Abstract
Brain tumor classification (BTC) from Magnetic Resonance Imaging (MRI) is a critical diagnosis task, which is highly important for treatment planning. In this study, we propose a hybrid deep learning (DL) model that integrates VGG16, an attention mechanism, and optimized hyperparameters to classify brain tumors into different categories as glioma, meningioma, pituitary tumor, and no tumor. The approach leverages state-of-the-art preprocessing techniques, transfer learning, and Gradient-weighted Class Activation Mapping (Grad-CAM) visualization on a dataset of 7023 MRI images to enhance both performance and interpretability. The proposed model achieves 99% test accuracy and impressive precision and recall figures and outperforms traditional approaches like Support Vector Machines (SVM) with Histogram of Oriented Gradients (HOG), Local Binary Pattern (LBP) and Principal Component Analysis (PCA) by a significant margin. Moreover, the model eliminates the need for manual labelling—a common challenge in this domain—by employing end-to-end learning, which allows the proposed model to derive meaningful features hence reducing human input. The integration of attention mechanisms further promote feature selection, in turn improving classification accuracy, while Grad-CAM visualizations show which regions of the image had the greatest impact on classification decisions, leading to increased transparency in clinical settings. Overall, the synergy of superior prediction, automatic feature extraction, and improved predictability confirms the model as an important application to neural networks approaches for brain tumor classification with valuable potential for enhancing medical imaging (MI) and clinical decision-making.
Similar content being viewed by others
Introduction
Accurate brain tumor classification is essential for the treatment plan and prognosis of patients, such imaging demands greater attention from medical personnel1. There are generally two types of brain tumors — benign and malignant — and they vary in how fast-growing, serious or treatable they are. The most common types of brain tumors are gliomas, meningiomas and pituitary tumors2. The non-invasive gold standard for brain imaging is magnetic resonance imaging (MRI), which allows the in vivo visualization of highly detailed images of brain structures. It is important to note that while human-based interpretation is quite thorough, it is also limited by radiologist variability, increased time demand, and difficulty distinguishing tumors on MRIs that appear very similar. This leads to the necessity for automated and yet reliable and explainable clinical decision support systems3,4. High risk tumors which are undertreated or low risk tumors which are overtreated can indirectly harm the patients if misclassified. Precise and correct multiclass diagnosis of brain tumors is therefore vital, as incorrect classifications can significantly impact on the choice of the treatment and prognosis5.
As a result, BTC has gone through several stages of advancement from conventional machine learning fundamentals to novel end to end deep learning architectures. More recently, MRI typification has been performed based on feature extraction methods such as HOG (Histogram of Oriented Gradients) and LBP (Local Binary Patterns), as well as on a dimensionality reduction method [PCA]. Traditional machine learning paradigms saw gains in performance when novel histogram-based features were applied. These conventional methods suffer from the major drawback of depending on hand-crafted imaging features that are typically not robust enough to accommodate the spatial complexity and variability in MRI scans6. In contrast, deep learning approaches such as CNNs which have enabled automatic hierarchical feature extraction directly from raw images, consequently reducing the need for manual feature engineering and achieving better classification performance7,8. Applying transfer learning approaches such as VGG16, and InceptionResNetV2 that obtains knowledge from large-scale image datasets, has been widely used in medical imaging9.
In addition, ensemble-based transfer learning methods have demonstrated that combining several pre-trained models effectively improve the classification performance also remedies the small data issue in medical fields10,11,12.These foundations are used in recent studies to take new directions in BTC. Dense CNN architectures are superior in feature representation leading to multiclass tumor classification improvement13. Differential evolution algorithms have produced stronger detection performance ensembles of networks without significantly increasing computational cost to the optimization14,15. Meta-heuristic optimization based on manta ray foraging and advanced residual blocks have also surpassed the previous methods. Hybrid deep network architectures which are based on combining different CNN designs have shown clear advantages in improving classification performance16.
An interesting reference is the application of residual skip block-based modified MobileNet models (ResMobileNet) that achieve a trade-off between diagnostic performance and computational efficiency for the case of MRI brain tumor detection. At the same time, there are still many hurdles that need to be addressed before these technologies see confident adoption into real world clinical practice. One of the biggest problems is the black-box nature of most deep learning models, which gives rise to a serious limitation—interpretability, essentially, a necessity in an accurate medical diagnosis. While, more than that, most of the existing methods consider all regions in MRI scans as uniformly important, because they are fed directly as model input, resulting in the majority of features becoming irrelevant for diagnosis. While attention mechanisms have the potential to emphasize diagnostically relevant regions, they have not yet been integrated sufficiently or effectively into transfer learning paradigms. This also leaves space for approaches that combine the data efficiency of classical machine learning and expressive modelling power of deep learning, a need for which we expect further hybrid solutions17.
To tackle these gaps, we propose a hybrid model that combines transfer learning and attention-based mechanisms to pay attention to tumor-related features in MRI images collected from Kaggle18. We use a VGG16 backbone and a Custom Attention (CA) layer that employs SoftMax-weighted attention to dynamically weigh tumor-specific features. This design takes advantage of the strengths of transfer learning where a network trained previously on a rich image domain can be repurposed to a narrowly-defined medical task, and enhances feature representation through attention. Our model is capable of highlighting clinically relevant areas, unlike regular CNNs, which pay equal attention to all features in the image. We further improve interpretability by using Grad-CAM visualizations to produce the heatmaps that show which parts of the image were most responsible for each prediction.
We pursue three objectives in this work. To address this issue, we firstly attempt to enhance classification accuracy through the integration of transfer learning and attention mechanisms that focus on tumor characteristics. Second, we assess whether techniques for visualization such as Grad-CAM could improve interpretability to yield human interpretable (and thus potentially actionable) model outputs for clinicians. Thirdly, we evaluate whether our hybrid CNN architecture achieves better performance compared to existing models in classifying glioma, meningioma, pituitary tumor, as well as no-tumor cases in terms of accuracy and inference metrics. In light of this, the primary aim of this work is fundamentally to bring the practice of obtaining high-quality AI models in line with what are often critical clinical expectations for transparency and reliability. Our model is evaluated on the publicly available Kaggle dataset that consists of MRI images of four distinct tumor classes. Integrating technical novelty with clinical relevance, our work provides a clinically useful and unique solution to brain tumor classification in medical images. In conclusion, we have laid out a methodologically sound and clinically applicable approach to brain tumor classification that provides a performance-interpretability trade-off. Our main contributions are: (i) a VGG16 transfer learning architecture combined with a SoftMax-weighted attention mechanism focusing on the most diagnostically relevant areas of the MRI scans, (ii) Grad-CAM visualizations improving interpretability of the model predictions validated by expert radiologists, and (iii) an assessment of our method against other recent approaches utilizing the same benchmark dataset. This work progresses towards reliable, explainable AI systems for neurodiagnostic applications through the alignment of algorithmic innovation with clinical insight.
The rest of the article is structured as follows: Brain tumor classification techniques have been reviewed in “Literature review”. In “Materials and methods”, we present the dataset and pre-processing (3.1), proposed model architecture (3.2) and evaluation metrics (3.3). Classification performance, visual interpretation, comparisons with previous models and detailed analysis is covered in “Results, discussions and evaluation”. Limitations and future scope are included in “Future work”, with the summary presented in “Conclusion”.
