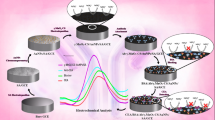
Abstract
Staphylococcal enterotoxin B (SEB) holds critical importance in disease diagnosis, food safety, and public health due to its high toxicity and potent pathogenicity. Traditional immunoassay methods for detecting SEB often exhibit insufficient accuracy and robustness. This study leverages machine learning technology to integrate the quantitative measurement advantages of electrochemical methods with the strong specificity of immunoassays, achieving high-precision coupled electrochemical immunodetection of SEB. Firstly, an electrochemical immunosensing system was developed to capture the target analyte SEB by immobilizing specific antibodies on the electrode surface. Cyclic voltammetry (CV) was utilized to accurately characterize the immune response process. Secondly, feature selection methodologies within machine learning are utilized to identify eight key parameters from CV curves that are highly related to SEB concentration. This enhancement significantly improves both the accuracy and interpretability of SEB measurement data. Lastly, a multivariate linear regression algorithm is employed to effectively train and fit the extracted feature data. This approach successfully mitigates noise introduced by variations in electrode batches, experimental conditions, and operational techniques-thereby enabling robust quantitative measurements of SEB concentration with high precision. The entire detection process requires only 20 μL sample and is accomplished in just two minutes. This method can detect antigen concentrations at both ng/mL and μg/mL levels, with a detection limit of 1 ng/mL. The \(R^2\) score for predicting SEB antigen concentration is approximately 0.999, accompanied by a mean absolute percentage error (MAPE) of 6.09% This approach achieves high precision, robustness, and specificity in SEB detection, offering extensive detection range, rapid response time, and cost-effectiveness, presenting new opportunities for identifying various pathogenic toxins.
Similar content being viewed by others
Introduction
Staphylococcus aureus is a prevalent foodborne pathogen commonly found on human skin and within nasal cavities. It is responsible for a variety of diseases due to its secretion of numerous virulence factors1. Among these, staphylococcal enterotoxin B (SEB) stands out as a highly heat-stable toxin frequently detected in contaminated food, which poses significant challenges for complete elimination during food processing2. Given SEB’s potent toxicity and high transmissibility, it can induce a range of food poisoning symptoms such as vomiting, diarrhea, and abdominal pain; in severe cases, it may lead to life-threatening complications like toxic shock syndrome3. Current methods for detecting SEB primarily include Enzyme-Linked Immunosorbent Assay (ELISA), Lateral Flow Immunoassay (LFIA), antibody engineering techniques, and electrochemical approaches. However, these methods exhibit limitations regarding sensitivity, specificity, response time, and cost-effectiveness. Such constraints present challenges in fully addressing the demands of modern food safety testing. Therefore, developing a detection method that integrates high specificity, precision, robustness, rapid response times, and low costs for quantifying SEB concentration remains an ongoing challenge.
ELISA is a technique utilized to determine the presence of a target substance through the binding of specific antibodies and the detection of colorimetric changes resulting from enzyme-labeled substances. Despite its remarkable sensitivity and accuracy, ELISA presents significant limitations regarding operational costs, complexity, and applicability4. Specifically, the high operational costs associated with ELISA stem from expenses related to reagents and equipment, as well as prolonged experimental durations. Furthermore, the complexity of the procedure necessitates experienced personnel to ensure reliable results5. Additionally, ELISA typically requires several hours to a full day for completion, rendering it less suitable for applications that demand rapid responses6. Its cumbersome operation-primarily conducted in laboratory settings7-also restricts its portability and field application.
LFIA employs nitrocellulose membranes as solid-phase carriers, enabling the movement of sample solutions through capillary action on test strips. This method incorporates specific antibodies or antigens for detection. In comparison to ELISA, LFIA presents several advantages, including ease of operation, minimal technical requirements, and enhanced portability for field testing8, making it particularly suitable for rapid detection applications. However, the sensitivity of LFIA is relatively low, typically yielding only semi-quantitative results. For example, Cho and Paek developed an immunochromatographic analysis system in their study that achieves semi-quantitative analysis via barcode-style signal reading9. This limitation restricts its application in contexts necessitating high-precision analysis10. Furthermore, LFIA still requires manual operation and result recording while offering limited data analysis capabilities and insufficient quantitative accuracy of results11.
Electrochemical methods analyze target molecules by monitoring changes in current or voltage at the electrode surface, providing advantages of high sensitivity and rapid response. Techniques such as cyclic voltammetry (CV) and electrochemical impedance spectroscopy (EIS) are commonly employed, enabling real-time tracking of electrochemical reactions to meet various analytical needs. However, these methods encounter several challenges in practice. First, the performance of electrochemical sensors can be compromised by factors such as electrode surface contamination, environmental interference, and the stability of electrode materials. These issues may lead to inconsistent detection results. For instance, Hanssen et al. demonstrated that electrode fouling typically forms an increasingly impermeable layer that hinders direct interaction between the analyte and the electrode surface, resulting in unstable detection outcomes12. Second, these techniques often require sophisticated equipment and precise operating conditions, imposing significant demands on both the operational environment and technical personnel13. Furthermore, the selectivity and specificity of electrochemical sensors can be adversely affected by interfering substances; thus rigorous optimization and calibration are essential14. Despite their notable advantages in terms of sensitivity and detection speed, the stringent requirements for operational conditions and equipment-along with stability concerns-limit the widespread application of electrochemical methods15.
To address the stability and selectivity challenges encountered in electrochemical methods, integrating these techniques with machine learning technology can optimize sensor performance and enhance detection accuracy and reliability through data-driven analysis. Machine learning models are capable of predicting and compensating for environmental factors (such as temperature, pH, impurities, etc.), thereby calibrating the electrochemical data output from sensors and improving detection reliability16. Furthermore, machine learning facilitates the automation of sensor data processing and analysis, which reduces human error, increases detection efficiency, and further optimizes sensor performance by automatically adjusting operational conditions. Most importantly, machine learning enables the development of more accurate models for interpreting electrochemical signals by leveraging extensive experimental data. This capability significantly enhances both detection precision and consistency17.
