Abstract
Early diagnostic assessments of neurodivergent disorders (NDD), remains a major clinical challenge. We address this problem by pursuing the hypothesis that there is important cognitive information about NDD conditions contained in the way individuals move, when viewed at millisecond time scales. We approach the NDD assessment problem in two complementary ways. First, we applied supervised deep learning (DL) techniques to identify participants with autism spectrum disorder (ASD), attention-deficit/hyperactivity disorder (ADHD), comorbid ASD + ADHD, and neurotypical (NT) development. We measured linear and angular kinematic variables, using high-definition kinematic Bluetooth sensors, while participants performed the reaching protocol to targets appearing on a touch screen monitor. The DL technique was carried out only on the raw kinematic data. The area under the receiver operator characteristics curve suggests that we can predict, with high accuracy, NDD participant’s conditions. Second, we filtered the high frequency electronic sensor noise in the recorded kinematic data leaving the participants’ physiological characteristic random fluctuations. We quantified these fluctuations by their biometric Fano Factor and Shannon Entropy from a histogram distribution built from the magnitude difference between consecutive extrema unique to each participant, suggesting a relationship to the severity of their condition. The DL may be used as complementary tools for early evaluation of new participants by providers and the new biometrics allow for quantitative subtyping of NDDs according to severity.
Similar content being viewed by others
Introduction
NDDs, including ASD, ADHD, and comorbid ASD + ADHD, have witnessed significant diagnostic growth over the last few years1. ASD prevalence in the US is increasing rapidly (CDC, 1/36 children, with male to female ratio of 4:1). 9.4% of American children 5–17 years old have been diagnosed with ADHD, with boys more likely than girls to be diagnosed (12.9% compared to 5.6%)2. ADHD is one of the most common comorbidities diagnosed in children with ASD, with rates of co-occurrence ranging up to 70%3,4,5,6,7,8,9,10. The growing awareness of co-occurring ADHD symptoms in youth with ASD warranted the addition of the co-morbid diagnoses in the DSM-53,11,12,13,14,15. Symptoms experienced by NDD individuals may include: social and speech communication impairments, deficits in response inhibition, hyperactivity, inattention, as well as restricted interests, and repetitive behaviors16,17,18. Current diagnostics criteria focus on the core behavioral symptoms mentioned above18. Due to the complexity of diverse symptoms characterizing these NDDs, it is a lengthy process requiring significant expertise to diagnose these conditions. This challenge impairs the development of successful treatments and timely as well as early interventions which have been shown to benefit treatment10,19,20,21. Different attempts have been made to overcome these diagnostic challenges, commonly relying on complex qualitative behavioral observations.
Daily movements can reveal a great deal about neuromotor control and cognitive abilities. Movement is an essential part of the sequence of behaviors that involve our social communications22,23,24,25. It has been recognized that atypical movements may interfere with the development of adaptive skills, in particular in children26,27. Individuals diagnosed with an NDD have been known to have deficits in motor sequencing of complex movements, including simple reaching motions22,23,24,25. Previous studies have also observed delayed preparation of coordinated movements in children with ASD relative to (NT) peers25. Clear motor signatures were also found in studies of infants found to have ASD28. Studies of development in ASD and ADHD participants have highlighted the importance of early motor deficits29,30,31. Recent studies using machine learning have characterized the crucial importance of movement in assessments of ASD participants, including gesture patterns32,33. Also, multimodal studies of ASD and ADHD participants were able to differentiate ASD from ADHD with high accuracy34.
The development of high-definition sensors allows study of movements at increasingly finer timescales. Our prior work identified clear movement differences between ASD versus NT participants35. Here we expanded the set of kinematic variables measured by using new wireless MEM sensors while participants performed the simple reaching task, while expanding the sample to include participants with ADHD and comorbid ASD + ADHD. The data was recorded using millisecond time resolution motion sensors. We followed a two-pronged approach to analyze the kinematic data sets. First, we applied Deep Learning (DL) techniques36,37 to the “raw” kinematic data without any preconceived assumptions about the nature or characteristics of the underlying content. The DL neural network is trained to predict a specific potential diagnostic NDD/NT output based on a random subsequence from a single reaching trial. We generated multiple complementary training sets by changing the kinematic signal (or combination of signals) being used. These datasets are then split into a training and testing set. We use K-Fold Cross Validation38 to identify hyperparameters and then retrained the network using the entire training set. We subsequently assessed the performance of the DL algorithm on the remaining, untrained test data sets. A similar baseline was performed by training a Support Vector Machine under the same paradigm. More details are discussed in Methods and in the Supplementary Information (SI). The DL neural network successfully identified the diagnosis associated with the trials in the test set. DL and other advanced techniques are powerful, and applicable to other classes of data aiming to characterize those with NDDs, such as retinal images and FMRI studies in those with ASD39,40,41,42, but it is difficult to understand how they work. To gain insight into the sources of information, we explored different combinations of kinematic variables and applied the Shapley Additive Explanations (SHAP) framework43 to visualize how certain input features contribute to certain predictions (shown in Note 4 of the SI). We find that certain kinematic variables play a larger role in NDD/NT classification.
