Abstract
The human milk microbiome is dominated by typical oral and skin bacteria, suggesting that bacterial communities from the infant mouth and maternal skin contribute to the development of the human milk microbiome. It is postulated that breastfeeding characteristics, such as breastfeeding frequency and duration, could lead to different levels of exposure to oral and skin bacteria, and subsequently, altered bacterial profiles in human milk. To investigate the associations between breastfeeding characteristics and the human milk microbiome, this study analysed milk samples collected from 56 participants at 3 months postpartum by full-length 16 S rRNA gene sequencing. Breastfeeding characteristics (breastfeeding frequency, total 24 h breastfeeding duration, and 24 h milk removal volume) were recorded by the participants using the 24 h test weighing protocol. The milk microbiome was largely robust to breastfeeding practices, with only one association detected between breastfeeding characteristics and the milk microbiome. Duration of breastfeeding from the sampled breast (per 24 h) was weakly positively associated with the relative abundance of Streptococcus salivarius (P = 0.035). No associations with alpha nor beta diversity were detected. In conclusion, variations in breastfeeding characteristics do not have a major impact on the composition or diversity of the human milk microbiome.
Similar content being viewed by others
Introduction
Human milk contains many microorganisms including Staphylococcus, Streptococcus, Acinetobacter, and Cutibacterium species1,2. The origins of the human milk microbiome are thought to include the bacterial communities from the infant oral cavity and maternal skin, since a large number of typical oral and skin bacteria are detected in human milk2. This hypothesis was further supported by a previous study which found that mothers who fed their infants with expressed milk had a less rich human milk microbiome compared to mothers who fed the infants directly at the breast3. Another study suggested that breastfeeding frequency and total 24 h breastfeeding duration could be associated with specific bacterial taxa including Bifidobacterium4. According to recommendations from the Centers for Disease Control and Prevention, the majority of fully breastfed infants need to be fed every 2–4 h, resulting in 8–12 breastfeeding sessions per day in the first few months5. However, even within fully breastfeeding dyads, there is a wide range of normal breastfeeding characteristics, including time spent breastfeeding, volume of milk produced, and number of breastfeeds. There is little evidence regarding the impact of such characteristics on the human milk microbiome. Therefore, this study aimed to investigate how breastfeeding frequency, total 24 h breastfeeding duration, and 24 h milk production were associated with human milk bacterial profiles.
Results
The characteristics of the mothers included in the analysis are presented in Table 1. The majority of mothers fully breastfed their infants at the time of sample collection (96.4%).
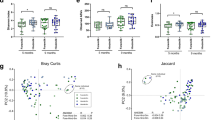
In these human milk samples (collected at 3 months postpartum), 573 bacterial genera and 30,290 operational taxonomic units (OTUs) were detected. A total of 20 bacterial taxa with relative abundance ≥ 0.5% were identified, representing 88% of the bacterial sequences recovered from the samples. The human milk microbiota was dominated by typical skin and oral taxa including Streptococcus mitis (mean relative abundance 15.6% ± 20.6%), Streptococcus salivarius (12.9% ± 19.3%), and Cutibacterium acnes (12.3% ± 17.6%) (Fig. 1a) with a high level of inter-individual variation (Fig. 1b).
(a) Composition of the human milk microbiota at 3 months postpartum. Operational taxonomic units (OTUs) with a mean relative abundance of ≥ 0.5% are displayed; (b) Genus-level inter-individual variation in milk bacterial profiles of the 56 participants. Genera that comprised < 2% are grouped together as “other”.
Given that the breastfeeding characteristics analysed here (breastfeeding frequency, breastfeeding duration, and volume of milk removed) are likely interrelated, we tested for correlations between these factors. Breastfeeding frequency was positively correlated with 24 h breastfeeding duration (r = 0.299, P = 0.003) and 24 h milk removal (r = 0.310, P = 0.002). However, there was no correlation between 24 h breastfeeding duration and 24 h milk removal (r = 0.117, P = 0.206). Since samples were only collected from one breast, we restricted our analysis of breastfeeding characteristics to the sampled breast only. However, it should be noted that breastfeeding characteristics did not differ between the sampled and non-sampled breast (Student’s t-test, total 24 h breastfeeding duration: P = 0.175; breastfeeding frequency: P = 0.297; 24 h milk removal volume: P = 0.265).