Literature review
Mathivanan et al.19 proposed a secure Brain-Tumor Detection Network (BTDN), which enhances the quality of MRI images and ensures secure data transmission with highly accurate tumor classification. On three datasets—Br35Hc, BraTS, and Kaggle MRI data—BTDN achieved accuracy rates of 99.68%, 98.81%, and 95.33%, respectively. To ensure secure data transmission, the study also incorporated Secure-Net (SN), making it a comprehensive and reliable package for medical diagnosis. Hun et al.20 presented DL-based classification of brain tumors using an ensemble of fine-tuned models included DenseNet121 and EfficientNet-B0. Incorporating fully connected layers to enhance classification performance produced a remarkable increase of 99.67% and 99.39% accuracy on the Figshare Dataset-I and the Kaggle Dataset-II, respectively.
Strika21 et al. published a narrative review on the use of artificial intelligence (AI), particularly large language models (LLMs), to reduce disparities in healthcare in “medical deserts.” Their work focused on AI-enabled telehealth, diagnostic chatbots, and eHealth platforms as a way to enhance access to quality care in under-resourced regions. They also described the applications of AI in medical teaching and real-time decision making, thereby underlining its role in overcoming systemic disparities in global health care delivery. Wang et al.22 presented a complete review to deep learning Model in medical image analysis by explaining CNN and its various generation, and transfer leaning approach. Structural characteristics of CNNs were considered as well as their use in various medical domains, and how transfer learning helps with performance in small-data conditions was discussed. Their results lend support for the growing use of hybrid architectures, and context-specific pre-trained models to enhance clinical decision making and diagnostic accuracy.
Sara Tehsin et al.23 proposed GATransformer, a novel transformer architecture built upon a Graph Attention Network (GAT) to enhance explainability in brain tumor detection (BTD). It makes an integration of attention mechanism, GAT, and transformers, in order to extracting deeper features and improving model representation. It offers interpretable attention maps that expose certain tumor regions, aiding clinical understanding. GATransformer achieved high classification accuracy on FigShare, Kaggle, BraTS2019 and BraTS2020 datasets by also improving model transparency.
Sachdeva et al.24 compared multiple pre-trained CNN models, concluding that Efficient Net performed the best. Experimental results show that, compared with the classical model, it improves the classification accuracy by 2.14% and 3.98% on Kaggle and BraTS respectively, and achieves 97.32% on the real-time datasets. Sandeep Kumar Mathivanan25 et al. 2024, examined the transfer learning model to detect brain Tumor from the Kaggle Brain MRI data set, and it comprises four transfer learning models ResNet152, VGG19, DenseNet169, MobileNetv3. Using image enhancement, cross-validation, and frontal images without rotation, MobileNetv3 outperforms comparative methods and achieves the highest accuracy at 99.75% over the categories pituitary, normal, meningioma, and glioma. Ahmed et al.26 proposed ‘Brain tumor detection and classification in MRI using hybrid ViT and GRU model with explainable AI in Southern Bangladesh’. In this study, a hybrid Vision Transformer (ViT) and Gated Recurrent Unit (GRU) model was proposed for BTD using MRI data collected from BSMMCH, Bangladesh. So, it extracted features with ViT and scrutinized relationships with GRU, resulting in an F1-score of 97% on BrTMHD-2023 dataset. The best achieved accuracy was 98.97% with AdamW. Attention Maps, SHAP, and LIME are explainable AI techniques that helped increase model interpretability. The model achieved 1.26% improvement over the existing transfer learning models.
Krishnan et al.27 in their work ‘Enhancing brain tumor detection in MRI with a rotation invariant Vision Transformer’ proposed to resolve various issues using rotated patch embeddings. When evaluated on the Kaggle Brain Tumor MRI dataset, RViT achieved an accuracy of 98.6%, significantly higher than traditional ViT models. The method increased robustness in tumor classification and may be generalized to additional MI tasks. Islam et al.28 in their work ‘Deep Fusion Model for BTC Using Fine-Grained Gradient Preservation’ proposed a fusion model which combined ResNet152V2 and modified VGG16 was presented, it was especially optimizing for deployment in areas with limited compute resource. Thereby, the model preserved gradients properly and yielded 98.36% accuracy on Figshare and 98.04% accuracy on Kaggle. For efficient deployment on edge devices, the model size was minimized using 8-bit quantization from 289.45 MB to 73.88 MB. Grad-CAM was added to improve interpretability. Aamir et al.29 proposed hyperparametric CNN model for BTD. The model was validated on three Kaggle MRI datasets, achieving an average accuracy of 97% by fine-tuning various hyperparameters—including, but not limited to, batch size, number of layers, learning rate, activation function, and filter size. The proposed optimized CNN model demonstrated strong generalization ability and outperformed state-of-the-art approaches in BTC. Rao et al.30in their work ‘An Efficient Brain Tumor Detection and Classification Using Pre-Trained CNN Models’ employed a DL approach for MRI-based BTC using pre-trained CNN models (ResNet50, EfficientNet). To enhance the model’s robustness, data augmentation techniques were applied. The proposed model outperformed existing methods in terms of accuracy, precision, and recall, as evaluated using metrics such as confusion matrices and validation loss.
Tin et al.31 developed the Xception Deep CNN (DCNN) for BTC, effectively addressing issues related to overfitting and computational complexity. The model was trained using TensorFlow on the Kaggle and BraTS datasets, achieving a training accuracy of 99 ± 0.005% and a validation accuracy of 98 ± 0.2%, while requiring relatively low computational resources. Natha et al.32 proposed a new multi-model ensemble DL model, referred to as SETL_BMRI, that includes a set of feature extractors (AlexNet and VGG19) to enhance the classification performance. Their model was tested on a Kaggle MRI dataset, which had images of meningioma, glioma, and pituitary tumor with an accuracy of 98.70%, and precision, recall and F1-scores were more than 98.6%. Güler et al.33in their study ‘Brain Tumor Detection with DL Methods: Classifier Optimization Using Medical Images’, focused on MRI-based BTC using various DL architectures, including VGG, ResNet, DenseNet, and SqueezeNet. The use of ensemble learning combined with parameter optimization significantly improved performance, with ResNet achieving 100% accuracy. Verma et al.34 investigates the significance of data management in BTD using CNNs and also emphasises on data optimization methodologies to obtain right class of classification. This highlights how much conversion the machine learning-based preprocessing, augmentation, and model tuning can do.
Shamshad et al.35 studied the performance of transfer learning models for MRI-based BTC, which includes VGG-16, VGG-19, Inception-v3, ResNet-50, DenseNet, and MobileNet. VGG-16 reached an accuracy of 97% and a 22% reduction in computational freeze. Gade et al.36 proposed an enhanced Lite Swin Transformer (OLiST) for BTD. The model leverages the global feature extraction capabilities of transformers combined with the strengths of CNNs. Additionally, the Barnacles Mating Optimizer (BMO) was employed for hyperparameter optimization, making both components more efficient. The OLiST model significantly improved classification performance and demonstrated faster processing speeds compared to other transfer learning approaches on the Kaggle MRI brain tumor dataset.
Wageh et al.37 demonstrated the development of an MRI image-based computer-aided detection (CAD) system for the early discovery of brain tumors. The model used pre-trained CNNs (VGG-16, Inception V3, ResNet-101, and DenseNet-201) to perform transfer learning. A genetic algorithm was used to select and concatenate these features. When applied to two open-access datasets (Navoneel and Br35H), it provided the accuracy of 99.7% and 99.8%, respectively, outperforming comparators. Rahman et al.38 proposed PDCNN (Parallel Deep Convolutional Neural Network) to enhance the extraction of both global and local features by applying dropout regularization and batch normalization in order to avoid overfitting considerations. The model reached 97% accuracy on several datasets.