Given a desire for greater understanding of what NDD effects might be observed in movement, we applied a second, more hypothesis-driven approach, to analyze our kinematic data. We examined the Fano Factor (FF)44 and the Shannon Entropy (S) of distributions of kinematic fluctuations of the movement signal. Our first DL approach allows us to make screening predictions about NDD (or NT) conditions. While the second provides a deeper biological understanding of the severity of each NDD participant’s condition. Our two approaches have the potential to be used as complementary diagnostic tools by providers.
Results
Deep learning data analysis
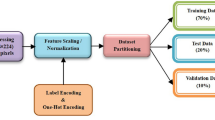
We hypothesize that movement contains latent hidden information about participants’ cognitive abilities35,45. Our goal is to move towards matching the diagnostic capabilities of providers with more efficiency. The DL analysis was performed on the raw kinematic data collected from participants reaching tasks (Fig. 1). The data was separated into trials consisting of one cycle of reaching their hand to the target and then returning it to rest. The type of kinematic data measured is discussed in Methods. The start and finish times for the reaching trials were determined from the filtered data, as discussed in Note 1 of the SI, and are used to identify the beginning and end of each trial in the raw data. We included trials that were at least 105 samples long (slightly under 1 s). The entire dataset has 6432 such trials, split into a training (80% of the entire dataset) and testing set (20% of the entire dataset). We maintained the approximate ratio of the number of trials between classes. A breakup of the number of trials in the dataset based on the NDD/NT category is provided in Table 1. Since the duration of the trials can be quite long, they can contain information specific to everyone. To avoid overfitting, stemming from such information, from each trial we only select a random sequence of only 40 samples in length.
Reaching paradigm plus raw and filtered kinematic data. (A) shows a participant performing the reaching task. Participants touch a target reappearing at random times on a touch screen while wearing a sensor on their hand, seated comfortably, retracting their hand to the edge of the laptop and repeating the process at their own pace. (B–D) show the raw kinematic data for two sets of forward–backward motions. (B) provides the 3D components of linear acceleration (m/s2), (C) for the angular velocity ω (°/s2), and (D) the roll, pitch, and yaw (RPY). (E) shows the raw (blue) and Gaussian kernel filtered (orange) of the magnitude of the linear acceleration data for one full trial.
To model the temporal movement signal in our neural network, we predominantly used Long Short-Term Memory (LSTM) cells46. LSTM cells are a set of artificial neurons that can retain information when input is provided in sequences. These neurons use learned parameters to maintain: (1) an internal state that summarizes the recent, short-term inputs; and (2) a state summarizing task-relevant long-term history. Collectively, these states enable LSTM cells to store information extracted from the kinematic variable temporal sequences. To transform the LSTM layers’ memory-based output into a representation capable of predicting a diagnosis, the output was passed one-to-one to a Linear Layer, modelling input–output relationships using the learned parameters in the corresponding hyperplane. The number of linear layer outputs corresponds to the four diagnostic categories. The predicted probability of the participant having a specific condition is proportional to the strength of the corresponding output neuron activation. To make a diagnostic prediction, the linear layer outputs are transformed into probability distributions by normalizing their activations using the SoftMax function (described in Note 2 of the SI) and selecting the diagnostic category with maximum probability. Using clinical diagnostic information provided by each participant or their care givers, we represented the desired label as a probability distribution with the entire probability amassed in their clinically diagnosed condition. We then applied the Cross Entropy loss function47 to quantify the output prediction error (see Note 2 in the SI for more details). To prevent overfitting, we applied an L2 regularization to the loss function that penalizes learned weights with large magnitudes. Additionally, we also added a dropout of 0.3 between the two LSTM layers as well as before the linear layer. This means that during training, 30% of those connections were randomly severed thereby reducing the reliance of the network on a small subset of connections and learning redundant representations that generalize. Figure 2 renders the DL network architecture, which takes as input the 3-dimensional components of each kinematic variable and is trained via backpropagation36,37.
