Abstract
The genus Andrias includes the largest extant salamanders, and is comprised of one Japanese species A. japonicus and four Chinese species. The fossil record of the giant salamander is incomplete and modern giant salamanders are not differentiated osteologically among species, making it difficult to identify bones at the species level. In this study, we re-examined a fossil series of giant salamander discovered from a cave on Shikoku Island, Japan. We obtained ancient DNA from the fossil and confirmed that the partial sequence of mitochondrial DNA was identical to that of extant A. japonicus. These remains were dated to the Late Pleistocene, however, the result of carbon-14 dating in this study estimated the age as more recent, approximately 3,500–4,100 years ago. Currently, there is a small population of A. japonicus in Shikoku, but it is far removed from the fossil discovery area. Our findings suggest that wild A. japonicus in western Shikoku may have been extirpated very recently.
Similar content being viewed by others
Introduction
Cryptobranchidae is a family of giant salamanders that includes the largest extant amphibians which has a total body length of over 1.8 m. Two extant genera are recognized: Andrias in East Asia and Cryptobranchus in North America. Andrias consists of four species from China and one species, A. japonicus, from Japan. According to the latest molecular phylogenetic analyses, the Cryptobranchidae appeared around 61.3 Ma, and Andrias japonicus and Chinese Andrias spp. are estimated to have diverged around 15.8 Ma1. Furthermore, molecular and morphological studies strongly suggest an Asian origin for cryptobranchids with subsequent expansions into Europe and North America2. However, the fossil record in Asia after the Miocene is not well documented. Thus, the time of the origin of these species is poorly understood.
The genus Andrias was first proposed for the fossil species A. scheuchzeri but now includes both fossil and extant species. This is because the morphological characters of fossil species from Europe and Asia are highly conserved. The extant species A. davidianus was once misclassified and synonymized with the fossil species A. scheuchzeri3. On the other hand, the extant Chinese giant salamander A. davidianus has recently been separated into four species: A. davidianus sensu stricto, A. jiangxiensis, A. sligoi, and A. cheni4,5,6, this revision is largely based on molecular data but also includes some minor differences in external morphology. However, molecular data was unavailable from fossil species, and comparative osteological studies on extant Andrias species are scant. Due to this situation, the taxonomy of Andrias has proven very confusing and it has been extremely difficult to taxonomically integrate fossil and extant species.
In this context, giant salamander remains were discovered in the Shikimizu bed and exposed at a quarry in Ozu City, Ehime Prefecture, Shikoku Island, Japan (Fig. 1B). The fissure deposits at the quarry of the Shikimizu bed are mainly composed of brown brecciated clay 14 m thick underlain by layers of black sand, yellow clay, and gravel, and overlain by red clay and black humus7. A total of nearly 1,900 fossils have been collected from this deposit, of which about 1,800 are vertebrates including fishes, amphibians, reptiles, birds, and mammals8. The exact age of the deposit is unknown, but it has been inferred to date from the Late Pleistocene or Early Holocene based on the mammalian species composition9.
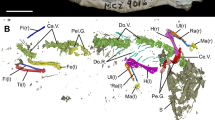
The giant salamander remains from Ehime contain the right parietal, a chunk of parasphenoid close to its right border, three right dentaries, and three trunk vertebrae (Fig. 1A). All specimens are incomplete and have yellowish-white color. In a previous study, these remains were identified as the extant species A. japonicus based on morphological comparisons only with A. japonicus skeletons, and at least four individuals can be identified based on size differences and duplicated elements; at least one individual is small-sized (approximately 500 mm long) and at least three are large-sized (approximately 900 mm long)7.