Overall, only a single breastfeeding characteristic was associated with a small, though potentially biologically relevant, alteration to the human milk microbiome (Supplementary Table 1). Twenty-four-hour breastfeeding duration from the sampled breast was weakly positively associated with the relative abundance of S. salivarius (coefficient = 0.026, P = 0.035; Fig. 2). No other associations were detected for the other 19 OTUs analysed here, nor for Shannon diversity or richness of the human milk microbiome (Supplementary Fig. 1). Similarly, breastfeeding characteristics were not associated with the Bray-Curtis distance between the human milk samples.
Discussion
This pilot study provides new data demonstrating that the human milk microbiome is largely unaffected by breastfeeding practices. We found that at 3 months postpartum, the total duration of breastfeeding per 24 h was positively associated with the relative abundance of S. salivarius. However, it is important to note the small effect size of this association (coefficient = 0.026). None of the breastfeeding characteristics analysed here were associated with Shannon diversity, richness, or the beta diversity of the human milk microbiome. Thus, our findings indicate that the human milk microbiome is largely robust to variations in breastfeeding characteristics, with no structural changes, despite a single minor variation in composition.
In our study, a longer duration of breastfeeding was associated with a greater relative abundance of S. salivarius in human milk. As S. salivarius has been demonstrated to be a part of the healthy infant oral microbiome2,6,7, and milk from the infant oral cavity may flow back into the mammary gland during breastfeeding8, this finding is biologically plausible. It is postulated that there is bidirectional exchange of bacteria between the infant oral cavity and the lactating mammary gland9,10. This theory is supported by data from a previous study of Guatemalan mothers, showing a higher level of typical oral bacteria, including Streptococcus species, in mothers who exclusively breastfed compared to those who mix-fed11. However, other strain-level evidence demonstrates that typical oral species, including Streptococcus species, are present in pre-colostrum prior to contact with the infant oral cavity12. Thus, these taxa are likely true colonisers of the mammary gland, with their abundance potentially bolstered by interaction with the infant oral cavity. Given the paired t-test results showing no difference in the breastfeeding characteristics between left and right breasts in the individual participants, these findings could be representative of the other breast which we did not take samples from. Breastfeeding duration and breastfeeding frequency were positively correlated in this study. It is therefore perhaps surprising that breastfeeding frequency was not associated with the relative abundance of S. salivarius. This observation may suggest that it is the amount of time that the mammary gland is exposed to the infant oral cavity, as opposed to the number of bouts of exposure, that matters. This theory makes sense from a microbial ecology standpoint. Future studies with larger sample sizes are needed to investigate further the associations between exposure to the infant oral microbiome and the abundance of oral bacteria in the milk microbiome.
Our study did not find any associations between breastfeeding characteristics and the diversity of the human milk microbiome. In a recent study on 46 mother-infant pairs living in the United States (US), breastfeeding frequency was found to be negatively associated with milk microbiome richness4. Given that longer time spent breastfeeding and higher breastfeeding frequency would lead to the mammary gland having more exposure to the bacterial communities from maternal skin and infant mouth, which may lead to a more diverse microbiome in human milk, these findings were unexpected. It is important to note that, although infants feed at night, the US study only observed breastfeeding from 7 am to 7 pm. In addition, the recording of breastfeeding ‘bouts’ (defined as breastfeeding sessions separated by > 30 seconds) could artificially increase the breastfeeding frequency analysed. Therefore, the findings of this study need to be interpreted with caution. Another factor to consider in interpreting both the results of the present study and the US study is the use of 16S rRNA gene sequencing. Since this methodology provides relative abundance rather than absolute quantification of microorganisms, it is possible that repeated exposure to skin and oral bacteria increases the absolute abundance of certain bacteria thereby reducing the likelihood of identifying associations with lower-abundance bacteria13. Therefore, quantitative methods such as qPCR may be useful to further investigate associations between breastfeeding practices and the structure of the human milk bacterial communities.