The network was initially trained using Stratified K-Fold Cross Validation38 (K = 5, discussed in Note 2 of the SI) applied to the training set (80% of the entire dataset). Each fold trains a network from scratch using the same hyperparameters, but on a slightly different subset of data while maintaining the proportion of data belonging to each class. The network was trained for a maximum of 750 epochs (passes over the entire dataset) on K-1 folds using a mini-batch of size 500. It is then evaluated on the Kth fold (or validation set). This process was repeated multiple times with each fold acting as the validation set and the remaining folds being used to train the model. To prevent overfitting, early stopping is implemented such that training is preemptively stopped when the loss on the validation set does not show improvement for 50 consecutive epochs (patience = 50) with a minimum improvement of at least 0.005 (min delta = 0.005). The network was trained using the Adam optimizer48. Hyperparameters such as the number of LSTM cells, the number of LSTM cells per layer, the regularization constant and the learning rate were tuned using the average performance metrics on the validation sets. Specifically, we use the macro-average of the AUC (the average AUC across all 4 output labels) as a metric to optimize. These hyperparameters result in a network that consists of 2 LSTM layers containing 10 LSTM cells each, a learning rate of 0.001 and a regularization constant of 0.01 when considering all kinematic variables. This process also highlights how the network is indeed able to consistently learn features relevant to the diagnosis task as the training data is varied. Finally, a single neural network was retrained using the best hyperparameters on the entirety of the training set and then evaluated on the remaining, unseen test set.
The DL analysis found improved diagnostic accuracy as a function of the number of training epochs on the validation set as demonstrated by the mean increases and the standard deviation reductions (where the mean and standard deviation were measured across the 5 folds) as well as on the final test set. We assessed multiple networks where the inputs were changed while all other network parameters were fixed.
Table 2 shows the accuracy for different kinematic signal combinations and how using multiple input signals results in higher classification accuracy. Figure 3 shows the learning curves and performance when using all kinematic signals for 5-Fold Cross Validation. We can see the training and validation loss decrease for each fold as well as a corresponding increase in the training and validation accuracy. Figure 4 shows the same metrics for the test set when using the best hyperparameters described above.
Deep Learning analysis results for 5-Fold Cross Validation. (A–D) show the results for each fold for the 5-Fold experiment for the DL training process using all kinematic signals. For (A) the Training Loss, (B) Validation Loss, (C) Training Set Accuracy, and (D) Validation Set Accuracy as a function of completed number epochs. The training loss decreases as the network learns, resulting in a corresponding increase in classification accuracy until convergence at a mean of 66.86% on the validation data. See Note 2 of the SI for additional details and analyses.
Deep Learning analysis results for final training. (A–D) show the results of the DL training process when training on all kinematic signals and the entire training set. For (A) the Training Loss, (B) Test Loss, (C) Training Set Accuracy, and (D) Test Set Accuracy as a function of completed number epochs. The training loss again decreases as the network learns, resulting in a corresponding increase in classification accuracy until convergence at a mean of 71.48% on the unseen test data.
To further assess the DL diagnostic result validity, we calculated the Area Under the Receiver Operating Characteristic (ROC) Curve (AUC) for the validation and test sets. The ROC curve was computed by plotting the true positive rate versus the false positive rate for different classification thresholds in a binary classification task. To extend this to a multi-class problem (the 4 diagnostic conditions we must consider), we plot the ROC for multiple one-vs-rest conditions (described in the Note 3 of the SI). These ROC curves are shown in Fig. 5 for the validation sets during cross validation and Fig. 6 for the final test set when using all kinematic signals. The AUC provides a scale-independent measure between 0 and 1 for the classifier aptitudes. Results for specific experiments are shown in Tables 3 and 4 for the validation sets and the test set respectively. Lastly, we also show the confusion matrix49 on the final test set in Fig. 7 when using all kinematic signals. This is a standard tool used to visualize the predictive performance in classification tasks in machine learning. The rows correspond to the diagnosed conditions of the trials in the test set while the columns correspond to the predicted outputs of the DL. Each cell in the matrix provides the number of times an input in the test set that belongs to a specific category yi was predicted to belong to a category yj. It reflects the potential for the classifications to be correctly or mistakenly predicted.
Receiver operating characteristics (ROC) during 5-Fold cross validation evaluated on the validation sets. The ROC curves for different conditions are shown in a one-vs-rest approach when using all kinematic variables. The corresponding mean and standard deviation (over folds) of the corresponding area under the curve (AUC) for each is shown in the legend. As the curves tend to the top right corner, they approach a perfect classifier area of the ROC (an AUC of 1.0). See Note 3 of SI for additional details.
Receiver operating characteristics (ROC) for the final training evaluated on the test set. The ROC curves for different conditions are shown in a one-vs-rest approach when considering 5-Fold cross validation and using all kinematic variables data sets. The corresponding Area Under the Curve (AUC) for each is shown in the legend. As the curves tend to the top right corner, they approach a perfect classifier area of the ROC (an AUC of 1.0).
The confusion matrix showing count of the true labels along with the predicted labels for trials in the test set. The rows correspond to the true labels of trials in the test set while the columns consist of the predictions made by the network. Each cell shows the number of times a trial with a true label was predicted with the corresponding predicted label. Larger magnitudes along the diagonal show a high degree of correct predictions. This also gives insight into what type of mistakes are made by the network. The network that made the predictions was trained on the entire training set using all the kinematic variables and evaluated on the test set.