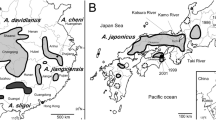
(A) The fossil remains of Andrias japonicus from Ehime Prefecture. a–e (OCM-10, OCM-14, OCM-13, OCM-12, OCM-11): left lateral view of trunk vertebrae; f (OCM-8): dorsal view of right parietal; g (OCM-9): ventral view of right parasphenoid; h–j (OCM-7, OCM-6, OCM-5); medial view of right dentaries. (B) Map of central and southwestern Japan showing localities of samples used in the study. The red star indicates the locality of the fossil sample of A. japonicus. For locality numbers, refer to Table 1. (C) Maximum likelihood phylogenetic reconstruction of partial cyt b gene sequences from giant salamanders. Asterisks on nodes indicate significant supports (Bootstrap value ≥ 70%). Locality numbers are concordant to Fig. (B). This figure was created using Adobe Photoshop 26.1 and Adobe Illustrator 29.1.
However, as mentioned above, given the taxonomic issues within Andrias, it is impossible to identify the fragmentary remains from Ehime Prefecture as A. japonicus without considering comparisons with other species. Moreover, unidentified Pliocene Cryptobranchid fossils were reported from Oita, Japan, which may represent a new species of Andrias or even a different genus10. So, it is also not appropriate to identify the remains from Ehime as A. japonicus solely because it was discovered in Japan. Identifying the giant salamander remains from Ehime has been challenging, but we believe that recent advances in ancient DNA methodology may overcome this problem. The aim of this study is to reexamine the taxonomic assignment of a rare series of Asian giant salamander remains through an integrated approach combining morphological comparisons, ancient DNA analysis, and radiocarbon dating.
Identification based on morphological characteristics of bones
The giant salamander from Ehime can be assigned to Cryptobranchoidea and Cryptobranchidae because of its large body size, absence of the spinal nerve foramina in trunk vertebrae, and unicapitate transverse processes in trunk vertebrae11. They are also assigned to Cryptobranchinae because the vertebrae are rectangular in lateral view, and the prezygapophyses usually project above the bases of the neural spine12. Furthermore, these remains can be assigned to the genus Andrias based on a larger angle (about 40 degrees) between the neural spine and the axis of the vertebral centrum (19–37 degrees in Andrias vs. 15–20 degrees in Cryptobranchus)3,13. The giant salamander from Ehime differs from the Miocene to Pliocene A. scheuchzeri3,14,15 by possessing a more slender and slightly curved dentary. Therefore, the morphological character of the Ehime giant salamander fossil suggests that it may be assignable to a recent extant species.
However, the diagnostic features that differentiate A. japonicus and A. davidianus sensu lato (maxilla and anterior part of dentary) were not preserved. We can provisionally assign the fossil only to the generic level until reliable data on the comparative osteology of the modern species of Andrias or new material from giant salamander remains appear from Ehime.
Radiocarbon dating and phylogenetic analysis
We obtained collagen and ancient DNA from one of the trunk vertebrae. This sample was dated to 4,140–3,587 cal BP by carbon-14 dating. The mitochondrial cytb sequence extracted from the remain of Andrias sp. from Ehime Prefecture, Shikoku Island, Japan is identical with A. japonicus samples collected from Ishikawa, Mie, Nara, Shiga, Kyoto, Osaka, Tokushima, Hyogo, Okayama, Tottori, Hiroshima, Shimane, Oita and Kumamoto prefectures in Japan. The phylogenetic analysis recovered the fossil as part of the Western Japan clade of the species (Fig. 1C), and hence we identify the fossil sample as the extant species, A. japonicus.
Most ancient DNA (aDNA) studies were conducted on mammals including humans, with very little focus on amphibians. This is because amphibian habitats are mainly wetlands, ponds and other watery places, where genetic information is often destroyed by DNase. Among the extremely few examples of amphibian aDNA, successful extraction has been achieved from frog remains in Australia16 and small salamander remains in the United States17, both discovered in caves. The remains we analyzed in this study were also found in caves. This suggests that the cave environment may be relatively suitable for preserving DNA.