This study has several strengths. Firstly, the majority of the participants fully breastfed their infants, which reduced the possibility of formula use altering the human milk microbiome as a confounder. We also prospectively collected real-time data on breastfeeding characteristics to avoid recall and observer bias. Further, the use of full-length 16 S rRNA gene sequencing allowed us to investigate the human milk microbiota at the species level. Despite these strengths, this study also has limitations. Despite best practice to limit reagent-based contamination14, we identified some taxa in our milk profiles that were likely to be contaminants, namely Burkholderia, Ralstonia, and Bradyrhizobium. Reagent-based contamination is ubiquitous in microbiome data and is especially problematic for low-biomass samples such as human milk. Secondly, the sample size of our analysis was relatively small and contained mainly Caucasian and multiparous women. Given that breastfeeding characteristics vary geographically4,15, the findings of this study may not be representative of other populations, especially in low- and middle-income countries. Thirdly, data on breastfeeding characteristics were only available at 3 months postpartum, so the findings of this study may not be generalised beyond the first few months of lactation. Finally, volume of milk removed from the sampled breast was used as a proxy for 24 h milk production of the breast, since milk production data were calculated from both breasts16. Therefore, the associations with milk removal from the sampled breast might not accurately reflect the associations between milk production and the human milk microbiome at the individual breast level. Additionally, there are other breastfeeding practices that were not analysed here that would be valuable to investigate, including hand/pump expression, pumping frequency, breast pump cleaning practices, and allomaternal feeding. Future longitudinal studies with larger sample sizes, human milk samples and milk production data collected for both breasts, as well as the inclusion of more feeding and milk removal practices are needed for in-depth investigations into the associations between breastfeeding characteristics and the human milk microbiome.
In conclusion, this study identified a single minor change in the composition of the human milk microbiome associated with breastfeeding characteristics, suggesting that variance in breastfeeding practices largely does not alter the milk microbiome.
Methods
Study design and sample collection
This study included mothers from the Breastfeeding Longitudinal Observational Study of Mothers and kids (BLOSOM) birth cohort. Cohort recruitment, inclusion and exclusion criteria have previously been reported17. Briefly, healthy mother-infant dyads were included, with all mothers in this cohort fully (n = 54) or predominantly (n = 2) breastfeeding at the 3-month sample time point. Written informed consent was obtained from all participants, and the study was approved by the Human Research Ethics Committee of The University of Western Australia (RA/4/20/4023). All methods were performed in accordance with the relevant guidelines and regulations.
As described previously2, the participants chose one breast to donate milk samples over the study period. The human milk samples were self-collected aseptically by the participants and stored in their home freezer for up to 18 h, before being transported to the laboratory and stored at -80⁰C until further analysis.
Breastfeeding characteristic data collection
Breastfeeding characteristics were recorded at 3 months postpartum by the participants using the 24 h test weighing protocol18. Given that milk was only sampled from one breast, our analysis focused on breastfeeding characteristic data from that breast only. However, milk production is necessarily calculated using data from both breasts, therefore we used total 24 h milk removal (g) from the sampled breast as a proxy for 24 h milk production of the breast. Twenty-four-hour milk removal from the sampled breast was calculated by summing the differences in weight of the infant before and after every breastfeed and differences in weights of milk collection bottles before and after every pumping session over 24 h. Each feed from a single breast was counted as one breastfeed. The total 24 h breastfeeding duration (minutes) was summed from the duration of all recorded breastfeeds from the sampled breast. Mothers with complete human milk microbiome and breastfeeding characteristic data available at 3 months postpartum were included in the analysis (n = 56).