For RPY, we find a mean validation accuracy of 65.42% (standard deviation 2.13), while the mean validation accuracy using linear acceleration only was 42.80% (standard deviation 1.24). However, combining these two kinematic variables increases validation accuracy to 66.06% (standard deviation 2.30). Combining all three kinematic variables yields a slightly better accuracy reaching a mean of 66.86% (standard deviation 1.80). It is unclear why the \(\omega\) does not perform as well as other kinematic variables. It consistently performs poorly and provides an extremely small benefit even when combined with other signals. The corresponding accuracies for the test set also shows a similar trend and highlight how RPY seems to have the largest capability to aide in prediction (test accuracy = 67.83%) followed by the linear acceleration (test accuracy = 44.44%) and finally the angular velocity (test accuracy = 32.17%).
The trained models have a mean AUC evaluated on the validation sets ranging from 0.50 to 0.92. If we do not consider experiments that use the angular velocity, the range of the mean validation AUC increases from 0.56 to 0.96 instead, showing a good discriminatory diagnostic ability. Similarly, the AUC on the held-out test set ranges from 0.5 to 0.95, but changes to 0.59 to 0.95 when ignoring any experiments that use the angular velocity. The AUC of NTs is consistently high and clearly distinct from the NDDs. Further, qualitatively, the AUC for the comorbid ASD + ADHD condition is relatively lower, highlighting the difficulty in its diagnosis, as seen often by clinicians. These results add further support to the hypothesis that the NDD conditions inherently affect motor function in a clearly detectable manner.
The confusion matrix highlights the predictive capabilities of the final model. It shows how correct predictions are frequently made (larger values along the diagonal). Additionally, it shows how when incorrect predictions are made for the ASD or ADHD classes, they are often predicted as ASD + ADHD. This is to be expected as the underlying features for ASD or ADHD would also be expected to be concurrent in ASD + ADHD. Interestingly, ASD + ADHD, when predicted incorrectly, is most often identified as NT and vice versa. We later show similar behavior observed in our biometrics.
There is an increase in performance (accuracy and AUC) as multiple signals are used together despite no change in the model structure beyond the number of input neurons. This finding suggests that the information present in the joint probability distribution over several kinematic variables is greater than each individual kinematic measurement. Indeed, visualizing the contributions of input features in a predictive label (Note 4 of SI) concurs with this result. This observation can be used to guide future DL work in this area.
Lastly, to ensure that features based on the underlying NDD/NT condition are being extracted, we performed additional experiments while shuffling the labels of a subset of the data. Shuffling the labels ensures that the proportion of each class is maintained while introducing noise into the dataset. Note 5 of the SI highlights how the performance (measured by both the accuracy and AUC) is negatively impacted by shuffling, suggesting the algorithm was sensitive to important elements of the conditions under study.
We conclude that DL, on the reaching paradigm data, can correctly identify the NDD conditions under study and it has important potential for screening participants for further assessment without a priori clinical information, in particular when several kinematic variables are used in combination39,40,41,42,50,51. We also compared our results to a simpler machine learning approach, demonstrating that our LSTM outperforms a similar Support Vector Machine based classification model (described in detail in Note 6 of the SI).
However, we wanted to go beyond the DL diagnoses to further understand the motor data measured more physiologically, as will be discussed in the next section.
Statistical analysis
To quantify and to further understand the inherent cognitive information in a participant’s movements, we filtered out the high frequency electronic noise present in the sensors from the kinematic data collected from the reaching protocol, as described in Note 7a of the SI. The filtered signal contains a smooth component plus the participant’s physiological motor noise. We found millisecond time scale kinematic fluctuations that show randomness in their frequency and magnitude that are characteristic to each participant. We compared changes in the amplitudes between subsequent, nearest neighbor (NN) local maxima and minima (∆ANN) that occur within each trial of a participant’s data (see Fig. 8A), focusing here on the linear acceleration while the linear jerk is discussed in Note 7b of the SI. We obtained the frequency histogram, P(∆ANN), from these amplitude changes, shown in Figs. 8B–E for participants of varying diagnosis and severity. Note that for ease of readability between the 3 NDD conditions, only for those participants whose severity was judged by a clinician, we used low-functioning (LF), mid-functioning (MF) and high-functioning (HF) corresponding to Level 3, Level 2, and Level 1 for ASD as per the DSM-518.
Statistical distributions of the amplitude change between nearest neighbor (NN) extrema. (A) shows data for an example of the local maxima-minima locations of the linear acceleration, highlighting two examples of the NN magnitude fluctuations (blue arrows). (B–E), show the histograms of the change in amplitude between NN extrema. In (B) for a NT, (C) mid-functioning ASD + ADHD, (D) a low-functioning ASD and (E) a high-functioning ADHD participant. Similar results for the linear jerk are shown in Fig. S6.