Zhang et al.39 MRC-TransUNet: MRC-TransUNet is state of the art segmentation model that utilizes residual transformer modules with efficient backbone to improve segmentation results in medical imaging. Their approach addresses the semantic gap caused by the U-Net skip connections using MR-ViT and reciprocal attention architectures. We found that the architecture achieved superior performance on breast, brain, and lung images, with evaluation metrics (Dice coefficient and Hausdorff distance) demonstrating its potential suitability for a clinical setting in a variety of imaging scenarios.
Vankdothu et al.40 in their study “Brain Tumor Detection and Classification: A CNN-LSTM based Approach” proposed a CNN-LSTM hybrid model for detecting and classifying brain tumors through MRI images. The Kaggle dataset on which the model was trained achieved greater accuracy than conventional CNN and RNN models. Among some of the recent studies Vankdothu, et al.41 in their study “Brain tumor MRI images identification and classification based on the recurrent CNN” proposed Identifying and classifying the MRI images of brain Tumors based on RCNN, the proposed method begins with a preprocessing step (removing background noise), followed by the segmentation mechanism (IKMC) and feature extraction (GLCM) to classify brain Tumors. The proposed method achieved an overall cross-validation accuracy of 95.17%, exceeding those of BP, U-Net and standard RCNN models.
Rasool et al.42 introduced a hybrid CNN-based model comprising GoogleNet al.ong with SVM and SoftMax classifiers. Kesav et al.43 proposed a novel low-complexity RCNN architecture incorporating a Two-Channel CNN for BTC and detection. Their approach achieved 98.21% accuracy in classifying glioma versus healthy MRIs using the Figshare and Kaggle datasets. Irsheidat et al.44 proposed a CNN-based model which uses mathematical operations and matrix analysis for BTD from MRI images. Their model demonstrated effective tumor detection with high performance when validated on different datasets. Choudhury et al.45 proposed a CNN based model to classify MRI images for tumor detection followed by a fully connected network for classification. This approach improved early detection compared to traditional manual diagnostic methods. Anil et al.46 model classify brain MRIs into tumor and non-tumor categories using transfer learning and DL-based techniques. This model accelerated the detection process by reducing manual effort and resource usage, which were significant limitations in earlier methods. It demonstrated better applicability compared to other existing approaches. Febrianto et al.47 model focuses on early diagnosis, comparing two CNN models to classify brain tumors in MRI images before diagnosis. It used DL for image classification without the need for expert input while having varied data. The model performed classification fairly well.
Materials and methods
This section explains the various steps involved in multiclass BTC using MRI scans, outlining dataset preparation, preprocessing, exploratory data analysis (EDA), model architecture, training, hyperparameter tuning, and evaluation. To achieve accuracy and interpretability to facilitate clinical diagnostics, we take an approach that leverages an attention mechanism, visualization tools and a pre-trained CNN. The methodological framework is designed to be rigorous, reproducible, and clinically meaningful.
Exploratory data analysis and preprocessing
Brain Tumor MRI Dataset, retrieved from Kaggle, a public repository known for offering various datasets aimed at machine learning studies was employed for model training. The dataset contains 7023 MRI images, which have been precisely classified into four categories namely glioma (1621 images), meningioma (1645 images), pituitary (1757 images), and a no tumor (2000 images) class.
Each class has its own pathology and it’s telling: gliomas are aggressive tumors that have infiltrative growth patterns which can create irregular and jagged edges; meningiomas are more benign and exophytic lesions that are slower-growing and more uniform in shape; pituitary tumors are usually found toward the base of the brain and thus have rounder contours; and “no tumor” scans show the brain’s natural symmetry without abnormal growths. To maintain consistency across this varied collection, each image was scaled down to a common resolution of 150 × 150 pixels. This size was selected both to promote computational efficiency during model training and to properly maintain resolution of diagnostic attributes, such as the subtle textural variations between tumor types or the absence of abnormalities in healthy tissue.
The exploratory part of data analysis (EDA) was used to examine the dataset composition and whether it was appropriate to classify. A bar chart (Fig. 1) plotted the distribution of images across the four classes, it was clear that there is a slight class imbalance: the no tumor category has about 28.5% in the dataset (2000 images), glioma 23.1% (1621 images), meningioma 23.4% (1645 images), and pituitary 25% (1757 images) make the middle. Such an imbalance, albeit small, indicated at least an inherent risk of model bias towards the “no tumor” class which we took into consideration for subsequent design decisions.
Additionally, visual inspection of representative sample images from each category (Fig. 2, bottom) provided qualitative confirmation of their distinguishing features. Gliomas exhibited chaotic, irregular edges indicative of their aggressiveness; meningiomas displayed organized, well-defined contours characteristic of benign tumors; pituitary lesions appeared smooth and rounded near the pituitary gland; and ‘no tumor’ images showed the brain’s natural symmetry, devoid of abnormal growths. These visual distinctions reinforced the dataset’s suitability for automated classification, as the morphological variations were sufficiently prominent to guide a machine learning model.
The dataset creators provided a predefined split consisting of 5,712 images for training and 1,311 for testing. This pre-allocated dataset was restructured to better support training and evaluation. From the original splits 5,712 training images and 1311 testing images, a total of 5,688 were retained for training and 703 images for testing dataset for final evaluation respectively, along with that 632 images (approximately 10%) were set aside as a validation set for monitoring model performance during development, ensuring that there was no overlap within the training testing and validation splits.
Each label was one-hot encoded, so “glioma” could be represented as [1, 0, 0, 0], “no tumor” as [0, 1, 0, 0], “meningioma” as [0, 0, 1, 0], and “pituitary” as [0, 0, 0, 1]. The pixel values, which originally ranged between 0 (black) and 255 (white), were normalized on a 0–1 scale by dividing by 255, assisting the neural network in numerical stability during training. We used augmentation techniques to enrich our relatively small training set, including rotations of up to 40 degrees, shifts of up to 20% in all directions, zooming in and out by 20%, and horizontal flipping. These transformations simulate real-world variability (such as patient motion during imaging). The validation and test sets were never augmented to ensure an unbiased evaluation.
For reproducibility and consistent results across experimental runs, we set a fixed random seed (i.e., a single-number random generator) for all stochastic processes, including data splitting and augmentation.
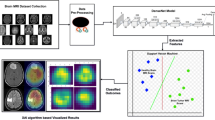
In this article, we implement a hybrid model that applies transfer learning by adapting the pre-trained VGG16 model—originally trained on generic image datasets—to the domain of BTC. By incorporating an attention mechanism that emphasizes critical regions (such as tumor boundaries) and Grad-CAM visualizations that generate heatmaps highlighting influential areas, our model offers both accurate and clinically transparent predictions of residual tumor volume.
Model workflow
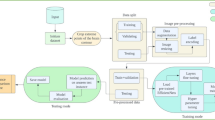
Figure 3 illustrates the model development workflow, encompassing the overarching framework used throughout the entire study. It provides a sequential structure designed to maximize performance and clinical utility, with careful attention to each step—from data preparation to evaluation.
First, it loads the dataset, which accesses the images of MRIs with labels via a Kaggle dataset that includes all four scans—glioma, meningioma, pituitary, no-tumor. Next, preprocessing: Images are resized to 150 × 150 pixels, pixel values scaled to 0–1 by dividing by 255, and the training set is augmented by up to 40-degree rotations, 20% shifts in any direction, 20% zooms and horizontal flips. This ultimately creates a training corpus containing a variety of important transformations and mimics how scans vary with everyday practice (such as patients who tilt their heads during scans), while the images are kept unchanged for validation and testing sets to allow for impartial performance evaluation. Nearest-neighbour fill values maintain the integrity of the input images; empty regions are filled from augmentations.