The integration of machine learning methodologies as a pivotal technology for predicting electrochemical data within immunological frameworks and identifying target analytes has been extensively investigated in the realm of biomedical data analysis and prediction. For instance, Jiang et al.18 proposed the application of support vector machines (SVM) to forecast survival rates among gastric cancer patients and assess the benefits of adjuvant chemotherapy, thereby underscoring its potential utility in the biomedical domain. Additionally, in neuroscience research, Aileni et al.19 employed artificial neural networks (ANN) to analyze electroencephalogram (EEG) data, successfully predicting the onset of epileptic seizures. Furthermore, Ludwig et al.20 utilized decision tree algorithms to examine gene expression data, achieving effective classification across various cancer types. In cardiovascular disease research, Alzahrani et al.21 applied logistic regression to investigate cardiovascular risk factors within transgender populations and predict heart attack risk factors. These algorithms proficiently manage complex data structures and facilitate the development of accurate predictive models. Moreover, recent studies, such as the one by Enginler et al.22, have integrated machine learning algorithms with electrochemical biosensors to predict biomarker concentrations for detecting canine mammary tumors. This research demonstrated the significant potential of combining electrochemical detection with machine learning for non-invasive, accurate diagnostics, offering a promising approach to biomarker prediction in medical diagnostics. However, thus far, there has been no literature reported on the accurate quantification of target analytes in electrochemical sensors based on immunological principles with the assistance of machine learning methods. We have explored the practical challenges faced in this detection.
This study presents a novel method that integrates machine learning, electrochemical techniques, and immunological principles for the determination of SEB. The experimental procedure is illustrated in Fig. 1. Initially, SEB samples with varying concentrations are prepared. These samples then undergo electrochemical analysis. Subsequently, features are extracted and analyzed visually. Finally, machine learning is employed to achieve precise regression of SEB concentration. This artificial intelligence (AI)-driven electrochemical system demonstrates high efficiency, cost-effectiveness, and rapid predictive capabilities. SEB was chosen as the target analyte due to its critical importance in food safety and public health, alongside the stringent requirements for sensitivity, accuracy, and speed in detection methods23. Our approach first addresses the challenge of specific recognition of SEB target analytes through immunodetection technology by achieving highly specific binding to SEB antigens via immobilization of SEB antibodies on the electrode surface. Following this step, CV is utilized to record current response changes associated with electrochemical reactions at the electrode surface; this provides high-precision electrochemical data pertinent to SEB concentration and establishes a foundation for accurate quantitative measurement. Lastly, machine learning algorithms-incorporating multi-feature parameters along with multivariate linear regression-effectively tackle the issue of high-precision data fitting. This enhances both data analysis accuracy and robustness while overcoming limitations inherent in traditional quantitative methods. Furthermore, this methodology offers a broad detection range along with rapidity and low-cost automated detection capabilities; it paves new research avenues for developing portable automatic detection devices as well as detecting other biomarkers24.
Materials and methods
The systematic workflow diagram (Fig. 2) comprehensively illustrates the operational pipeline of our detection system, encompassing critical modules including electrode modification, electrochemical measurement, feature engineering, and data analysis.
Chemicals and materials
The SEB antigen antibodies were supplied by the State Key Laboratory of Pathogen and Biosecurity. \(\beta\)-Mercaptoethylamine was procured from Shanghai Macklin Biochemical Co., Ltd. Ultrapure water was obtained from China National Pharmaceutical Group Chemical Reagent Co., Ltd. 2.5% Glutaraldehyde was purchased from Shanghai Aladdin Biochemical Technology Co., Ltd. Potassium ferricyanide \(({\hbox {K}}_{3}[{\hbox {Fe}}{\hbox {(CN)}}_{6}])\) was acquired from Xilong Scientific Co., Ltd. Phosphate-buffered saline (PBS) used in this study was sourced from Sigma-Aldrich. A 5 mM solution of \({\hbox {K}}_{3}[{\hbox {Fe}}{\hbox {(CN)}}_6]\) served as the electrolyte in the experiments, owing to its favorable redox properties that are easily measurable.25. All other chemicals utilized were of analytical grade and did not require further purification. Antibodies and antigens were prepared in PBS buffer, while all other solutions were formulated using ultrapure water.
Instruments
Electrochemical experiments were conducted using a CHI660e Electrochemical Workstation (Shanghai Chenhua Co., Ltd., Shanghai, China). Screen-printed electrode (SPE) were obtained from Ercon Inc. (Wareham, MA, USA). As depicted in Fig. 3A, the electrodes measure 34 mm in length, 12 mm in width, and 0.3 mm in thickness; the working electrode has a diameter of 4 mm. The substrates for both the working and auxiliary electrodes are composed of gold, while the reference electrode is made of silver. All experiments were carried out in a 5 mL beaker.
Electrode modification
The quantitative detection of SEB was conducted utilizing electrochemical immunoassay techniques. The modification and measurement process of the SPE is illustrated in Fig. 3. This procedure consists of four key stages: electrode pretreatment and modification, antibody immobilization, antigen capture, and subsequent electrochemical measurement and analysis. A series of chemical modification steps were implemented to functionalize the surface of the working electrode, thereby enhancing its biocompatibility, stability, and sensitivity.
A Dimensions of the SPE. B The cross-linking reaction between cysteamine and glutaraldehyde on the surface of the working electrode. C Following antibody incubation, ethanolamine is introduced for blocking, after which antigen binding to the antibody occurs. D An electrochemical reaction takes place when the electrode is immersed in a 5 mM \(\hbox {K}_{3}[\hbox {Fe}\hbox {(CN)}_6]\) solution and connected to an electrochemical workstation.
Chemical modification and functionalization
Pretreatment of the electrodes significantly enhances their voltammetric characteristics and activity26. The pretreatment steps for the SPE are as follows: Initially, 20 μL of a 10 mM cystamine solution was applied precisely to the surface of the working electrode, ensuring uniform coverage across the entire area. This cystamine solution introduced \(-\hbox {NH}_{2}\) groups, which serve as functional groups for subsequent chemical reactions. The working electrode was then incubated at room temperature, allowing cystamine to react with the electrode surface and form covalent amino groups. After 2 h of incubation, the electrode was washed with phosphate-buffered saline (PBS) and then allowed to air-dry naturally. Subsequently, 20 \(\upmu\)L of a 2.5% glutaraldehyde solution was applied precisely to the cystamine-modified surface of the electrode, again ensuring uniform coverage. Glutaraldehyde acts as an electrophilic reagent that reacts with the amino groups introduced by cystamine (nucleophilic reagents), resulting in stable imine bonds27, as illustrated in Fig. 3B. Following a 15-minute incubation period, the electrode surface was washed with PBS and allowed to air-dry naturally to remove any unreacted glutaraldehyde and other contaminants, thereby minimizing their impact on subsequent measurements.
Immobilization of antibodies on the electrode and specific binding of antigens
The introduction of functional groups and the formation of chemical bonds during the pre-treatment process not only enhance the chemical reactivity and specificity of the electrode surface but also provide active sites for subsequent antibody immobilization, thereby improving the performance of the biosensor28.
