol. 30p. 635–649 (Mary Ann Liebert Inc., 2019). Antioxidants and Redox Signaling.
Zea, A. H. et al. Arginase-producing myeloid suppressor cells in renal cell carcinoma patients: a mechanism of tumor evasion. Cancer Res [Internet]. Apr 15 [cited 2025 Feb 26];65(8):3044–8. (2005). Available from: https://pubmed.ncbi.nlm.nih.gov/15833831/
Yu, Y. et al. Arginase-II promotes melanoma migration and adhesion through enhancing hydrogen peroxide production and STAT3 signaling. J. Cell. Physiol. 235 (12), 9997–10011 (2020).
Su, X. et al. Breast cancer-derived GM-CSF regulates arginase 1 in myeloid cells to promote an immunosuppressive microenvironment. J. Clin. Invest. ;131(20). (2021).
Bednarz-Misa, I. et al. Esophageal squamous cell carcinoma is accompanied by local and systemic changes in L-arginine/NO pathway. Int. J. Mol. Sci. 21 (17), 1–26 (2020).
Wang, X. et al. Arginase-1 inhibition reduces migration ability and metastatic colonization of colon cancer cells. Cancer & Metabolism 2022 11:1 [Internet]. 2023 Jan 13 [cited 2025 Feb 24];11(1):1–14. Available from: https://cancerandmetabolism.biomedcentral.com/articles/https://doi.org/10.1186/s40170-022-00301-z
Rotondo, R. et al. Arginase 2 is expressed by human lung cancer, but it neither induces immune suppression, nor affects disease progression. Int. J. Cancer. 123 (5), 1108–1116 (2008).
Umemura, S. et al. Arginase pathway markers of Immune-Microenvironment in thymic epithelial tumors and small cell lung Cancer. Clin. Lung Cancer. 23 (2), e140–e147 (2022).
Mussai, F. et al. Targeting the arginine metabolic brake enhances immunotherapy for leukaemia. Int J Cancer [Internet]. Oct 15 [cited 2025 Feb 24];145(8):2201. (2019). Available from: https://pmc.ncbi.nlm.nih.gov/articles/PMC6767531/
Panina, S. B., Pei, J. & Kirienko, N. V. Mitochondrial metabolism as a target for acute myeloid leukemia treatment. [cited 2025 Feb 24]; Available from: https://doi.org/10.1186/s40170-021-00253-w
Weis-Banke, S. E. et al. Arginase-2-specific cytotoxic T cells specifically recognize functional regulatory T cells. J. Immunother Cancer ;10(10). (2022).
Mlynarski, S. N. et al. Discovery of (2R,4R)-4-((S)-2-Amino-3-methylbutanamido)-2-(4-boronobutyl)pyrrolidine-2-carboxylic acid (AZD0011), an actively transported prodrug of a potent arginase inhibitor to treat Cancer. J. Med. Chem. ; (2024).
Doshi, A. S. et al. Novel Arginase Inhibitor, AZD0011, Demonstrates Immune Cell Stimulation and Antitumor Efficacy with Diverse Combination Partners. Mol Cancer Ther [Internet]. 2023 May 1 [cited 2025 Feb 26];22(5):630–45. Available from: /mct/article/22/5/630/726100/Novel-Arginase-Inhibitor-AZD0011-Demonstrates.
Ye, P. H. et al. A novel combination therapy of arginine deiminase and an arginase inhibitor targeting arginine metabolism in the tumor and immune microenvironment. Am. J. Cancer Res. [Internet]. 13 (5), 1952–1969 (2023). Available from: www.ajcr.us/.
Zhu, S. et al. Combination Strategies To Maximize the Benefits of cancer ImmunotherapyVol. 14 (Journal of Hematology and Oncology. BioMed Central Ltd, 2021).
Zaytouni, T. et al. Critical role for arginase 2 in obesity-Associated pancreatic cancer. Nat. Commun. ;8(1). (2017).
Acknowledgements
The authors would like to thank EU-OPENSCREEN for providing access to advanced research infrastructure, which enabled the MALDI‒MSI analyses conducted at CeMOS, Mannheim University of Applied Sciences (Mannheim, Germany). Their support was instrumental in visualizing the metabolic alterations induced by OATD-02.
Funding
The MSI part of the study was funded by the European Commission Horizon 2020 Project EU-OPENSCREEN-DRIVE (Grant Agreement No. 823893) and by the German Federal Ministry of Education and Research (BMBF; grant 13FH8I09IA – M2Aind-DrugsData) within the FH-Impulse framework (to CH)
Author information
Authors and Affiliations
Contributions
MMG, YU, AM, TR, RB, CH, and ZZ designed the study. MMG, MM, AK, and AZ performed the in vivo experiments. MMG, AZ, and PP conducted the in vitro experiments. AK, KG, MK, and MS-R carried out the flow cytometry analyses. MALDI–MSI analyses were performed by YU and TB. AT and TR conducted the LC–MS analyses, whereas AKJ performed the HPLC analyses. MMG, AM, AK, YU, and TB analyzed the data. MMG wrote the initial version of the manuscript, while ZZ, CH, and YU revised and edited it. MG, ZZ, and CH supervised the project and coordinated the study. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Several authors (MMG, AM, AK, KG, MS-R, MK, AZ, PP, MM, AT, TR, RB, ZZ) are employees of Molecure SA, which holds proprietary rights to OATD-02. The remaining authors declare no competing interests.
Ethical approval
All procedures complied with the Public Health Service Policy on Humane Care and Use of Laboratory Animals and the Guidelines for the Welfare and Use of Animals in Cancer Research. Ethical approval for the study was granted by the 1 st Local Ethics Committee for Animal Experiments in Warsaw, Poland (approval no. 891/2019). All experiments were performed in accordance with relevant institutional and national guidelines and regulations. The study is reported in accordance with the ARRIVE guidelines (https://arriveguidelines.org). Procedures involving animals, including anesthesia and euthanasia, were conducted in accordance with veterinary best practice and followed the recommendations outlined in the American Veterinary Medical Association (AVMA) Guidelines for the Euthanasia of Animals (2020).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Grzybowski, M.M., Uçal, Y., Muchowicz, A. et al. Metabolomic reprogramming of the tumor microenvironment by dual arginase inhibitor OATD-02 boosts anticancer immunity. Sci Rep 15, 18741 (2025). https://doi.org/10.1038/s41598-025-03446-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-03446-1