EDA then evaluates the composition of the dataset. A bar chart presents the class distribution — ‘no tumor’ images alongside glioma, meningioma, and pituitary tumors positioned toward the centre — revealing a slight imbalance that should be considered during model design. Sample images confirm distinctive characteristics: gliomas have jagged, irregular borders; meningiomas have well-defined edges; pituitary tumors exhibit smooth, rounded forms; and ‘no tumor’ scans display organic symmetry. These visual cues help validate the dataset for automated classification, supporting confidence in proceeding to model development. Training lies at the centre of the workflow, where the hybrid model is built and optimized. We begin with VGG16, which processes 150 × 150 × 3 RGB inputs through its layers and extracts features such as edges and textures. An attention mechanism is embedded to emphasize diagnostically salient regions—such as tumor margins—by dynamically amplifying these areas over less informative background noise, thereby improving classification accuracy. Additional layers are then added to this base model to customize it for predicting four classes: glioma, meningioma, pituitary, and no-tumor cases. The model is trained over multiple phases on 5,688 augmented images, learning patterns associated with each class through repeated iterations and weight updates. The diversity of the augmented data ensures that the model becomes sensitive to different presentations of tumors and healthy tissue.
Hyperparameter tuning refines the training stage by adjusting settings to maximize performance on our dataset. For each configuration, we vary the number of VGG16 layers that are retrained, along with the learning rate, dropout rate, and intermediate layer sizes. Outcomes are assessed using a 632-image validation set. A computer program automatically explores numerous configurations—retraining different layers of VGG16, adjusting the learning rate to accelerate convergence, and applying dropout to prevent overfitting—and selects the setup that yields the best validation accuracy.
This process provides the model with a final fine-tuning step, helping it strike a balance between retaining earlier-learned knowledge and adapting to task-specific features. Evaluation and Grad-CAM visualization are then employed to measure the model’s performance and interpret its predictions. The model is tested on a separate 703-image test set, previously unseen during training, to assess its accuracy and generalization across the four classes. Metrics such as precision and recall quantify performance, while Grad-CAM generates heatmaps that highlight regions influencing predictions—for example, emphasizing glioma’s irregular borders or meningioma’s defined edges.
Crucially, these visualizations align with clinical reasoning, providing a transparent foundation for trust in the model’s outputs. The workflow concludes with a final testing phase to evaluate the model’s reliability on new data and its potential to assist radiologists in real-world diagnostic scenarios. This systematic process—comprising data loading and preprocessing, model training, hyperparameter tuning, and evaluation—produces a classifier that is both accurate and clinically actionable.
Model architecture and hyper-parameter tuning
In order to design a classification method that focuses on a balance between reliability and interpretability of brain MRI analysis, we introduce a hybrid model using VGG16 model to facilitate both high accuracy as well as easily interpretable classification of brain MRI. Pre-trained on ImageNet data, the VGG16 is well suited for extracting low-level features and is adapted here to the brain tumor classification task. The architecture incorporates a novelty attention mechanism that reprioritizes feature activations according to their diagnostic importance, and increases the model ability to detect subtler changes across different tumors. Dropout is used in the dense layers to help avoid overfitting and improve generalization across imaging samples. Keras Tuner makes hyperparameter tuning ideal for exploring the architecture options, so that the performance is optimized without any manual tuning. This architecture represents a concerted effort to marry computational efficiency with clinical interpretability.
The model architecture, summarized in Fig. 4, employs VGG16 for BTC, incorporating customized enhancements to improve precision and clarity. We use a variant of VGG16—a DCNN originally pre-trained on a large dataset of natural images—for transfer learning to classify brain tumor images in MRI scans. This approach leverages the network’s existing ability to capture visual features and fine-tunes it for the specific task of recognizing tumor characteristics in medical images.
A detailed summary of the model’s layers is illustrated in Fig. 3, where input MRI scans are read as RGB images and fed into the model—an organized series of convolutional and pooling layers designed to progressively extract features. These layers transform the input step by step, enabling the extraction of both low- and high-level patterns necessary for tumor detection. As mentioned earlier, the VGG16 architecture includes stages that enhance the interpretation of MRI data. Early convolutional layers detect lower-level features (e.g., edges or texture changes) that are essential for delineating tumor boundaries. These initial layers capture distinct patterns that help differentiate tumor types—for example, the characteristic features of a glioma versus a meningioma—while deeper layers learn increasingly complex attributes specific to each tumor class.
Pooling layers between the convolutional layers reduce the dimensionality of the feature maps, retaining critical information while minimizing computational cost. In the final stage, the model extracts features from the MRI input in a high-dimensional form and compresses them into a low-dimensional, sparse representation, which serves as a robust foundation for accurate tumor classification.
This framework is further enhanced with an attention mechanism added on top of the convolutional stages. This layer focuses on high-level features that are more critical for tumor diagnosis—such as regions with rapid abnormal tissue growth—while minimizing the influence of less relevant background features. This refinement improves both the diagnostic focus and the clinical relevance of the model.
Additionally, a dropout layer is incorporated to prevent overfitting by randomly deactivating a portion of the features, making the model more robust to variations in MRI samples. These enhancements ensure that the architecture remains both accurate and adaptable. The processed features are then passed to a dense layer, which integrates the extracted features and emphasizes those most indicative of specific tumor categories.
This consolidation enables the SoftMax layer to output probabilities across the four classes—as mentioned above—during the final classification step. The resulting output provides a predictive score for tumor presence and type, offering insights that are both clinically interpretable and actionable.
The effectiveness of the architecture relies on its large number of trainable parameters, which allow it to adapt to the specific characteristics of brain tumor MRI data. Hyperparameter tuning maximizes performance by systematically adjusting key variables such as the number of VGG16 layers retrained, dropout rate, number of neurons in the dense layer, and learning rate. Using Keras Tuner, this optimization is performed on a validation set through an automated loop powered by the Hyperband algorithm, which identifies the parameter combinations that yield the highest classification accuracy. This process fine-tunes the model to achieve optimal accuracy while avoiding overfitting, ultimately establishing a strong foundation for reliable and clinically relevant diagnostic pathways in BTD.
Model training
A Tesla P100 GPU with 16 GB of RAM was used to train the model on Kaggle’s cloud infrastructure. In this case, the development environment was built using Python and TensorFlow and Keras libraries. To ensure that the best hyperparameters were selected in a suitable search space that made sense (i.e., drop out rate, number of dense layers, learning rate, and how many unfrozen layers in the VGG16 base), we used Keras Tuner feature, so that we were indeed using the optimal configuration for training, utilizing Hyperband. We compiled the model using the Adam optimizer and categorical cross-entropy loss. We utilized the standard training callbacks (early stop, model checkpoint and learning rate scheduling) to bolster stability and generalization. During training, TensorBoard was integrated to keep track of accuracy and loss trends. Creating a controlled and reproducible environment to ensure ease of model convergence and performance evaluation.
The proposed model was trained over 20 epochs on a dataset of 5688 augmented MRI images, with an epoch being one complete pass of the dataset to update the weights of the model. Images are sent in batches of 32 to improve computational time and processing of the images. The batch size limits the amount of data that the model has to work with at any one time, so after making predictions, updating weights based on those predictions, and updating with the new information, the model progresses to the next batch.