On the chemically modified SPE working electrode surface, 20 μL of a 100 μg/mL SEB antibody solution was applied and incubated at room temperature for 2 h. Following incubation, the electrode surface was washed with PBS buffer to remove any unbound antibodies. Subsequently, 20 μL of 0.5M ethanolamine was applied to the working electrode area and incubated at 37 °C for 15 min to react with any unreacted glutaraldehyde. This step deactivates residual electrophilic reagents and prevents non-specific cross-linking reactions with subsequently added biomolecules (such as antibodies)29. The electrode was then washed again with PBS buffer and allowed to air-dry at room temperature. At this stage, the immunoelectrode was successfully obtained.
A series of antigen solutions with varying concentrations were prepared, with 20 μL of each solution applied to the surface of the working electrode and incubated at 37 °C for 15 min. Following incubation, the electrode was washed with PBS buffer to remove any unbound antigens from the antibody-coated surface and then thoroughly dried (as illustrated in Fig. 3C). Subsequently, the Cys-NH2/GA/SEB Ab(SEB)/SEB (where Cys-NH2 refers to cystamine; GA denotes glutaraldehyde; SEB Ab(SEB) represents staphylococcal enterotoxin B antibody; and SEB stands for staphylococcal enterotoxin B) electrode was immersed in a 5 mM K3[Fe(CN)6] solution for electrochemical measurement (as depicted in Fig. 3D).
To ensure adequate recognition of antigens by antibodies immobilized on the electrode surface, it is essential that the quantity of antibody molecules significantly exceeds that of antigen molecules. In this study, the concentration of anti-SEB solution utilized ranged from 10 to 100,000 times greater than that of the SEB solution. Furthermore, all chemical reagents employed in this experiment do not interfere with SEB activity, thereby preventing degradation or other alterations to SEB.
Electrochemical measurement parameter settings
When the immunoelectrode adequately captures antigen molecules from the sample, it generates a specific electrochemical response. Quantitative analysis of this phenomenon through electrochemical measurements allows the detection of minute amounts of antigen content within the sample.
The SEB antigen-antibody complex generally exhibits minimal electrical activity and redox properties. It influences the oxidation-reduction reaction of \(\hbox {K}_{3}[\hbox {Fe}\hbox {(CN)}_6]\) during the reaction process, manifesting as varying degrees of inhibition depending on the SEB concentration30. CV experiments are typically conducted in a \(\hbox {K}_{3}[\hbox {Fe}\hbox {(CN)}_6]\) electrolyte solution with a potential range of − 0.3 to 0.5 V, a scan rate of 100 mV/s, and a sampling interval of 1 mV. The sensitivity is set at 1e−4 A/V.
Antibody characteristics and challenges
Antibodies, also known as immunoglobulins, are highly specific proteins synthesized by the immune system. They are characterized by their intricate structure and distinct functions. As depicted in Fig. 4A, antibodies consist of two heavy chains and two light chains, which together form an antigen-binding site. This binding site is situated between the variable regions of both the light and heavy chains and is stabilized by multiple disulfide bonds, as illustrated in Fig. 4B. Each heavy chain and light chain comprises a variable region and a constant region; it is the variable region that determines the antibody’s specificity31 through its ability to recognize and bind to particular antigen molecules. The binding process relies not only on structural complementarity between the antibody and the antigen but also on the formation of a stable antigen-antibody complex. This complex modifies the biochemical properties of both the antigen and antibody, thereby triggering various biological effects such as activation of immune cells or suppression of antigen function32.
However, the intricate structure and specificity of antibodies present considerable challenges to traditional immunoassay methods. Conventional techniques typically depend on the signal intensity generated by antibody-antigen interactions for measurement; however, this signal is often influenced by the complexity of antigen structures and experimental conditions. The high specificity and sensitivity of antibody binding sites imply that even minor structural alterations or environmental fluctuations can significantly affect detection outcomes, leading to data instability and poor reproducibility. For instance, factors such as impurities in the sample, temperature variations, or pH changes may disrupt the signal from antibody-antigen interactions, resulting in inconsistencies in detection signals that compromise both accuracy and reliability of results.
Artificial intelligence methods present substantial advantages in addressing these challenges. Machine learning algorithms adeptly process and analyze complex data patterns, enabling the identification and correction of noise and interference within signals. By training on extensive experimental datasets, AI can effectively model the intricate characteristics of antibody-antigen interactions, thereby enhancing detection accuracy. Moreover, AI technology facilitates the integration of data from diverse experimental conditions, mitigating variability in antibody-antigen interactions and yielding more consistent and stable results. This capability for data integration allows AI to compensate effectively for differing experimental environments and conditions, thus improving result consistency and reproducibility. Furthermore, AI algorithms possess the ability to optimize and adjust the detection process in real-time, which enhances automation levels while reducing errors associated with manual operations. Consequently, this leads to faster and more accurate immunoassays.
Dataset
The dataset utilized in this study is derived from electrochemical experiments. Following the completion of all modification steps, the electrode was immersed in a potassium ferricyanide solution, and cyclic voltammetry curves were recorded using an electrochemical workstation to obtain experimental data. The experimental setup is illustrated in Fig. 1. SEB was categorized into six concentration gradients based on the common detection range: 0, 1 ng/mL, 10 ng/mL, 100 ng/mL, 1 μg/mL, and 10 μg/mL (as depicted in Fig. 5A). In accordance with immunoassay principles, a uniform antibody concentration of 100 μg/ml was employed throughout the study to ensure adequate capture of the target substance. Different concentrations of SEB antigen were added dropwise to the modified electrodes; after a incubation period of fifteen minutes, they were rinsed with PBS buffer and dried. Subsequently, the electrodes were submerged in a 5 mM K3[Fe(CN)6] solution where six cyclic voltammetry curves were recorded. Figure 5B presents the cyclic voltammetry curves obtained from antigen-antibody complexes formed by exposure to varying concentrations (0 ng/mL, 10 ng/mL, and 1 μg/mL) of SEB antigen combined with a constant antibody concentration of 100 μg/mL.
The experiment employed chemical reagents as shown in Fig. 4 for electrode modification. We conducted multiple cyclic voltammetry tests over a span of six months utilizing a concentration of 100 ng/mL SEB antigen. The cyclic voltammetry curve displayed in Fig. 5C demonstrates the stability achieved through electrode modification. To evaluate reproducibility within our experimental framework, five consecutive cyclic voltammetry scans were performed using a concentration of 10 ng/mL SEB antigen; resulting curves exhibited remarkable similarity-thereby confirming repeatability (Fig. 5D). The entire experimental procedure was completed within seventy-five seconds.
A Concentration gradient of SEB in the experiment. B CV curves for SEB concentrations of 0 ng/mL, 10 ng/mL, and 1 μg/mL. C Reproducibility assessment: CV curves for 100 ng/mL SEB at various time points over a period of six months under identical experimental conditions. D Stability assessment: CV curve for 10 ng/mL SEB scanned consecutively five times.