Currently, there are small populations of A. japonicus in Shikoku18, but they are not in the area where these fossils were discovered. This suggests that wild A. japonicus in Ehime Prefecture likely became extinct very recently. Giant salamander bones were also found, along with fish and mammal bones, in a cave site inhabited by humans in Hiroshima19. In the Ehime caves, several individuals were also found together with fishes and mammals. Therefore, it is highly likely that humans consumed giant salamanders as a food or a ritual. Although direct evidence like a cut mark was not found on the present bones, human activities might have contributed to the local extirpation of A. japonicus in this area.
Materials and methods
Samples
The fossil of Andrias japonicus is designated as the cultural property of Ozu City and is deposited at the Ozu City Museum. These specimens include three right dentary fragments (OCM-5, OCM-6, OCM-7), a right parietal fragment (OCM-8), a right parasphenoid fragment (OCM-9), and five trunk vertebrae (OCM-10, OCM-11, OCM-12, OCM-13, OCM-14).
Radiocarbon dating
We extracted collagen fraction with > 30k Da from a small fragment (~ 100 mg) of each bone for C dating following the procedures20,21, and the 14C ages were measured using the HVEE Tandetron accelerator mass spectrometer at Nagoya University22. We used CALIB23 v8.2 for 14C age calibration.
DNA extraction
DNA extractions were conducted in a purpose-built positive-pressure ancient DNA (aDNA) laboratory located within the Museum of Natural and Environmental History, Shizuoka. This laboratory is physically isolated from other molecular laboratories with independent air intake, and there has been strict compliance with the ‘one-way rule’24 upon using these laboratories. We followed procedures employed20 for aDNA extraction. A negative extraction was included in each aDNA extraction procedure.
Mitochondrial DNA sequencing and phylogenetic analysis
We amplified the partial mitochondrial gene cytochrome b using the newly designed primers andrias_cytb4 F (ACAGGGTCAAGCAATCCAAC) and andrias_cytb4R (TGGGAGTAGCAGTGAAATCAAT). The expected amplification length of the targeted gene fragment is 93 bp (excluding primers). The total volume of the PCR reaction was 25 µl containing 0.48 µM of each primer, 0.16 mM of each dNTP, 4 mM MgCl2, 1 M betaine, 1 U of AmpliTaq Gold 360 DNA Polymerase (Applied Biosystems), 1× PCR buffer, and 2 µL of extracted DNA. The PCR amplification conditions included initial denaturation at 95℃ for 9 min, followed by 45 cycles at 95℃ for 20 s, 53℃ for 30 s, and 72℃ for 30 s, with a final extension at 72℃ for 4 min. A negative control was included in each PCR. The PCR mixtures were prepared in the aDNA laboratory, and a thermal cycler installed in the post-PCR laboratory was used for PCRs. PCR products were purified using ExoSAP-IT Express (Applied Biosystems). The PCR products were sequenced using the PCR primers and the BigDye v3.1 Cycle Sequencing kit and visualized in an ABI 3130xl Genetic Analyzer or 3730xl DNA Analyzer (Applied Biosystems, Waltham, Massachusetts, USA). In addition to the newly produced sequence data, we used previously published sequence data for Andrias japonicus, A. davidianus, A. cheni, and A. jiangxiensis for comparisons and Cryptobranchus alleganiensis and Hynobius nebulosus sequences were used for outgroup (Table 1). Sequences were aligned using the MUSCLE algorithm27 in MEGA X28 with default parameters. We used Maximum likelihood (ML) methods to conduct phylogenetic analyses. Suitable models were selected using Modeltest-NG29 with the corrected Bayesian Information Criterion (BIC). The ML analysis was conducted using RAxML-NG30. Uncorrected p-distances among the sequences were calculated using MEGA X.