A validation set of images was used to monitor the model’s performance during training, providing real-time feedback on how well the model generalizes the task without overfitting to the training data. By transforming the data via rotations, shifts, zooms and flips, the dataset is augmented to give the model greater exposure to tumor and healthy tissues with diverting presentations. Such augmentation improves robustness and simulates the difference between test and training MRI given an inherent variation of patient orientation and angle of imaging in real-life clinical settings.
Multiple strategies are employed to guide and optimize the training process. Key metrics—accuracy (the ratio of correct predictions) and loss (a measure of how different the predicted labels are from the true labels)—are logged and visualized over the course of 20 epochs using TensorBoard. These metrics are dynamically plotted to monitor the model’s learning behavior (e.g., whether it is improving steadily or beginning to plateau), allowing adjustments to the training strategy as needed.
A checkpointing mechanism is used to save the model’s weights whenever the validation accuracy reaches a new peak, ensuring that the best-performing version of the model is preserved. This provides a reliable fallback in case performance degrades in later epochs due to issues like overfitting.
When the validation loss has not improved after two epochs, the learning rate scheduler is applied to decrease the learning rate with a factor of 0.1 (e.g. 0.001 →0.0001), so that training can continue to be fine-tuned. This change allows for smaller weight updates, which prevents the model from getting caught in local minima and helps the model converge more accurately (especially when it plateaus for a long while). Finally, an early stopping mechanism stops training if the validation loss does not improve for five epochs, which suggests additional epochs will unlikely offer more improvements. This mechanism is triggered by a lack of improvement in val_acc and it restores the best model weights from a checkpoint with the best val_acc, preventing the waste of computing resources on unproductive epochs, and keeps us focused on performing well.
The proposed model employed categorical cross-entropy loss function for training, which is appropriate for multi-class classification task and computes the difference between the predicted probability distributions and one-hot encoded ground truth values. The Adam algorithm for the optimization to update the weights according to the computed gradient for each batch was used. Adam utilizes momentum and dynamic learning rates to speed up convergence by helping high-magnitude weights to be updated slower than small-magnitude weights during training while preventing overshooting and further ensures sufficiently large magnitudes from altering training. In these 20 epochs, the model learns to differentiate between the four classes of glioma (jagged edges), meningioma (symmetric shapes), pituitary tumor (round shapes), and no tumor (regular shapes). This iterative mechanism of learning improves the ability of the model to differentiate these unique traits which is clinically useful for diagnosis.
At the end of training, the best model is restored, containing the weights that give the best accuracy on the validation dataset, and is used to evaluate performance. Combining data augmentation, metric monitoring, and careful optimization techniques, proposed model shows better classification & generalization. This framework is readily applicable to develop a reliable system for classifying tumors using MRI data intended for usage in clinical brain tumor markers.
Mathematical justification and representational efficiency
Incorporating these architectural and optimization strategies, the proposed model achieves a compromise among feature richness, parameter efficiency, and interpretability. The bottom network VGG16 takes in an image \(\:{x=\mathbb{\:}\mathbb{R}}^{150\:\times\:\:150\:\times\:3\:}\), as input to produce a feature map \(\:{z=\mathbb{\:}\mathbb{R}}^{h\:\times\:\:w\:\times\:c\:}\), with c being the number of channels in the last convolutional block. This is followed by the global average pooling layer that calculates the average activations across all channels:
This results in a fixed size feature vector \(\:\text{v}\:\in\:\:{\mathbb{R}}^{c}\), which closes the semantic gap and allows for efficient compression.
A channel-level attention mechanism encodes normalized (using SoftMax) trainable weights \(\:\text{w}\in\:\:{\mathbb{R}}^{c}\) in order to modulate the relative importance of those extracted features that are given by:
Here, \(\:\odot\) denotes element-wise multiplication. The attention-weighted vector \(\:{v}_{att}\) selectively enhances informative features while down-weighting irrelevant ones—improving discriminative power without increasing model complexity.
The final dense classification layers apply a linear transformation followed by a SoftMax activation to produce output probabilities:
The model is trained by minimizing the categorical cross-entropy loss between the predicted distribution, \(\:\widehat{y}\) and the ground truth y, using Adam which employs adaptive learning-rate per parameter. We perform hyperparameter tuning that finds best combination of hyperparameters like dropout, learning rate and dense layer size to be robust and generalize well over data.
A last convolutional layer used in the differentiable pipeline with Grad-CAM is then applied post-hoc to obtain class-specific saliency maps by weighting the feature maps from the final convolutional layer with the corresponding gradients with respect to our target class, yielding interpretable heatmaps co-localized with radiological features. This formulation allows the model to focus on medically interpretable salience maps whilst retaining a lightweight architecture—providing improved generalisability, reduced overfitting, and greater clinical compatibility over standard CNN classifiers.
Results, discussions and evaluation
In this section, a comprehensive analysis of proposed hybrid DL model performance in classifying brain tumors using MRI images is presented. We assess the model’s training history, its classification accuracy on the test set, and the interpretability of its predictions using Grad-CAM visualizations. As a result, we able to assess the model’s accuracy and its possible use in medicine. We also unlock comparative comparisons to previous models to show the superiority of prosed model in BTC.
Training progress
We tracked two major sources of information, loss and accuracy for both the training set (5688 images) and the validation set (632 images) over 20 epochs (training iterations) to determine how well the model learned during training. These metrics were used to plot the model’s learning behaviour, as shown in Fig. 5.
(a) Training and validation loss over 20 epochs. The sharp initial drop followed by convergence in both curves suggests effective learning and minimal overfitting. The model quickly learns to minimize error, reaching stable performance early. (b) Training and validation accuracy across epochs. Both curves show a steady rise, with close alignment after epoch 5, indicating strong generalization and consistent performance across unseen MRI data.
The plot in Fig. 5a shows the training and validation loss, a metric that quantifies the difference between the model’s predictions and the actual labeled data. A stable decrease in loss indicates that the model’s predictions are becoming more accurate. The training loss (blue line) starts around 0.8, reflecting significant initial errors, but drops rapidly—falling below 0.2 by epoch 5 and below 0.1 by epoch 15. This steady, monotonic decrease suggests that the model quickly learned to classify the training images effectively.
The validation loss (orange line) begins at approximately 0.4 and follows a similar downward trend, reaching 0.1 by epoch 10, with slight fluctuations afterward while remaining relatively stable. These fluctuations are expected, as the validation set comprises only 10% of the entire dataset. However, the overall trend closely mirrors the training loss, indicating that the model generalizes well to unseen data. The close alignment of the two curves suggests the model is not overfitting; in other words, it is learning meaningful patterns that can be applied to new MRI images, rather than simply memorizing the training data.
The plot in Fig. 5(b) shows the accuracy of the training and validation sets, representing the percentage of correct predictions. The training accuracy (blue line) starts at around 65%, meaning that in the initial epochs, the model correctly classified 65% of the training images. It increases sharply, reaching 90% by epoch 5 and nearly 100% by epoch 10, where it remains for the rest of the training. This indicates that the model quickly learned the patterns in the training data.
The validation accuracy (orange line) begins slightly higher, around 80%, likely because the validation set is smaller and less diverse. It rises steadily to reach 95% by epoch 10 and stabilizes around 96% by epoch 15. The small gap between training and validation accuracy (less than 5%) suggests strong generalization to unseen data, confirming that the model is not overfitting.