Microbiome analysis
The 56 samples used in the microbiome analysis were a subset of those from a broader cohort (539 human milk samples collected at 9 time points from 84 mothers). All these samples were analysed in 96 well-format batches, consisting of 94 samples, one negative extraction control, and one negative amplification control. In total, seven plates (batches) were used. To avoid batch effects, samples were randomised across the plates. Total DNA was extracted from 1 mL aliquots of milk using the QIAGEN MagAttract Microbial DNA Isolation Kit on the Kingfisher Flex platform. The full-length 16 S rRNA gene was amplified in 30 µL reactions consisting of: 1X AccuStart II PCR ToughMix, 0.3 µM each of the PacBio barcoded forward (27 F) and reverse (1492R) primers, 0.75 µL each of dsDNase and DTT from the ArcticZymes PCR decontamination kit, 3 µL nuclease-free water, and 6 µL of template. The PCR cycling conditions consisted of an initial heating stage of 94 °C for 3 min, followed by 38 cycles of 94 °C for 30 s, 52 °C for 30 s, and 72 °C for 2 min 30 s and a final extension stage of 72 °C for 5 min. Amplicons were multiplexed in equimolar concentrations, and Macherey Nagel NucleoMag NGS beads were used to concentrate and purify the multiplexed pools. Sequencing was performed on the PacBio Sequel II platform at The Australian Genome Research Foundation (AGRF).
Sequence data processing was performed using mothur v.1.48.019. Sequence quality filtering was performed based on length (1336–1742 bp), alignment (position 1044–43116), and homopolymers (≤ 9). The SILVA reference alignment v132 was used to align the sequences20. VSEARCH was used to exclude the chimeric sequences after alignment21. OTUs were generated using the cluster split method with a cutoff distance of 0.03. Subsampling was performed at 1999 reads, resulting in an average sequence coverage of 97%. Our subsampling method resulted in the exclusion of 3 low-yield samples. The remaining samples had an average sequence read count of 11,878 ± 7,754 and a median of 10,538. Alpha diversity was assessed by richness and Shannon diversity, and beta diversity was assessed using Bray-Curtis distance. Compositional analysis was performed at the OTU level, with OTUs with a mean relative abundance of ≥ 0.5% included in the analysis (n = 20). OTU abundance data was centred log ratio (CLR) transformed. Initial genus-level taxonomic assignment was mapped using the SILVA taxonomy database (v132), with species-level assignment manually performed using BLAST22. To assign species-level taxonomy with BLAST, the top hit for sequences generated using whole genome or long-amplicon 16 S rRNA gene sequencing was selected, resulting in sequence identity score of ≥ 98.25% for all assignments. Genus-level identity is provided in cases of OTUs having an equally good match for more than one bacterial species, or in cases where all good matches (> 90% identity score) were at the genus-level or above. BLAST-derived taxonomy including percent ID and coverage data are provided in Supplementary Table 2.
Reads recovered from negative extraction and amplification controls are listed in Supplementary Table 3.
Statistical analysis
Kendall’s Tau correlations were used to assess correlations between breastfeeding characteristics. Paired t-tests were used to analyse the difference in the breastfeeding characteristics between left and right breasts in the individual participants.
In this small investigative study, we modelled associations between the human milk microbiome and breastfeeding characteristics using linear models individually fitted to Shannon diversity, richness, and CLR-transformed abundance of each OTU as the response variables. Fixed effects were breastfeeding frequency, 24 h breastfeeding duration, and 24 h milk removal from the sampled breast. Full model outputs are reported in Supplementary Table 1. Associations between these variables and beta diversity were assessed by PERMANOVA using the adonis2 function of the vegan R package23 with a backward selection model with 999 permutations. P-values of < 0.05 were considered significant for all statistical analyses. R (version 4.3.1) was used for all statistical analysis and data visualisation24.
Data availability
Sequence data have been deposited in NCBI’s SRA (BioProject accession: PRJNA1229851).
References
Kordy, K. et al. Contributions to human breast milk Microbiome and enteromammary transfer of Bifidobacterium breve. PLoS One. 15, e0219633 (2020).
Cheema, A. S. et al. Exclusively breastfed infant microbiota develops over time and is associated with human milk oligosaccharide intakes. Int J. Mol. Sci 23, 859 (2022).
Moossavi, S. et al. Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell. Host Microbe. 25, 324–335e324 (2019).
Holdsworth, E. A. et al. Breastfeeding patterns are associated with human milk Microbiome composition: the Mother-Infant microbiomes, behavior, and ecology study (MIMBES). PLoS One. 18, e0287839 (2023).
Centres for Disease Control and Prevention. How much and how often to breastfeed (2024, accessed 7 Jul 2024). https://www.cdc.gov/nutrition/infantandtoddlernutrition/breastfeeding/how-much-and-how-often.html.