According to research observations, significant changes in the redox peaks of the cyclic voltammetry curve were noted with increasing concentrations of SEB antigen. At a zero antigen concentration, the cyclic voltammetry curve displayed distinct redox peaks, indicating a prominent redox reaction involving potassium ferricyanide at the electrode surface. However, as the concentration of SEB antigen increased, these redox peaks gradually diminished and ultimately disappeared. This phenomenon may be attributed to the formation of antigen-antibody complexes. With rising antigen concentrations, the binding between antigens and antibodies leads to several effects:
-
1.
Competitive Blocking Effect: Antigens competitively occupy active sites on the electrode surface, thereby reducing contact between potassium ferricyanide molecules and the electrode and diminishing the intensity of the redox reaction.
-
2.
Electrochemical Shielding Effect: The formation of antigen-antibody complexes impedes electron transfer by creating a shielding layer that weakens the current signal associated with the redox reaction.
As further increases in antigen concentration occur, an escalation in antibody-antigen complex formation takes place, occupying nearly all active sites on the electrode surface, which hinders effective interaction between potassium ferricyanide molecules and electrodes. Consequently, this results in either a reduction or complete disappearance of observable redox peaks.
Data preprocessing
In traditional electrochemical measurements, specific points on the CV curve are typically utilized to indicate the concentration of reactants, rather than relying on the curve as a whole. In this study, we selected a series of representative points from the CV curve as input features for electrochemical reactions to train our machine learning model. As illustrated in Fig. 6A, eleven features were extracted from the CV curves obtained from the aforementioned experiments (Mathematical definitions are detailed in Table 1.): maximum current (maxI), maximum voltage (maxV), minimum current (minI), minimum voltage (minV), beginning current (beginI), ending current (endI), the area of the curve (area), oxidation integral difference (OID), max-min current slope (k), zero current voltage (zeroIV), and shortest top-bottom curve distance (distance).
Subsequently, we employed a model training approach based on the Random Forest algorithm to rank feature importance as depicted in Fig. 6B. In our subsequent analysis, three features with low importance (maxV, zeroIV, beginI) were excluded. The remaining eight features were then selected as inputs for training the machine learning model. By screening and distinguishing more significant features within cyclic voltammetry curves across different concentrations and establishing a regression model accordingly; we aimed to focus on signals pertinent to our target while minimizing interference from nonspecific or irrelevant signals.
Ultimately, the dataset generated by these electrodes comprises 42 samples and 8 features that are used to predict SEB concentration. A total of 36 samples were designated as the training set while 6 samples served as the test set.
Notwithstanding the notable advancements made by metaheuristic algorithms in structural optimization in recent years33,34. Exemplified by the greylag goose algorithm that effectively addresses complex structural optimization problems through simulating the coordinated flight patterns of geese flocks35, their transference to feature selection tasks in electrochemical detection presents substantial challenges. Innovative approaches such as the 2-archive multi-objective cuckoo search algorithm36,37,38, which enhances multi-objective optimization efficiency through a novel dual-archive mechanism, and the multi-objective brown bear optimization algorithm that solves constrained optimization problems by emulating brown bear foraging behaviors39, demonstrate formidable global search capabilities in engineering optimization contexts. However, their application to electrochemical feature selection exposes inherent limitations.
Regarding computational complexity, metaheuristic algorithms typically necessitate extensive iterative global search procedures, with certain implementations requiring hundreds of complete objective function evaluations per iteration40,41,42. This engenders substantial computational overhead, potentially creating severe efficiency constraints in resource-limited applications. In marked contrast, random forest algorithms generate feature rankings through a single training cycle, enabling computationally economical and expeditious feature screening. Concerning result stability, metaheuristic outcomes exhibit pronounced sensitivity to initial parameter configurations and random seed values, frequently yielding divergent results across executions-a manifestation of their inherent instability. The random forest approach, conversely, delivers superior stability by aggregating results from multiple decision trees, thereby producing consistent and reliable feature selection outcomes. Most critically, in terms of interpretability, metaheuristic feature selection processes rely on intricate optimization mechanisms to determine optimal feature subsets, resulting in opaque decision pathways. The random forest methodology, by contrast, constructs an electrochemically meaningful feature space incorporating temporal characteristics of CV curves (e.g., redox peak potentials/currents), with feature importance metrics enabling quantifiable tracing of individual feature contributions.
When compared to conventional multi-objective metaheuristic methods prevalent in structural optimization, the proposed methodology demonstrates three principal advantages:
-
(1)
Systematic incorporation of electrochemical theory into feature selection to preclude mathematically optimal yet electrochemically inconsistent solutions;
-
(2)
Establishment of an interpretable evaluation framework for feature contributions enabling hierarchical traceability of critical signal sources;
-
(3)
Implementation of a lightweight computational workflow that maintains accuracy while significantly reducing processing time, thereby better accommodating on-site rapid detection requirements.
This establishes a novel feature selection paradigm for electrochemical analysis that harmonizes operational efficiency, interpretability, and domain-specific adaptability.
Principal Component Analysis (PCA) was utilized to conduct dimensionality reduction on eight features within the electrochemical data, thereby enhancing our understanding of the intrinsic structure of the data and facilitating visualization analysis. PCA, a widely recognized technique for dimensionality reduction, effectively transforms high-dimensional data into a lower-dimensional space while preserving the most significant feature information.
The eight-feature dataset was projected onto a two-dimensional plane through PCA dimensionality reduction, resulting in an intuitive visualization of the data, as illustrated in Fig. 6C. The clustering observed within the data clearly delineates distinct formations in feature space, indicating a specific classification structure corresponding to different concentrations of SEB features. Furthermore, no outliers or anomalies were detected, underscoring the strong consistency and reliability of the dataset. This consistency not only validates the efficacy of both feature extraction and preprocessing methods in accurately representing SEB characteristics at varying concentrations during electrochemical reactions but also highlights the stability and repeatability inherent in the electrochemical measurement process. This provides a robust foundation for subsequent regression analyses.
Machine learning model analysis
In the field of machine learning, regression models serve as a fundamental yet powerful tool for modeling and predicting continuous output variables. The primary objective is to generate predictions by understanding the relationship between input features and the corresponding output. Typically, it is posited that the output variable can be represented through either a linear or nonlinear combination of one or more input features. For example, in simple linear regression, the output variable \(y\) is expressed as a linear function of the input variable, accompanied by an error term, as illustrated in Eq. (1):
Here, we aim to estimate the parameters \(\beta _0\) and \(\beta _1\), which facilitates predictions and interpretations of the dependent variable \(y\). The primary objective of linear regression is to adjust the training data so that the predicted values closely align with the actual observed values. Simple linear regression employs a single independent variable in its predictive framework, whereas multivariate or multiple linear regression incorporates several independent or predictor variables into the forecasting process.