An early stopping mechanism was employed to halt training if no improvement in validation loss was observed over five epochs, ensuring retention of the best-performing model. Additionally, a learning rate reduction—dividing the rate by a factor of 0.1 after two consecutive epochs without improvement—helped the model adjust and contributed to this balanced performance.
Classification performance
To test its performance on unseen data, we evaluated the model on the test set of 703 images. The test set consists of images from all of the four categories: No Tumor, Glioma images, Meningioma, Pituitary. We used classification report, confusion matrix and ROC curves to help evaluate the effectiveness of the model.
Classification report
In addition to accuracy, Table 1 shows detailed metrics including precision, recall, F1-score and support for each category, as well as overall averages. The metrics are defined as:
Precision: It is the number of correct predictions for a category divided by the total number of predictions made for that category. In other words, it answers the question: ‘Of all the images the model labelled as this category, how many actually belonged to it?
Recall: The fraction of correctly identified instances of a category. It answers the question: ‘Out of all the images that actually belonged to this category, how many did the model predict correctly?
F1-Score: The harmonic mean of precision and recall, providing a single metric that balances the two. It is particularly useful when precision and recall values are close but not exactly the same.
Support: The total number of test images per category, representing the sample size for each class.
On the test set, the proposed model achieved an average accuracy of 99% across the four classes—no tumor, glioma, meningioma, and pituitary tumor—indicating that the model correctly classified nearly all images. The model exhibited excellent performance for the no-tumor class, with a precision of 1.000, meaning that all predicted non-tumor cases were indeed non-tumorous. However, there was slightly lower recall, suggesting that a small minority of actual non-tumor cases were missed, possibly due to rare imaging artifacts resembling tumor features. Despite this, the model achieved a high F1-score, reflecting trustworthy identification of healthy brain tissue.
In glioma detection, the model maintained exceptional accuracy, with only limited positive and negative errors due to the infiltrative nature of gliomas, which spread into neighboring tissue. This resulted in a balanced F1-score, reflecting the model’s ability to handle these challenging cases. For meningiomas, the model performed flawlessly, correctly identifying all ground-truth cases and confirming the vast majority of predictions. This is likely due to the uniquely homogeneous appearance of meningiomas on MRI scans, making them easily distinguishable, as reflected by the best F1-score. Regarding pituitary tumors, the model excelled once again, achieving precision and recall scores of 0.94 and 0.91, respectively—demonstrating perfect recognition of their rounded borders. Errors resulted in lower F1-scores, likely due to overlapping characteristics with other tumor types (e.g., meningiomas).
The precision-recall-F1-score macro-average of 0.99 reflects consistent performance across all classes, despite the varying tumor morphologies. Similarly, the weighted average of 0.99 further supports this stability, indicating that the distribution differences among classes did not impact the model’s performance. This reinforces the model’s ability to generalize across different tumor presentations, highlighting its potential as a reliable and accurate tool for brain tumor diagnosis in clinical settings.
The precision, recall, and F1-score comparison (Fig. 6) reinforces the model’s strong performance, with consistently high scores observed across all data splits—approximately 0.98–0.99 for training, and 0.98 for both validation and test. These results indicate a high degree of reliability and generalization. Nonetheless, minor performance variations point to potential areas for enhancement. This evaluation highlights the model’s diagnostic strengths, particularly its capacity to generalize across diverse tumor morphologies, while also emphasizing the need for further optimization in feature extraction methods or training dataset composition to support its clinical applicability in brain tumor assessment.
This class-wise performance gives very important information about the behaviour of the model in diagnostics. The model performs very well on meningiomas with a high F1-score which is likely due to their homogenous morphological characteristics and clear tumor borders are more readily captured through convolutional and attention-based mechanisms. In comparison, recall is a little lesser for gliomas due to their diffuse and infiltrative behaviour which makes their spatial boundaries less clear, resulting in them being more susceptible to partial classification error. The few false positives in the no tumor group indicate that incidental anatomical asymmetries or other image artifacts may be mistakenly identified as pathology in some cases. A few patients may be misclassified between glioma and pituitary tumor due to overlapping grayscale characteristics, particularly when the pituitary lesions are atypically growing in extra-axial compartment (25). These results emphasize that while the macro-averaged scores from the model are high, they are not simply a function of class balance, but indicative of its variable performance across complex tumor morphologies. Understanding driven by such interpretation is key for putting numerical metrics into perspective and truly evaluating whether the model is ready for the clinic.
Confusion matrix
The confusion matrix provides a detailed breakdown of the model’s predictions, including the number of images correctly classified and where mistakes occurred (Fig. 7). The matrix helps interpret the classification performance of the model for the four classes—no tumor, glioma, meningioma, and pituitary tumor—by mapping predicted labels to actual labels. A clear diagonal line indicates that the model is capable of accurately predicting each class, learning the differences between specific MRI features, including the irregular, infiltrative patterns of gliomas, the well-defined, homogeneous structures of meningiomas, the round, compact shapes of pituitary tumors, and the balanced, symmetrical characteristics of non-tumor samples.
Non-diagonal entries represent classification errors, presumably due to the homogeneity of visual features in MRI scans, where overlapping tissue densities or confusion in boundary detection between grading types may prevent separation. These errors highlight potential limitations in the model’s ability to discriminate subtle differences between classes, possibly due to image noise or variations in how tumors present across different patients. This analysis reveals valuable diagnostic strengths of the model, particularly its ability to generalize across tumor morphologies, as well as challenges that may need to be addressed (e.g., feature extraction and/or the training dataset) to further enhance the model’s clinical deployment for brain tumor evaluation.
Confusion matrices (Fig. 7—Training, Test, and Validation) discuss how each matrix provides different, insightful angles on how a model performs over the course of development. Figure 7 presents confusion matrices for (a) training, (b) test, and (c) validation sets, showing the class-wise prediction breakdown. Diagonal dominance across all matrices suggests effective learning and low misclassification rates. Training matrix (Fig. 7a) that shows strong initial ability of model to identify different MRI features, e.g. infiltrative patterns of glioma versus homogeneous structures of meningioma, but also highlights some challenging misclassifications. This visual breakdown reinforces that the model not only performs well numerically, but also maintains clinically relevant class separability, which is essential for its use in diagnostic setting.
(a) Confusion matrix for the training set. The model shows high class-wise accuracy with minor confusion between glioma and pituitary, reflecting their shared grayscale and boundary features. Diagonal dominance confirms strong initial learning. (b) Confusion matrix for the test set. Maintains high accuracy with sparse misclassifications, particularly between glioma and meningioma—consistent with real-world morphological overlaps in MRI. (c) Confusion matrix for the validation set. Performance is consistent with the training and test sets, confirming generalization and robustness. Misclassifications are minimal and class-specific patterns are well preserved.
The test matrix (Fig. 7b) provides an evaluation of performance in circumstances closer to real-world scenarios, since an accurate classification indicates that features are effectively representing the underlying biology, whereas residual pairwise misclassifications (e.g., confusion between glioma and meningioma) indicate inter-class feature overlap that may be subtle.
Finally, the model generalization capacity is assessed via a validation matrix (Fig. 7(c)) in which stable learning is quantified by correct predictions while off-diagonal errors are caused by the varying effects of the dataset or the presence of imaging artefacts.