We used the Fano Factor44 (FF) of the P(∆ANN) to quantify the randomness of a participant’s movements. The Shannon entropy (S) also provides an added biometric to represent the information about the random kinematic physiological fluctuations present in the movement histograms. These biometrics were selected to provide summarizing information about the amount of randomness in P(∆ANN) and thus reflect the irregular nature of the fluctuations. Both biometrics are explained further in Note 8 of the SI.
We find that the FF biometrics are less than one, indicating under-dispersion or sub-Poisson variability in the P(∆ANN). Figure 9 provides the results for the FF (left column) and S (right column) for all participants by diagnostic category. ASD + ADHD participants show more overlap with NT participants for all functioning levels compared to the other diagnoses (Fig. 9A). This matches the results obtained in the DL, where trials from ASD + ADHD and NT also showed similarities in the confusion matrix. ASD shows trends between different clinical severities and a clear separation from NT (Fig. 9C). This is an important consideration for providers when testing a participant for the first time. Participants with high functioning ADHD have FF results close to those of ASD + ADHD participants, but MF and LF ADHD participants had very low FF values (Figure 9E). Figs. 9B, D, and F provide the corresponding S associated with the \(P\left({\Delta A}_{NN}\right)\) for the linear acceleration showing how an increase in severity correlates well with larger entropy in all NDDs.
Linear acceleration Fano Factor (FF, Left Column) and Shannon entropy (S, Right Column). Rows include (A, B) ASD + ADHD comorbidity, (C, D) ASD, and (E, F) ADHD organized by functional level and compared to NT. Both metrics show consistent discrimination between NDDs, although each provides a different type of statistical information. Linear jerk results are shown in Fig. S7.
We also tested the stability or habituation for a participant’s biometrics as a function of the number of trials (discussed in detail in Note 9 of the SI). We found that, in most cases, habituation occurs well within 100 trials. This allows us to study how reliable the biometrics are for a single participant as more reaching trials are collected. For example, the FF for the linear acceleration reaches 100% habituation at an average of 27.41 trials and demonstrates stability with a mean and standard deviation of the variance after habituation of only 5.85 × 10−5 and 1.21 × 10−4, respectively. The S results are consistent to those of the FF. It was found that 95.65% of participants habituated at an average of 64.30 trials. After habituation, the mean variance of S was 3.40 × 10−3 with a standard deviation of 4.8 × 10−3. Similar results for the linear jerk are provided in Table S5 and S6 in the SI.
Discussion
The goals of this research were two-fold. First, to develop Deep Learning solutions for rapid ASD, ADHD, ASD + ADHD, and NT assessments that yielded high diagnostic predictive accuracy. Secondly, to analyze the unique physiological information present in each participant’s data about their diagnostic severity information. Motor data was gathered from hand-reaching experimental protocols using high-definition, high sampling rate wireless sensors. Our results support the idea that important discriminative clinical information is embedded in the way a person moves, if considered at timescales invisible to the naked eye. Our cohort had competition between three NDD conditions and NT. We tested the stability and accuracy of the DL prediction results through 5-Fold Cross Validation and evaluation of the accuracy and ROC analysis on a held-out test set. The predictive power of the DL increased when more than one kinematic variable was used but had limited information provided by the angular velocity. Subsequent SHAP analysis concurred. We also evaluate DL performance while shuffling the labels of different proportions of the dataset as well as in comparison to a more traditional Support Vector Machine classifier (details in SI). We studied the unique millisecond random fluctuations in terms of two biometrics characterizing their distributions: the Fano Factor and the Shannon Entropy that quantitatively characterized the severity of each NDD condition. We analyzed the stability of the biometric results by evaluating the habituation of these biometrics as a function of trial number, helping to establish a baseline for future testing.
After training on a larger and more comprehensive dataset, the Deep Learning approach could play an important role as an early screening tool for participants suspected of having a neurodivergent disorder, not only in the clinic but also in schools and other non-medical settings. With rapid improvements in sensor technology, MEM sensors are becoming more affordable, reliable and ubiquitous (such as in smartphones and smartwatches) making the study of kinematic data for applications such as this increasingly relevant. Of importance in the DL study was that we used only the raw kinematic data, which included the electronic sensor noise. It is also unknown if the participants had taken their prescribed medication (if any) for their NDD condition. Despite that complexity, the DL analysis seems to extract the inherent cognitive information inherent in the participants’ movement. Other architectures may also be informative when applied to this problem. For example, transformer-based models can capture long-range characteristics. Another future area for exploration is the use of ensemble methods, such as Random Forests, that combine the predictions from multiple (relatively) weak models to mitigate overfitting or bias.
This study is a proof of concept that applying Deep Learning methods can identify NDD conditions in new participants by using high precision motor data. Future longitudinal studies may also use these metrics to assess how they change over time or as a function of clinical intervention. Larger complementary data sets will be needed to further test the conclusions and potential extensions of the results presented in this paper.