Dashper, S. G. et al. Temporal development of the oral Microbiome and prediction of early childhood caries. Sci. Rep. 9, 19732 (2019).
Ferretti, P. et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut Microbiome. Cell. Host Microbe. 24, 133–145e135 (2018).
Ramsay, D. T., Kent, J. C., Owens, R. A. & Hartmann, P. E. Ultrasound imaging of milk ejection in the breast of lactating women. Pediatrics 113, 361–367 (2004).
Stinson, L. F. et al. The human milk microbiome: who, what, when, where, why, and how? Nutr. Rev. 79, 529–543 (2021).
Demmelmair, H., Jiménez, E., Collado, M. C., Salminen, S. & McGuire, M. K. Maternal and perinatal factors associated with the human milk Microbiome. Curr. Dev. Nutr. 4, nzaa027 (2020).
Lopez Leyva, L., Gonzalez, E., Solomons, N. W. & Koski, K. G. Human milk Microbiome is shaped by breastfeeding practices. Front. Microbiol. 13, 885588 (2022).
Ruiz, L. et al. Microbiota of human precolostrum and its potential role as a source of bacteria to the infant mouth. Sci. Rep. 9, 8435 (2019).
LeMay-Nedjelski, L. et al. Maternal diet and infant feeding practices are associated with variation in the human milk microbiota at 3 months postpartum in a cohort of women with high rates of gestational glucose intolerance. J. Nutr. 151, 320–329 (2021).
Stinson, L. F., Keelan, J. A. & Payne, M. S. Identification and removal of contaminating microbial DNA from PCR reagents: impact on low-biomass Microbiome analyses. Lett. Appl. Microbiol. 68, 2–8 (2019).
Fouts, H. N., Hewlett, B. S. & Lamb, M. E. A biocultural approach to breastfeeding interactions in central Africa. Am. Anthropol. 114, 123–136 (2012).
Jin, X. et al. Maternal breast growth and body mass index are associated with low milk production in women. Nutrients 16, 745 (2024).
Cheema, A. S. et al. Human milk lactose, insulin, and glucose relative to infant body composition during exclusive breastfeeding. Nutrients 13, 1456 (2021).
Ramsay, D. T. et al. Milk flow rates can be used to identify and investigate milk ejection in women expressing breast milk using an electric breast pump. Breastfeed. Med. 1, 14–23 (2006).
Schloss, P. D. et al. Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2012).
Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahé, F. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584 (2016).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930 (2003).
R Core Team, R. R: a language and environment for statistical computing (2013).
Acknowledgements
This work was funded by an unrestricted research grant from Medela AG (Switzerland) administered by The University of Western Australia. Ruomei Xu was funded by the International Research Training Program Tuition Fees Offset Scholarship and University Postgraduate Award. The authors would like to acknowledge all of the participants and Ms. Erika van den Dries for collecting the samples, as well as Dr Matthew Payne for the use of his laboratory space and equipment.
Funding
This research was funded by unrestricted research grant from Medela AG. The funder had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Author information
Authors and Affiliations
Contributions
Conceptualisation, L.F.S., M.P.N. and D.T.G.; methodology, L.F.S.; formal analysis, R.X. and L.F.S.; investigation, R.X. and A.S.C.; resources, D.T.G.; data curation, A.H.W., Z.G., J.L.M., S.L.P. and R.X.; writing—original draft preparation, R.X.; writing—review and editing, A.H.W., M.P.N., L.F.S., A.S.C., J.L.M., S.L.P., Z.G. and D.T.G.; visualisation, R.X.; supervision, L.F.S., D.T.G. and M.P.N.; funding acquisition, D.T.G. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
D.T.G. declares past participation in the Scientific Advisory Board of Medela AG. D.T.G., J.L.M., Z.G. and L.F.S. receive a salary from an unrestricted research grant from Medela AG administered by The University of Western Australia. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results. R.X., M.P.N., A.S.C., A.H.W. and S.L.P. declare no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xu, R., Nicol, M.P., Cheema, A.S. et al. The human milk microbiome is minimally associated with breastfeeding practices. Sci Rep 15, 19308 (2025). https://doi.org/10.1038/s41598-025-03907-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-03907-7