Receiver operating characteristic (ROC) curves
ROC curves (Fig. 8) test the model performance using the True Positive Rate (Recall) vs. the False Positive Rate as we change the decision threshold using a receiver operator curve plot. The ROC curve of each category (No Tumor, Glioma, Meningioma and Pituitary) is a straight line from (0,0) to (0,1) and (1,1), which means perfect performance. This is the point at which the True Positive Rate (along the y-axis) is 1.0 (i.e., 100% recall) while the False Positive Rate (x-axis) is 0.0, which means there are no false positives in the proposed model. The AUC for all categories is 1.00, which is the maximum value possible. An AUC value of 1.00 indicates that the model perfectly separates each category by a wide margin, without any overlap between classes. Overall, such an extraordinary AUC score for all the classes demonstrates the accuracy of the model whereby it can rightly classify the images in to the categories of No Tumor, Glioma, Meningioma, and Pituitary without any indecisions at all.
Additionally, we used Grad-CAM to visualize where the model focused when making its predictions, ensuring that the model’s predictions were interpretable and clinically relevant. This is a vital step in medical applications, where a physician needs to understand the reasoning behind the model’s conclusions in order to trust its recommendations.
There is a representative classification results between four different diagnostic categories (Fig. 9). Here, each sub-image presents its respective ground truth label along with the predicted class, serving as a visual verification of the model classification ability. True and predicted labels matches (top) across multiple MRI orientations (axial, coronal, sagittal) and contrast conditions demonstrate the model robustness to a wide range of tumor morphologies and normal brain structures.
The distinction from meningioma is also correct, as glioma cases often feature diffuse and irregular infiltrative regions while meningiomas appear as homogeneously dense and well-circumscribed masses. In addition, the model lands consistently as zero tumor for cases with obvious midline cysts containing symmetric, structurally similar brain. while also correctly labelling pituitary tumors. The similarity of these predictions reinforces the quantitative performance characteristics and indicates that the model generalizes appropriately to real clinical data. This further confirms its possibilities in helping radiologists with non-invasive, MRI-based brain tumor screening and diagnosis.
Furthermore, multiple MRI images were tested as shown in Fig. 10(top) to understand how the model makes decisions across different tumor type. For each case, the model was able to classify the image into its respective tumor type (glioma, pituitary, meningioma). We used Grad-CAM to highlight the regions that contributed most to this prediction. In the Grad-CAM visualizations, areas of high model attention are shown in red and yellow, while the areas of low attention are shown in blue.
The corresponding MRI slices demonstrate clearly visible tumor, such as a rounded homogeneous tumor which will often displace brain. This Grad-CAM heatmap also confirms that the model was looking mostly in the tumour region and especially around its borders which are clinically important area. This suggests that the model is using clinically relevant features by aligning its attention to the tumor location in a manner that imitates the radiologist interpretative strategies used.
Model transparency is greatly improved with this visualization in addition to the fact that it also reflects that the predictions made by the model are based upon tumor-related anatomical landmarks rather than irrelevant image artefacts. This not only aids in the validation of the model but is also important towards building confidence and thereby unlocking the utility of AI systems in the clinical workflows of MI.
Discussion
We also combined VGG16 with a custom-made attention model and Grad-CAM, and the results show an accuracy of 99% on the test set with 703 images in total. This suggests that our proposed hybrid model combines the best of existing techniques to achieve superb results in classifying brain tumors. Despite class imbalance among the categories in the dataset, the average precision, recall, and F1-scores are high, indicating good reliability of the model across all categories. The confusion matrix for training, validation and test set shows that the model is giving correct labelling with low mislabelling. Most errors are between glioma and pituitary tumor, and this is expected given the similarities in their appearance on MRI. This specific tumor type tends to share the characteristics of texture patterns and boundary features which at times can cause confusion. Even so, it consistently separates all four classes—with good overall reliability, supporting the robustness and generalizability of the model over the datasets. However, these discrepancies are negligible and do not undermine the general robustness of the model. While the model achieved an AUC of 1.00 across all classes, which reflects excellent separability, it is acknowledged that such perfect classification is rare in medical imaging. These results may be influenced by the dataset’s limited ambiguity and inter-class separability. Future validation on more heterogeneous clinical datasets will be important to verify generalizability.
The training and validation plots provide insights into the learning process of our model. The fact that the loss has decreased and accuracy has increased significantly shows that the model learned well. We can also observe that the training and validation metrics are very close, indicating that the model is generalizing well on out-of-sample data. This prevents overfitting, meaning the model performs well on new images. Dropout layers were used to discourage co-adaptation, early stopping was used to prevent overtraining, and a learning rate scheduler was used to refine weight updates at convergence. Data augmentation – 40° rotation, translations, zoom and flipping – also helped to prevent overfitting by mimicking real world variations. However, we acknowledge that a formal ablation study quantifying the individual contributions of each augmentation strategy was not conducted. This remains a relevant direction for future analysis.
The Grad-CAM visualization introduces an interpretability element that is very important for clinical use. In addition to quantitative analysis, qualitative analysis using Grad-CAM also has clinical value. These maps consistently lit up diagnostically relevant areas of the brain, and corresponded well with established radiological heuristics. This visual verification confirms that the model is able to attend to relevant anatomical structures - a critical feature in clinical context where trust and transparency are paramount. In the case of meningiomas, for instance, the area of focus on the tumor mass and its edges corresponds with the observations radiologists make when diagnosing meningiomas, which typically involves looking for well-defined, rounded masses that push aside adjacent brain tissue. By aligning with medical logic, this model is a promising tool for helping radiologists because it not only provides accurate predictions but also explains its decisions in a way that doctors can understand and trust. The interpretability provided by Grad-CAM might help doctors confirm the model’s predictions, detect possible errors, and confidently incorporate the model into their diagnostic workflow. This interpretability claim is further supported by expert opinion from a Consultant Radiologist at Phoenix Hospital, Pune, who examined the Grad-CAM outputs. An independent radiologist (Consultant, Phoenix Hospital, Pune) confirmed that the attention maps were well-aligned with diagnostically significant regions across all tumor categories, consistent with standard clinical interpretation. This validation contributes confidence to the medical relevance of the model and supports its argument for translation use.
Such classification models differentiate brain tumor types from non-tumorous cases, and their performance is monitored using relevant metrics at the multi-class level (Table 2). Initial attempts laid a robust foundation by identifying broad imaging paradigms but frequently lacked specificity regarding the individual complexities of anatomic tumor details. Gradually, deeper networks and improved feature extraction have refined the output, generalizing across cases with increasing robustness. Additionally, the new approach combines a convolutional base with a custom attention layer and interpretability capability, increasing precision and accuracy—particularly for non-tumoral cases and challenging tumor types—compared to previous methods. Although all compared models utilize the same Kaggle MRI dataset, differences in preprocessing—such as image resolution, normalization, augmentation protocols, and train-validation splits—limit the fairness of strict one-to-one benchmark comparisons. This underscores the robustness of our model under a clearly defined and reproducible experimental setup. This highlights the robustness of the performance of our model across a tightly controlled experimental paradigm, increasing its reliability and comparability.
Looking at the big picture, DL-based classification approaches have advanced from simple methods to complex frameworks addressing multi-class classification problems (Table 2). Early work used pre-trained models for simple tasks, and from there, the field evolved to include multi-scale methods and hybrid systems for more complex tasks. Our model optimizes this trajectory by relying on an attention-based design and visualization to sharpen its performance across all measures, surpassing previously developed multi-class approaches by better targeting salient features in the image, yielding a model that generalizes robustly (Table 3).