In this study, we utilized a multivariate linear regression model to analyze and predict SEB concentration. This method is widely recognized in both statistics and machine learning for exploring the linear relationships between multiple independent variables and a continuous dependent variable. Its fundamental form can be expressed by Eq. (2):
In this equation, \(y\) represents the dependent variable (output variable) being predicted, while \(x_1, x_2, \ldots , x_p\) denote the independent variables (input features). The parameters of the model, \(\beta _0, \beta _1, \beta _2, \ldots , \beta _p\), represent the slopes, associated with each independent variable. Additionally, \(\epsilon\) is the error term that accounts for random errors not explained by the model. The multivariate linear regression model estimates these parameters using training data to accurately predict new observations. In practical applications, various statistical methods-such as Ordinary Least Squares (OLS)-can be employed to fit the model and determine optimal parameter estimates.
Linear regression is particularly well-suited for handling small to medium-sized datasets due to its rapid modeling speed, strong interpretability, and high computational efficiency. Figure 6D illustrates the complete workflow for predicting SEB concentration using a linear regression algorithm. This process begins with experimental data preprocessing followed by feature extraction and selection; it ultimately culminates in both training and testing phases of the model.
Initially, electrochemical CV experiments are conducted on SEB electrode sheets with established concentration gradients to obtain raw data. This data undergoes preprocessing steps, including standardization and cleaning, to ensure consistency and quality. Subsequently, a series of key features are extracted from the preprocessed CV curves, capturing the essential characteristics of the electrochemical reactions. Feature selection is then performed to identify those features that significantly influence SEB concentration prediction, thereby enhancing the model’s accuracy and robustness. During the model training phase, the dataset is divided into training and test sets. The training set is utilized to develop the regression model and conduct hyperparameter tuning. Regression coefficients are estimated from this training data, allowing the model to effectively capture the relationship between input features and SEB concentration. This process typically involves optimizing a loss function; regression coefficients are determined when this loss function reaches its minimum value, ultimately resulting in a linear regression equation. Once trained, the model can be employed to predict SEB concentrations within the test set. The test set serves as a means of validating both the generalization capability and predictive performance of the model. A range of evaluation metrics (e.g., \(R^2\), MAPE) are used to assess performance; predicted results are compared against actual values to visually demonstrate both effectiveness and predictive accuracy. Finally, the derived regression model is applied for predicting SEB concentrations in unknown samples.
Results and discussion
Analysis of feature impact on SEB concentration prediction across varying concentrations
The Random Forest algorithm was utilized to examine the impact of various features on predictions across different categories of SEB concentration. This analysis is essential for comprehending the role of each feature in the electrochemical reaction process and for clarifying the contribution of distinct features to variations in SEB concentration (see Table 2).
As illustrated in Fig. 7, the maxI holds significant importance across all concentration categories, particularly within the control group (0 ng/mL) and at lower concentrations, where its sensitivity is notably high and critical for accurate prediction. However, its significance diminishes slightly at intermediate to high concentrations, likely due to coverage effects and reaction kinetics inherent in the electrochemical reaction. Conversely, the importance of the minI increases with rising SEB concentrations. This trend indicates that within the intermediate to high SEB concentration range, the SEB antigen-antibody complex exerts a substantial influence on the oxidation process, thereby rendering the reduction current a primary distinguishing feature. Consequently, at these levels of concentration, the minI plays a crucial role in predicting SEB concentration. The distancedemonstrates considerable importance across most concentration categories, especially within both control group conditions and those involving high concentrations. The k exhibits comparable significance to other features under intermediate to high concentration conditions; this reflects the notable impact of current change rates on the reaction process due to intensified competition between diffusion and reaction rates at these elevated concentrations.
The OID is of greater significance in the control group and at high concentrations, while its relevance diminishes at intermediate concentrations. In the control group, OID primarily reflects background variations in baseline current, thus demonstrating heightened importance under baseline conditions. At elevated concentrations, the substantial formation of antigen-antibody complexes significantly influences the redox process, leading to an increased relevance of OID for differentiating high levels of SEB. The area exhibits considerable importance within the control group and specifically at a concentration of 10 ng/mL. It maintains relatively consistent significance across other concentration levels, indicating its reliability in assessing overall reaction activity and stability. Conversely, the endI shows limited importance within the control group (0 ng/mL), but its significance markedly increases at 1 ng/mL due to pronounced changes in electrochemical reactions associated with minimal SEB antigen-antibody complexes. The minV demonstrates relatively stable importance across various concentrations, reflecting its dependable influence on the reduction threshold voltage during electrochemical reactions.
By analyzing shifts in feature importance rankings across different concentrations, it can be inferred that varying SEB concentrations may induce distinct electrochemical reaction mechanisms; this variation subsequently affects each feature’s impact on SEB concentration. A deeper understanding of these features’ significance and trends enhances comprehension of their mechanistic roles at differing concentrations, thereby providing valuable insights for designing electrochemical sensors and investigating electrochemical reaction mechanisms.
Prediction of SEB concentration
In this study, the concentration gradient of SEB spans several orders of magnitude, ranging from 0 ng/mL to 10,000 ng/mL. Directly modeling these raw values poses challenges due to the extremely wide numerical range, which can lead to instability during model training and increased computational complexity. To mitigate these issues, we standardized the concentration unit to pg/mL and applied a base-10 logarithmic transformation to the SEB concentrations (with 0 corresponding to 10 pg/mL). This approach reduces the numerical range, enhances data linearity, and minimizes skewness, as illustrated in Fig. 8A. Specifically, the logarithmic transformation of SEB concentration labels is detailed in Table 3.
To evaluate the feasibility of employing a linear regression algorithm for predicting SEB concentration, we analyzed SEB at various concentrations through cyclic voltammetry (CV) curves. After extracting relevant features from these CV curves, we partitioned the data samples into training and testing sets to perform multiple predictions of the logarithmic labels corresponding to SEB concentration. For a comprehensive evaluation of model performance, six machine learning regression methods (including linear regression, support vector regression, and random forest) were comparatively analyzed. The coefficient of determination (\(R^2\)) for each model on the testing set is presented in Table 4. Subsequently, we calculated the discrepancies between the predicted logarithmic values and their true counterparts. Figure 8B presents the prediction results for SEB concentration in logarithmic form. It is evident that the linear regression model provides reliable predictions, achieving a mean absolute percentage error (MAPE) of 6.09%. Furthermore, as illustrated in Fig. 8C, utilizing leave-one-out cross-validation yielded a mean relative standard deviation (RSD) of 6.32% for SEB concentrations (in log form; unit: pg/mL). These methodologies ensure both accurate model evaluation and dependable practical applications.