Ancient DNA authenticity
We amplified and sequenced the mtDNA cytB partial sequence two times independently, and confirmed consistency between two independently-amplified sequences. A negative control (i.e., a PCR mixture using the negative extract as a DNA template) was included in each PCR, and no amplification from the negative control was detected. We performed all aDNA extraction and amplification procedures in the Museum of Natural and Environmental History, Shizuoka where amplification of modern amphibian DNA has never been conducted since its inception.
Data availability
The newly obtained sequence is deposited in figshare (10.6084/m9.figshare.28295672).
References
Marr, M. M. et al. What’s in a name? Using species delimitation to inform conservation practice for Chinese giant salamanders (Andrias spp). Evol. J. Linn. Soc. 3, kzae007 (2024).
Browne, K. R. et al. The giant salamanders (Cryptobranchidae): part A. palaeontology, phylogeny, genetics, and morphology. Amphib Reptile Conserv. 5, 17–29 (2012).
Estes, R. in Caudata. In Handbuch Der Paläoherpetologie Part 2. 1–115 (eds Wellnhofer, P.) (Fischer, G., 1981). Gymnophyona.
Turvey, S. T. et al. Historical museum collections clarify the evolutionary history of cryptic species radiation in the world’s largest amphibians. Ecol. Evol. 9, 10070–10084 (2019).
Chai, J. et al. Discovery of a wild, genetically pure Chinese giant salamander creates new conservation opportunities. Zool. Res. 43, 469–480 (2022).
Gong, Y. A. et al. A new species of the giant salamander of the genus Andrias from Qimen, Anhui, China (Amphibia: Cryptorchiidae). Chin. J. Zool. 58, 651–657 (2023).
Shikama, T. & Hasegawa, Y. Discovery of the fossil giant salamander (Megalobatrachus) in Japan. Trans. Proc. Palaeontol. Soc. Jpn., N. S. 45, 197–200 (1962).
Hasegawa, Y. et al. Late pleistocene fossil vertebrate assemblage from the limestone fissure deposits (Shikimizu Bed) of the Karaiwadani quarry, Hijikawa-cho, Ozu-shi, Ehime Prefecture. Bull. Gunma Mus. Natl. Hist. 19, 17–38 (2015). [in Japanese].
Iwamoto, M. On a skull of a fossil macaque from the Shikimizu limestone quarry in the Shikoku district, Japan. Primates 16, 8394 (1975).
Matsui, M., Kitabayashi, E., Takahashi, K. & Sato, S. A fossil giant salamander of the genus Andrias from Kyushu, Southern Japan. Res. Rep. Lake Biwa Mus. 18, 72–28 (2001). [in Japanese].
Gao, K. Q. & Shubin, H. N. Late jurassic salamandroid from Western Liaoning, China. Proc. Natl. Acad. Sci. 109, 5767–5772 (2012).
Cope, E. D. The batrachia of North America. Bull. U S Natl. Mus. 34, 1–525 (1889).
Meszoely, C. North American fossil cryptabranchid salamanders. Am. Midl. Nat. 75, 495–515 (1966).
Westphal, F. Die tertiären und Rezenten Eurasiatischen Riesen salamander (Genus Andrias, urodela, Amphibia). Palaeontogr Abt A. 110, 20–92 (1958).
Böttcher, R. Neue Funde von Andrias scheuchzeri (Cryptobranchidae, Amphibia) Aus der Süddeutschen molasse. Stuttgarter Beiträge Zur Naturkunde B. 131, 1–38 (1987).
Seersholm, F. V. et al. Ancient DNA from bulk bone reveals past genetic diversity of vertebrate fauna on Kangaroo Island, Australia. Quatern Sci. Rev. 262, 106962 (2021).
McMenamin, K. S. & Hadly, A. E. Ancient DNA assessment of tiger salamander population in Yellowstone National park. PLoS ONE. 7, e32763 (2012).
Matsui, M., Tominaga, A., Liu, W. Z. & Tanaka-Ueno, T. Reduced genetic variation in the Japanese giant salamander, Andrias japonicus (Amphibia: Caudata). Mol. Phylogen Evol. 49, 318–326 (2008).