Methods
Study design
We tested a total of 109 participants, including NDD and NT; 17 of them were removed from the final dataset for reasons described below. Participants were recruited from outpatient child and adolescent psychiatry clinics at Indiana University School of Medicine (IUSM) and from students at its Bloomington campus, between 2019 and 2023. Our protocols were approved by the IRB at Indiana University and all participants provided informed consent or assent from them or their legal guardian, in compliance with the Helsinki Declaration. Eligibility was limited to ages 5 + , who were able to perform the reaching task for the duration of the study. The 17 participants that were excluded from the analysis were either because of an inability to finish the test, additional disabilities affecting their motor skills, or issues with the data collection device. Of the 92 participants who contributed to the dataset, 17 had ASD, 15 had ADHD, 26 had comorbid ASD + ADHD, and 34 were NT. Table S8 and S9 in the SI provide more details about the participants’ demographics.
People with and without NDDs were encouraged to participate. Participants with NDDs had their condition and its severity diagnosed by a provider and reported by themselves. For children recruited from the IUSM clinic, diagnosis was confirmed by their psychiatrist. We did not ask if the NDD participants had or had not taken medication for their NDD condition; it appears, however, that the DL analysis can perform well despite this complexity.
Kinematic variables measured
We used XSENS MTw Awinda high-definition Bluetooth 120 Hz motion capture sensors (www.xsens.com/products/mtw-awinda). The sensors were inserted in a glove used on the dominant hand of the participant as they performed the reaching protocol, shown in Fig. 1A. The participant was instructed to begin with their hand placed at the corner of the laptop and to touch the target on the touchscreen and to return their hand to the starting position at a pace that was comfortable for them and to repeat the process when the target reappeared. The target appeared roughly 100 times, giving 100 forward and 100 backward trials, taking about 15 min. The measured kinematic data included the roll, pitch, and yaw (RPY), the angular velocity (ω), and the linear acceleration (a). The linear jerk (j)52,53 was obtained from the time derivative of the acceleration. Figure 1B–D illustrate one second unfiltered or “raw” RPY, ω, and results from a participant with ASD + ADHD. Figure 1E illustrates the effect of filtering the noise from the linear acceleration signal (discussed in Methods and in Note 7a of the SI). Not all kinematic signals show the same utility for NDD identification using DL. As discussed in the main body of the paper, generally there is an increase in performance (accuracy and AUC) when multiple signals are used together despite no change in the DL model structure beyond the number of input neurons.
Electronic filtered data analysis
The raw data collected from the sensors included a considerable amount of high frequency electronic sensor noise. To suppress this high frequency noise, we convolved the raw sensor data signals with a Gaussian Filtering kernel (GF). The GF is a low pass filter minimizing the effects of electronic noise. The filtered signal contains a smooth motor component plus its physiological noise produced during the motion. We describe the GF calculational details in Note 7a of the SI.
Deep learning data analysis
We implemented a supervised deep neural network to predict NDD conditions using the raw kinematic sensor data. We used the Long Short-Term Memory (LSTM) cells as the neurons of the network. LSTM cells are a form of artificial neurons that can retain information when the input is provided in sequences. We discuss the implementation details in Note 2 of the SI.
Statistical analysis
The smoothness of the trajectories depends on the number and size of the nearest neighbor extrema separations. For each kinematic variable of each participant, a histogram was made from the frequency of the amplitude changes between consecutive extrema. The histograms were characterized by their Fano Factor and Shannon Entropy, providing a measure of the participants’ NDD severity.
Data availability
All code and anonymized data associated with the study is available on GitHub (https://github.com/Neurodivergent-Motor-Testing-Lab/NDD_motor_diagnosis_meta_repository)
References
Developmental Disabilities, CDC.
Danielson, M. L. et al. ADHD prevalence among U.S. children and adolescents in 2022: Diagnosis, severity, co-occurring disorders, and treatment. J. Clin. Child Adolesc. Psychol. 53(53), 343–360 (2024).
Antshel, K. M., Zhang-James, Y., Wagner, K. E., Ledesma, A. & Faraone, S. V. An update on the comorbidity of ADHD and ASD: A focus on clinical management. Expert Rev. Neurother. 16, 279–293 (2016).
Joshi, G. et al. Symptom profile of ADHD in youth with high-functioning autism spectrum disorder: A comparative study in psychiatrically referred populations. J. Atten. Disord. 21, 846–855 (2017).
Lee, D. O. & Ousley, O. Y. Attention-deficit hyperactivity disorder symptoms in a clinic sample of children and adolescents with pervasive developmental disorders. J. Child Adolesc. Psychopharmacol. 16, 737–746 (2006).