The findings suggest that by applying a logarithmic transformation to SEB concentration labels and implementing a linear regression model, precise predictions of SEB concentration can be attained. This approach demonstrates particularly commendable performance within higher concentration ranges while exhibiting lower prediction errors. Such insights offer a robust strategy for early diagnosis of SEB and contribute to further enhancing the detection capabilities of electrochemical sensors.
Notably, while nonlinear models such as SVR and random forest can capture intricate feature relationships, linear regression exhibits superior fitting performance (\(R^2\)=0.999715) in SEB concentration prediction. This finding simplifies model deployment complexity.
Discussion
This study presents a novel assay method that integrates machine learning, electrochemical, and immunological principles, targeting SEB as the analyte of interest. Compared to existing methodologies, this approach exhibits exceptional performance and numerous advantages, as outlined in Table 5. For instance, while the immunochromatographic test strips (LFA) developed by Gholamzad et al. and Tsui et al. provide rapid detection within minutes, they exhibit relatively lower sensitivity with a detection limit of 10 ng/mL43,44. In contrast, our method achieves superior sensitivity with a detection time of merely two minutes. Moreover, by utilizing machine learning algorithms for data processing and analysis, our approach reduces errors associated with manual readings, thereby enhancing both accuracy and reliability.
ELISA methods offer high sensitivity but typically require longer detection times. For example, Jin et al. developed an IgY-based sandwich ELISA for detecting SEA, SEB, SEC, SED, and SEE with a detection limit (LOD) ranging from 1 to 5 ng/mL in PBS and a total detection time of 8–10 h45. Some advancements in ELISA have achieved pg/mL level detection limits; for instance, Ji et al. proposed a nanobody-based sandwich immunoassay that effectively avoids interference from protein A by employing nanobodies without Fc tails-achieving a remarkable detection limit as low as 0.3 ng/mL. Soto et al. introduced a sliding microtube-based sandwich immunoassay featuring a detection range of 0-100 ng/mL with an LOD of 0.5 ng/mL. Singh et al., on the other hand, developed an ELISA-based sandwich method yielding an LOD between 0.5 and 0.75 ng/mL; however it necessitates several hours for completion. These conventional methods are generally complex and costly while being time-consuming-primarily suited for laboratory chip assays46,47,48. In contrast to these approaches, our method offers optimized conditions regarding both detection time and cost while maintaining high sensitivity.
It is essential to recognize that in double antibody sandwich immunoassays, elevated concentrations of pathogens can result in false-negative outcomes due to the saturation of antigen-antibody binding. This saturation ultimately hinders signal generation. Consequently, the linear range of the assay becomes critical, particularly when addressing high concentrations of pathogens or toxins. The method proposed in this study-by integrating immunoassay techniques, electrochemical analysis, and machine learning-significantly enhances the detection linear range and effectively resolves the bottleneck issues encountered with traditional methods during high-concentration sample analysis.
Traditional linear regression methods are frequently employed for statistical analysis; however, these approaches typically depend on single features, which may constrain model precision and applicability in specific contexts. For instance, a simple linear regression analyzing the relationship between oxidation peak current and SEB concentration yielded an \(\hbox {R}^{2}\) value of only 0.954, as illustrated in Fig. 8D.
This study significantly enhances model accuracy by extracting multiple feature parameters from cyclic voltammetry curves and employing the random forest algorithm to select features that exhibit a strong correlation with SEB concentration. Subsequently, techniques such as PCA were utilized for dimensionality reduction, effectively mapping high-dimensional data into a lower-dimensional space. This approach facilitated the observation of data clustering and validated classification structures to ensure consistency and reliability. Ultimately, the selected parameters and labels were input into a multivariate linear regression model for training, fitting, and prediction, resulting in an impressive \(R^2\) value of 0.999. This outcome demonstrates that integrating various feature parameters with artificial intelligence algorithms can yield a more precise electrochemical experimental data model.
Moreover, this method exhibits significant advantages in terms of robustness. By combining multiple feature parameters with machine learning algorithms, it maintains consistent performance across different experimental batches while effectively addressing noise and interference within the experimental environment. This ensures stability and reliability in detection results. Additionally, this methodology extends the detection dynamic range by accurately measuring SEB concentrations from 1 ng/mL to 10 μg/mL without compromising performance stability. The minimal sample requirement of only 20 μL further enhances detection convenience and broadens the practical application of electrochemical immunoassay methods, providing an innovative solution for advancements in immunoassay technologies.
In practical applications, this method also presents opportunities for further optimization. As the device is employed in hospitals or other real-world environments, the model can be retrained with continuously acquired new data to adapt to variations in operators and environmental factors. This capacity for ongoing optimization ensures that the method remains efficient and stable under dynamic real-world conditions, thereby providing more reliable support for detection in practical applications.
However, the current experiment has certain limitations. Firstly, due to the limited amount of data available, the model’s generalizability may be compromised, potentially leading to instability across different datasets or application scenarios. This limitation also restricts its universality and effectiveness in detecting other biomarkers. Furthermore, this study primarily relies on cyclic voltammetry without incorporating additional electrochemical detection methods such as differential pulse voltammetry and electrochemical luminescence; this somewhat constrains both detection sensitivity and diversity. These limitations indicate that while significant progress has been made in SEB detection using this method, further research and validation are necessary to enhance adaptability for other biomarkers and diversify electrochemical detection technologies.
Conclusion
This study presents an innovative approach that integrates immunoassay methods, electrochemical techniques, and machine learning algorithms to address the challenges associated with traditional SEB quantification. A highly specific and precise platform for SEB quantification has been established through this methodology. This approach not only resolves issues related to low detection accuracy and insufficient robustness in existing SEB concentration measurement techniques but also significantly enhances both the detection range and efficiency while reducing detection time and costs. The method demonstrates exceptional accuracy in model fitting, showcasing remarkable robustness alongside a broad detection range. In comparison to conventional rapid immunochromatographic methods, the detection time is reduced to just 2 min-seven times faster-with a sample requirement of only 20 μL. Furthermore, the simplicity of this method effectively mitigates noise from environmental factors, manual operations, and instability of immunocomplexes, thereby eliminating the necessity for highly skilled operators. The artificial intelligence model can autonomously execute detection tasks, substantially lowering overall detection costs. Data processing is enhanced through PCA, which facilitates data visualization while verifying consistency and reliability. Additionally, this study provides an in-depth analysis of how feature parameters influence electrochemical reactions at varying concentrations; this aids in understanding electrochemical mechanisms within biomolecular interactions and offers robust support for optimizing sensor design-thereby further improving detection accuracy. Experimental results indicate that this method excels in predicting SEB antigen concentrations across a range from 1 ng/mL to 10 μg/mL, achieving an \(R^2\) value of 0.999 along with a mean absolute percentage error 6.09% and a mean relative standard deviation of 6.32% across different concentrations-surpassing traditional methodologies. Overall, this integrated approach combining immunoassay methods, electrochemical techniques, and machine learning algorithms offers new opportunities for high-precision, robust, accurate, rapid, low-cost, and automated quantitative detection of biomarkers. This method effectively addresses the limitations of traditional technologies in quantitative analysis and provides an efficient platform for the specific quantification of multiple targets, demonstrating significant potential for practical applications.