Matsui, A. Kankyo Kokogaku E No Shotai: Hakkutsu Kara Wakaru Shoku, Toire, Senso [An Invitation To Environmental Archaeology: Food, Toilets, and War as Revealed by Excavation]4–6 (Iwanami Shoten, 2005). [in Japanese].
Kishida, T., Namigata, S., Nakanishi, T., Niiyama, Y. & Kitagawa, H. Dolphins from a prehistoric midden imply long-term philopatry of delphinids around Tokyo Bay. Biol. J. Linn. Soc. 143, blad159 (2024).
Kishida, T. et al. Hidden population turnover of small odontocetes in the Northwestern North Pacific during the holocene. Biol. Lett. 21, 20240525 (2025).
Nakamura, T. et al. The HVEE Tandetron AMS system at Nagoya university. Nucl. Instrum. Methods Phys. Res. B. 172, 52–57 (2000).
Stuiver, M. & Reimer, P. J. Extended 14C data base and revised CALIB 3.0 14C age calibration program. Radiocarbon 35, 215–230 (1993).
Fulton, T. L. et al. Setting up an ancient DNA laboratory. In Ancient DNA: Methods and Protocols (eds. Shapiro, B. 1–13 (2019).
Yan, F. et al. The Chinese giant salamander exemplifies the hidden extinction of cryptic species. Curr. Biol. 28, 590–592 (2018).
Nishikawa, K. et al. Discovery of ex situ individuals of Andrias sligoi, an extremely endangered species and one of the largest amphibians worldwide. Sci. Rep. 14, 2575 (2024).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucle Acids Res. 32, 1792–1797 (2004).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Darriba, D. et al. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 37, 291–294 (2020).
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B. & Stamatakis, A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455 (2019).
Acknowledgements
We are grateful to Souichi Okazaki (Ozu City, Ehime Prefecture) for his support during this work. We thank Nagisa Tasaki for helping us design the primers. We are grateful to Christine Kaiser and David Beamer for their contributions to improving our manuscript. This research was generously supported by a philanthropic gift from Y. Arai, K. Haruki, M. Imai, A. Kunihiro, K. Nishikawa, K. Okamoto, T. Watahiki, K. Yamashita, GS Craft Co. Ltd., and 513 supporters donated to the crowdfunding organized by READYFOR Inc. All experimental procedures in this study followed the experimental animal guidelines of Kyoto University (approval no. 29-A-7 and 30-A-7). The present study was conducted under the permissions issued by the Japan Agency of Cultural Affairs to Kanto Nishikawa for research in Kyoto City (no. 420) and in Kyoto Prefecture (no. 710).
Funding
This work was partly supported by the Environment Research and Technology Development Fund (JPMEERF20204002) of the Environmental Restoration and Conservation Agency of Japan to Kanto Nishikawa and by Fukada Grant-in-Aid of Fukada Geological Institute and JST SPRING Grant Number JPMJSP2110 to Masahiro Noda. Radiocarbon dating was carried out as a part of the joint-research program of the Institute for Space-Earth Environmental Research, Nagoya University. The ancient DNA extraction experiments were supported by JSPS KAKENHI Grant Number 23K23956 to Takushi Kishida.
Author information
Authors and Affiliations
Contributions
M. N. conducted morphological examinations; T. K. conducted ancient DNA analysis; H. K. conducted radiocarbon dating; I. F. conducted molecular analyses; M. N., T. K., H. K., I. F., and K. N. wrote the manuscript. The first draft was written by M. N., and all authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Noda, M., Kishida, T., Kitagawa, H. et al. Ancient DNA integrates fossil and modern giant salamander taxonomy. Sci Rep 15, 18642 (2025). https://doi.org/10.1038/s41598-025-03496-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-03496-5