Leitner, Y. The co-occurrence of autism and attention deficit hyperactivity disorder in children—What do we know?. Front. Hum. Neurosci. 8, 268 (2014).
Leyfer, O. T. et al. Comorbid psychiatric disorders in children with autism: Interview development and rates of disorders. J. Autism Dev. Disord. 36, 849–861 (2006).
Simonoff, E. et al. Psychiatric disorders in children with autism spectrum disorders: Prevalence, comorbidity, and associated factors in a population-derived sample. J. Am. Acad. Child Adolesc. Psychiatry 47, 921–929 (2008).
Sinzig, J., Walter, D. & Doepfner, M. Attention deficit/hyperactivity disorder in children and adolescents with autism spectrum disorder: Symptom or syndrome?. J. Atten. Disord. 13, 117–126 (2009).
Taurines, R. et al. ADHD and autism: Differential diagnosis or overlapping traits? A selective review. ADHD Atten. Deficit Hyperact. Disord. 4, 115–139 (2012).
Colombi, C. & Ghaziuddin, M. Neuropsychological characteristics of children with mixed autism and ADHD. Autism Res. Treat. 2017, 5781781 (2017).
Goldstein, S. & Schwebach, A. J. The comorbidity of pervasive developmental disorder and attention deficit hyperactivity disorder: Results of a retrospective chart review. J. Autism Dev. Disord. 34, 329–339 (2004).
Russell, G., Rodgers, L. R. & Ford, T. The strengths and difficulties questionnaire as a predictor of parent-reported diagnosis of autism spectrum disorder and attention deficit hyperactivity disorder. PLoS ONE 8, e80247 (2013).
Sprenger, L. et al. Impact of ADHD symptoms on autism spectrum disorder symptom severity. Res. Dev. Disabil. 34, 3545–3552 (2013).
Yoshida, Y. & Uchiyama, T. The clinical necessity for assessing attention deficit/hyperactivity disorder (AD/HD) symptoms in children with high-functioning pervasive developmental disorder (PDD). Eur. Child Adolesc. Psychiatry 13, 307–314 (2004).
Christensen, D. et al. Prevalence and characteristics of autism spectrum disorder among 4-year-old children in the autism and developmental disabilities monitoring network. J. Dev. Behav. Pediatr. JDBP 37, 64 (2015).
Sharma, S. R., Gonda, X. & Tarazi, F. I. Autism spectrum disorder: Classification, diagnosis and therapy. Pharmacol. Ther. 190, 91–104 (2018).
American Psychiatric Association. DSM-5, Diagnostic and Statistical Manual of Mental Disorders (2013).
Santosh, P., Baird, G., Pityaratstian, N., Tavare, E. & Gringras, P. Impact of comorbid autism spectrum disorders on stimulant response in children with attention deficit hyperactivity disorder: A retrospective and prospective effectiveness study. Child Care Health Dev. 32, 575–583 (2006).
Jang, J. et al. Rates of comorbid symptoms in children with ASD, ADHD, and comorbid ASD and ADHD. Res. Dev. Disabil. 34, 2369–2378 (2013).
Posserud, M., Hysing, M., Helland, W., Gillberg, C. & Lundervold, A. J. Autism traits: The importance of “co-morbid” problems for impairment and contact with services. Data from the Bergen Child Study. Res. Dev. Disabil. 72, 275–283 (2018).
Fabbri-Destro, M., Cattaneo, L., Boria, S. & Rizzolatti, G. Planning actions in autism. Exp. Brain Res. 192, 521–525 (2009).
Cattaneo, L. et al. Impairment of actions chains in autism and its possible role in intention understanding. Proc. Natl. Acad. Sci. 104, 17825–17830 (2007).
Mari, M., Castiello, U., Marks, D., Marraffa, C. & Prior, M. The reach–to–grasp movement in children with autism spectrum disorder. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 358, 393–403 (2003).
Gernsbacher, M. A., Sauer, E. A., Geye, H. M., Schweigert, E. K. & HillGoldsmith, H. Infant and toddler oral-and manual-motor skills predict later speech fluency in autism. J. Child Psychol. Psychiatry 49, 43–50 (2008).
Iverson, J. M. & Goldin-Meadow, S. Gesture paves the way for language development. Psychol. Sci. 16, 367–371 (2005).
Gentilucci, M., Campione, G. C., De Stefani, E. & Innocenti, A. Is the coupled control of hand and mouth postures precursor of reciprocal relations between gestures and words?. Behav. Brain Res. 233, 130–140 (2012).
Denisova, K. & Wolpert, D. M. Sensorimotor variability distinguishes early features of cognition in toddlers with autism. iScience 27, 110685 (2024).
Minshew, N. J., Sung, K., Jones, B. L. & Furman, J. M. Underdevelopment of the postural control system in autism. Neurology 63, 2056–2061 (2004).