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files). The Python implementation of the proposed method is publicly available on GitHub at https://github.com/sunt06/Electrochemical_Analysis.git.
References
Tong, S. Y., Davis, J. S., Eichenberger, E., Holland, T. L. & Fowler, V. G. Jr. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28, 603–661 (2015).
Foster, T. J. Staphylococcus aureus. In Molecular medical microbiology 839–888 (2002).
Hennekinne, J.-A., De Buyser, M.-L. & Dragacci, S. Staphylococcus aureus and its food poisoning toxins: Characterization and outbreak investigation. FEMS Microbiol. Rev. 36, 815–836 (2012).
Ellington, A. A., Kullo, I. J., Bailey, K. R. & Klee, G. G. Antibody-based protein multiplex platforms: Technical and operational challenges. Clin. Chem. 56, 186–193 (2010).
Toley, B. J. et al. A versatile valving toolkit for automating fluidic operations in paper microfluidic devices. Lab Chip 15, 1432–1444 (2015).
Lequin, R. M. Enzyme immunoassay (EIA)/enzyme-linked immunosorbent assay (ELISA). Clin. Chem. 51, 2415–2418 (2005).
Wild, D. The immunoassay handbook: theory and applications of ligand binding, ELISA and related techniques (Newnes, 2013).
Di Nardo, F., Chiarello, M., Cavalera, S., Baggiani, C. & Anfossi, L. Ten. years of lateral flow immunoassay technique applications: Trends, challenges and future perspectives. Sensors 21, 5185 (2021).
Cho, J.-H. & Paek, S.-H. Semiquantitative, bar code version of immunochromatographic assay system for human serum albumin as model analyte. Biotechnol. Bioeng. 75, 725–732 (2001).
Posthuma-Trumpie, G. A., Korf, J. & van Amerongen, A. Lateral flow (immuno) assay: Its strengths, weaknesses, opportunities and threats. a literature survey. Anal. Bioanal. Chem. 393, 569–582 (2009).
Parolo, C., de la Escosura-Muñiz, A. & Merkoçi, A. Enhanced lateral flow immunoassay using gold nanoparticles loaded with enzymes. Biosens. Bioelectron. 40, 412–416 (2013).
Hanssen, B. L., Siraj, S. & Wong, D. K. Recent strategies to minimise fouling in electrochemical detection systems. Rev. Anal. Chem. 35, 1–28 (2016).
Tajik, S. et al. Recent developments in polymer nanocomposite-based electrochemical sensors for detecting environmental pollutants. Ind. Eng. Chem. Res. 60, 1112–1136 (2021).
Simoska, O. et al. Recent trends and advances in microbial electrochemical sensing technologies: An overview. Curr. Opin. Electrochem. 30, 100762 (2021).
Bard, A. J., Faulkner, L. R. & White, H. S. Electrochemical methods: fundamentals and applications (John Wiley & Sons, 2022).
Giordano, G. F. et al. Machine learning toward high-performance electrochemical sensors. Anal. Bioanal. Chem. 415, 3683–3692 (2023).
Nashruddin, S. N. A. B. M., Salleh, F. H. M., Yunus, R. M. & Zaman, H. B. Artificial intelligence—powered electrochemical sensor: Recent advances, challenges, and prospects. (Heliyon, 2024).
Jiang, Y. et al. Immunomarker support vector machine classifier for prediction of gastric cancer survival and adjuvant chemotherapeutic benefit. Clin. Cancer Res. 24, 5574–5584 (2018).
Aileni, R. M., Pasca, S. & Florescu, A. EEG-brain activity monitoring and predictive analysis of signals using artificial neural networks. Sensors 20, 3346 (2020).
Ludwig, S. A., Picek, S. & Jakobovic, D. Classification of cancer data: Analyzing gene expression data using a fuzzy decision tree algorithm. In Operations Research Applications in Health Care Management 327–347 (2018).
Alzahrani, T. et al. Cardiovascular disease risk factors and myocardial infarction in the transgender population. Circ. Cardiovasc. Qual. Outcomes 12, e005597 (2019).
Enginler, S. Ö. et al. Enhancing electrochemical detection through machine learning-driven prediction for canine mammary tumor biomarker with green silver nanoparticles. Anal. Bioanal. Chem. 416, 5071–5088 (2024).
Schelin, J., Wallin-Carlquist, N., Thorup Cohn, M., Lindqvist, R. & Barker, G. C. The formation of staphylococcus aureus enterotoxin in food environments and advances in risk assessment. Virulence 2, 580–592 (2011).
Hassan, R. Y. Advances in electrochemical nano-biosensors for biomedical and environmental applications: From current work to future perspectives. Sensors 22, 7539 (2022).
Mouselly, M., Alawadhi, H. & Thangavel, S. S. Current status of ferro-/ferricyanide for redox flow batteries. Curr. Opin. Electrochem. 48, 101581 (2024).
Wang, J. Electrochemical glucose biosensors. Chem. Rev. 108, 814–825 (2008).
Ionescu, R. E. Use of cysteamine and glutaraldehyde chemicals for robust functionalization of substrates with protein biomarkers-an overview on the construction of biosensors with different transductions. Biosensors 12, 581 (2022).
Hermanson, G. T. Bioconjugate Techniques (Academic Press, 2013).
Barattin, R. & Voyer, N. Chemical modifications of atomic force microscopy tips. Atomic Force Microsc. Biomed. Res. Methods Protocols 457–483 (2011).
Gupta, G. et al. Molecularly imprinted polymer for the recognition of biological warfare agent staphylococcal enterotoxin b based on surface plasmon resonance. Thin Solid Films 519, 1115–1121 (2010).
Torres, M. & Casadevall, A. The immunoglobulin constant region contributes to affinity and specificity. Trends Immunol. 29, 91–97 (2008).
Janeway, C. A. Jr. The immune system evolved to discriminate infectious nonself from noninfectious self. Immunol. Today 13, 11–16 (1992).