Saad, J. F., Griffiths, K. R. & Korgaonkar, M. S. A systematic review of imaging studies in the combined and inattentive subtypes of attention deficit hyperactivity disorder. Front. Integr. Neurosci. 14, 31 (2020).
Surgent, O. et al. Microstructural neural correlates of maximal grip strength in autistic children: The role of the cortico-cerebellar network and attention-deficit/hyperactivity disorder features. Front. Integr. Neurosci. 18, 64 (2024).
Crippa, A. et al. Use of machine learning to identify children with autism and their motor abnormalities. J. Autism Dev. Disord. 45, 2146–2156 (2015).
Anzulewicz, A., Sobota, K. & Delafield-Butt, J. T. Toward the autism motor signature: Gesture patterns during smart tablet gameplay identify children with autism. Sci. Rep. 6, 31107 (2016).
De Francesco, S. et al. A multimodal approach can identify specific motor profiles in autism and attention-deficit/hyperactivity disorder. Autism Res. 16, 1550–1560 (2023).
Wu, D., José, J. V., Nurnberger, J. I. & Torres, E. B. A biomarker characterizing neurodevelopment with applications in autism. Sci. Rep. 8, 1–14 (2018).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Schmidhuber, J. Deep learning in neural networks: An overview. Neural Netw. 61, 85–117 (2015).
Allen, D. M. The relationship between variable selection and data agumentation and a method for prediction. Technometrics 16, 125–127 (1974).
Lai, M. et al. A machine learning approach for retinal images analysis as an objective screening method for children with autism spectrum disorder. EClinicalMedicine 28, 100588 (2020).
Wolfers, T., Buitelaar, J. K., Beckmann, C. F., Franke, B. & Marquand, A. F. From estimating activation locality to predicting disorder: A review of pattern recognition for neuroimaging-based psychiatric diagnostics. Neurosci. Biobehav. Rev. 57, 328–349 (2015).
Orru, G., Pettersson-Yeo, W., Marquand, A. F., Sartori, G. & Mechelli, A. Using support vector machine to identify imaging biomarkers of neurological and psychiatric disease: A critical review. Neurosci. Biobehav. Rev. 36, 1140–1152 (2012).
Yahata, N. et al. A small number of abnormal brain connections predicts adult autism spectrum disorder. Nat. Commun. 7, 1–12 (2016).
Lundberg, S. M. & Lee, S.-I. A unified approach to interpreting model predictions. In Advances in Neural Information Processing Systems vol. 30 (2017).
Fano, U. Ionization yield of radiations. II. The fluctuations of the number of ions. Phys. Rev. 72, 26 (1947).
Torres, E. B. et al. Autism: The micro-movement perspective. Front. Integr. Neurosci. 7, 32 (2013).
Hochreiter, S. & Schmidhuber, J. Long short-term memory. Neural Comput. 9, 1735–1780 (1997).
Cox, D. R. The regression analysis of binary sequences. J. R. Stat. Soc. Ser. B Methodol. 20, 215–232 (1958).
Kingma, D. P. & Ba, J. Adam: A Method for Stochastic Optimization (2017).
Stehman, S. V. Selecting and interpreting measures of thematic classification accuracy. Remote Sens. Environ. 62, 77–89 (1997).
Washington, P. et al. Data-driven diagnostics and the potential of mobile artificial intelligence for digital therapeutic phenotyping in computational psychiatry. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 5, 759–769 (2020).
Alves, C. L. et al. Diagnosis of autism spectrum disorder based on functional brain networks and machine learning. Sci. Rep. 13, 8072 (2023).
Hogan, N. An organizing principle for a class of voluntary movements. J. Neurosci. 4, 2745–2754 (1984).
Todorov, E. & Jordan, M. I. Smoothness maximization along a predefined path accurately predicts the speed profiles of complex arm movements. J. Neurophysiol. 80, 696–714 (1998).
Acknowledgements
KPD thanks Dr. Roderic Grupen for his continuous support of this work. Thanks to N.W. Parris for his assistance in some of the clinical testing. This work was partially funded by the National Science Foundation grant 1640909 (JVJ, DW) and grant 1735095, Interdisciplinary Training in Complex Networks (CM).
Author information
Authors and Affiliations
Contributions
Project was proposed by JVJ and JIN. JIN, MHP, CM and AP, helped recruit the participants tested. KPD, DW and CM developed programs and software to analyze the data gathered. AP and CM performed most of the clincial testing, with help from N.W. Parris. The manuscript was written by JVJ, KPD, MHP, and CM with important intelectual contributions and approval from all authors. The whole project was supervised by JVJ.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Doctor, K.P., McKeever, C., Wu, D. et al. Deep learning diagnosis plus kinematic severity assessments of neurodivergent disorders. Sci Rep 15, 20269 (2025). https://doi.org/10.1038/s41598-025-04294-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-04294-9