Panagant, N., Kumar, S., Tejani, G. G., Pholdee, N. & Bureerat, S. Many-objective meta-heuristic methods for solving constrained truss optimisation problems: A comparative analysis. MethodsX 10, 102181 (2023).
Mashru, N., Tejani, G. G., Patel, P. & Khishe, M. Optimal truss design with MOHO: A multi-objective optimization perspective. PLoS One 19, e0308474 (2024).
Mashru, N., Tejani, G. G. & Patel, P. Reliability-based multi-objective optimization of trusses with Greylag goose algorithm. Evol. Intel. 18, 25 (2025).
Tejani, G. G., Mashru, N., Patel, P., Sharma, S. K. & Celik, E. Application of the 2-archive multi-objective cuckoo search algorithm for structure optimization. Sci. Rep. 14, 31553 (2024).
Tejani, G. G., Sharma, S. K., Mashru, N., Patel, P. & Jangir, P. Optimization of truss structures with two archive-boosted MOHO algorithm. Alex. Eng. J. 120, 296–317 (2025).
Kumar, S. et al. A two-archive multi-objective multi-verse optimizer for truss design. Knowl.-Based Syst. 270, 110529 (2023).
Mehta, P., Kumar, S. & Tejani, G. G. MOBBO: A multiobjective brown bear optimization algorithm for solving constrained structural optimization problems. J. Optim. 2024, 5546940 (2024).
Kumar, S., Jangir, P., Tejani, G. G. & Premkumar, M. A decomposition based multi-objective heat transfer search algorithm for structure optimization. Knowl.-Based Syst. 253, 109591 (2022).
Kumar, S., Tejani, G. G., Pholdee, N. & Bureerat, S. Multiobjecitve structural optimization using improved heat transfer search. Knowl.-Based Syst. 219, 106811 (2021).
Kumar, S. et al. Optimization of truss structures using multi-objective cheetah optimizer. Mech. Based Des. Struct. Mach. 53, 1494–1515 (2025).
Gholamzad, M., Khatami, M. R., Ghassemi, S., Malekshahi, Z. V. & Shooshtari, M. B. Detection of staphylococcus enterotoxin B (SEB) using an immunochromatographic test strip. Jundishapur J. Microbiol. 8, e26793 (2015).
Tsui, P.-Y. et al. Development of staphylococcal enterotoxin b detection strips and application of SEB detection strips in food. J. Med. Sci. 33, 285–91 (2013).
Jin, W. et al. Application of IgY to sandwich enzyme-linked immunosorbent assays, lateral flow devices, and immunopillar chips for detecting staphylococcal enterotoxins in milk and dairy products. J. Microbiol. Methods 92, 323–331 (2013).
Ji, Y. et al. Nanobodies based on a sandwich immunoassay for the detection of staphylococcal enterotoxin b free from interference by protein a. J. Agric. Food Chem. 68, 5959–5968 (2020).
Soto, C. M., Martin, B. D., Sapsford, K. E., Blum, A. S. & Ratna, B. R. Toward single molecule detection of staphylococcal enterotoxin b: mobile sandwich immunoassay on gliding microtubules. Anal. Chem. 80, 5433–5440 (2008).
Singh, M. et al. Development and evaluation of sandwich ELISA for detection and quantification of staphylococcal enterotoxin-a in food. J. Food Saf. 44, e13114 (2024).
Sharma, A. et al. Enzyme free detection of staphylococcal enterotoxin b (SEB) using ferrocene carboxylic acid labeled monoclonal antibodies: An electrochemical approach. New J. Chem. 40, 8334–8341 (2016).
Kumari, S., Tiwari, M. & Das, P. Multi format compatible visual and fluorometric detection of SEB toxin in nanogram range by carbon Dot-DNA and acriflavine nano-assembly. Sens. Actuators B Chem. 279, 393–399 (2019).
Sharma, A., Kameswara Rao, V., Vrat Kamboj, D. & Jain, R. Electrochemical immunosensor for staphylococcal enterotoxin B (SEB) based on platinum nanoparticles-modified electrode using hydrogen evolution inhibition approach. Electroanalysis 26, 2320–2327 (2014).
Morissette, C., Goulet, J. & Lamoureux, G. Rapid and sensitive sandwich enzyme-linked immunosorbent assay for detection of staphylococcal enterotoxin b in cheese. Appl. Environ. Microbiol. 57, 836–842 (1991).
Nagaraj, S., Ramlal, S., Kingston, J. & Batra, H. V. Development of IgY based sandwich ELISA for the detection of staphylococcal enterotoxin g (SEG), an egc toxin. Int. J. Food Microbiol. 237, 136–141 (2016).
Yang, M., Sun, S., Bruck, H. A., Kostov, Y. & Rasooly, A. Electrical percolation-based biosensor for real-time direct detection of staphylococcal enterotoxin b (SEB). Biosens. Bioelectron. 25, 2573–2578 (2010).
Rasooly, A. Surface plasmon resonance analysis of staphylococcal enterotoxin b in food. J. Food Prot. 64, 37–43 (2001).
Acknowledgements
This work was supported by the National Key R&D Program of China (Grant No. 2021YFD1800500), the National Natural Science Foundation of China (Grant No.62306068), the Fundamental Research Funds for the Central Universities (Grant No. N2123004, 2022GFZD014), the Hebei Natural Science Foundation (Grant No. F2021203070, F2022501031), the Administration of Central Funds Guiding the Local Science and Technology Development (Grant No.206Z1702G), the Hebei Province Higher Education Teaching Reform Research and Practice Project (Grant No. 2020GJJG310).
Author information
Authors and Affiliations
Contributions
Yuliang Zhao: Writing - review & editing, Resources, Project administration, Methodology, Investigation, Funding acquisition. Tingting Sun: Writing - original draft, Validation, Methodology, Investigation, Data curation. Huawei Zhang: Formal analysis, Data curation, Conceptualization. Chao Lian: Writing - review & editing, Supervision. Zhongpeng Zhao: Methodology, Investigation. Yongqiang Jiang: Methodology, Investigation. Huiqi Duan: Methodology, Investigation. Yuhao Ren: Methodology, Investigation. Xuyang Sun: Methodology, Investigation. Zhikun Zhan: Writing - review & editing, Supervision. Mingyue Qu: Writing - review & editing, Project administration, Investigation. Shaolong Chen: Writing - review & editing, Resources, Project administration, Investigation, Funding acquisition.
Corresponding authors
Ethics declarations
Competing interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhao, Y., Sun, T., Zhang, H. et al. Artificial intelligence enhanced electrochemical immunoassay for staphylococcal enterotoxin B. Sci Rep 15, 19602 (2025). https://doi.org/10.1038/s41598-025-03930-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-03930-8