In contrast with traditional CNN-based methods, the proposed hybrid model gains an additional advantage in terms of their diagnostic accuracy and interpretability. Attention enables the model to generalize and dynamically focus on diagnostically relevant regions, while the Grad-CAM overlays deliver interpretable and transparent visual justifications for its predictions—a vital requirement for medical imaging, where black-box decisions are often not acceptable Conversely, many baseline models—while achieving high accuracy—lack interpretability, which limits their clinical applicability in sensitive diagnostic contexts. These advantages however have their costs. The integration of attention layers and Grad-CAM, while beneficial for interpretability, increases model complexity and inference time—posing deployment challenges in resource-constrained or real-time clinical environments. Also, although our model was designed to be robust based on the chosen dataset, how readily this model could be scaled to multi-modality (imaging) or incorporated with volumetric (3D) MRI, will be a potential focus in the future.
These advancements are influenced by the complexity of the dataset. Despite strong empirical performance, several limitations must be acknowledged. Although the dataset was well-annotated and diverse, it was derived from a single publicly available source and may not reflect the entire clinical heterogeneity. Grad-CAM provides intuitive interpretability; however, its coarse spatial granularity limits its precision in localizing fine anatomical structure. Moreover, the discrete impact of model components—such as the attention mechanism and specific augmentation strategies—was not empirically isolated, which will be addressed in future ablation studies. Clearly, non-tumorous cases always perform well, but some tumors—with all their heterogeneity—push older models to their limits. By taking advantage of its attention-enhanced structure, the proposed approach overcomes these challenges more efficiently, resulting in state-of-the-art scores for each individual class and overall performance. It strengthens an existing architectural foundation with contemporary techniques to deliver a new standard of accuracy and reliability in MI, transcending and building on its predecessors.
The model performance metrics validate the technical quality of the model, but the greater value will come from its clinical implementation. Built as a tool that is light and interpretable in nature and thus can assist the radiologists during the daily routine by highlighting the diagnostically relevant region through Grad-CAM. The visual clarity that comes from this explains helps to make the diagnosis more confident and faster, which is important in high-volume imaging centres or centres with limited specialist access. Due to its architecture, it can be deployed flexibly either within hospital PACS systems or cloud-based diagnostic platforms, with applicability to real-time screening, radiology training education, and telemedicine remote diagnostics.
Limitations
Although this work introduces a high-performing and interpretable hybrid deep learning model to classify brain tumor, several contextual insights are needed to influence future research and broader clinical adoption. This model was built using a single, well-annotated public dataset (Kaggle), facilitating controlled experimentation and reproducibility. Nevertheless, the dataset does not represent the entire clinical diversity available in multi-centre institutions due to large variances in imaging protocols, equipment vendors and patient populations. So, while the results are encouraging, external validation in diverse cohort is a necessary next stop to determine the model’s generalizability in real-world use.
Furthermore, although the joint design incorporates various improvements, including attention mechanism, Grad-CAM visualisation, and target augmentation, the latter are not fully decoupled and analysed separately in dedicated ablation studies. While this choice is in line with the focus on end-to-end performance, it restricts interpretability at the component level. Publications seeking to improve or simplify the structure may find granular analysis and discovery of highest impacting modules useful. In addition, the included interpretability blocks in the form of Grad-CAM and attention further improve clinical deliverability at the cost of moderate additional computational expense. While not ideal, this would impact the scalability for resource-constrained or low-latency applications. Investigating the use of lightweight alternatives or optimization methods that can better trade off interpretability and efficiency in more general deployment settings could be worthwhile.
The interpretability in Grad-CAM is beneficial in locating diagnostically significant regions, which is proven by expert reviewing. On the other hand, its low spatial resolution might be limited with respect to precision in applications needing fine anatomical localization (e.g., surgical planning, refinement of lesion boundary). Lastly, a direct comparison with transformer-based architectures such as MRC-TransUNet and hybrid architectures with large language models (which have gained significant attention in the recent literature).
In any case, these limitations do not diminish the contributions offered by the proposed method, but define valuable guidelines for further development and translational applicability.
Future work
The model demonstrates good performance; however, certain targeted directions not only exist to enhance its clinical utility, but also to increase its technical sophistication. Initially, we will compare the architecture proposed to Inception X-ray models for comparative accuracy, inference speed, and generalization compared to state-of-the-art deep learning models such as ResNet, EfficientNet, and DenseNet.
Secondly, we would like to perform ablation studies to evaluate the independent effects of augmentation methods like rotation, zoom, flip and translation on classification performance for every tumor subtypes. This would help us to identify the best augmentation strategies with capabilities of generalized robust well. Third, last but not least, we will discuss the incorporation of sophisticated attention mechanisms, such as self-attention layers as well as transformer-based architectural designs including those of ViTs as well as MRC-TransUNet. These techniques might help the model extract long-range spatial dependencies and minute structural differences between tumor types.
Its extension to multimodal data—e.g., patient demographics, clinical reports, genomic markers etc.—will be our next goal for patient-specific and context-specific brain tumor diagnosis. We will high prioritize external validation by using datasets from a broad array of institutions to test the robustness of these models across imaging modalities, scanner types, and the distribution of demographic characteristics.
Finally, in order to enable deployment in real-world resource-limited environments, we will explore model optimization techniques, including pruning, quantization, and knowledge distillation, to reduce computational requirements and make the model suitable to run on edge-devices.
Conclusion
An interpretable, effective deep learning for brain tumor classification based on MRI data: A case study We present this hybrid architecture—an attention mechanism with a pre-trained VGG16 backbone using Grad-CAM visualization—which achieves a high diagnostic accuracy while preserving clinical transparency.
Despite the presence of a mild class imbalance, the model attained a 99% accuracy on the test set with an average precision, recall, and F1-score of 0.99 per tumor class. Confusion matices showed negligible misclassifications (Supplementary Fig. 8), and ROC curves confirmed perfect discriminative capacities (AUC = 1.00; Fig. 3). During the training process, regularization methods, including data augmentation, dropout, and early stopping, were introduced to improve generalization and reduce overfitting.
Crucially, the model’s interpretability was validated using Grad-CAM heatmaps consistently identifying tumor-relevant areas consistent with the radiological expectations. Such explainability is vital for building clinician trust and aiding integration into the diagnostic pipeline as a decision-support tool.
Data availability
The data that support the findings of this study are available from the corresponding author, [S.P], upon reasonable request.
Change history
08 January 2026
A Correction to this paper has been published: https://doi.org/10.1038/s41598-025-29967-3
References
Amran, G. A. et al. Brain tumor classification and detection using hybrid deep tumor network. Electron.. 11(21), 3457. https://doi.org/10.3390/ELECTRONICS11213457 (2022).
Sujatha, S. & Reddy, T. S. 3D Brain Tumor Segmentation with U-Net Network using Public Kaggle Dataset. In Proceedings of the 3rd International Conference on Artificial Intelligence and Smart Energy, ICAIS. Published online 2023:829–835. https://doi.org/10.1109/ICAIS56108.2023.10073895 (2023).
Asif, S., Qurrat-ul-Ain, Khan, S. U. R., Amjad, K. & Awais, M. SKINC-NET: an efficient Lightweight Deep Learning Model for Multiclass skin lesion classification in dermoscopic images. Multimed. Tools Appl.. https://doi.org/10.1007/S11042-024-19489-X (2024).
Khan, S., Asim, M., & Vollmer, S. AI-Driven Diabetic Retinopathy Diagnosis Enhancement through Image Processing and Salp Swarm Algorithm-Optimized Ensemble Network. arxiv.org. Published online 2025. Accessed May 22, 2025. (2025). https://arxiv.org/abs/2503.14209.
Ullah, N. et al. An effective approach to detect and identify brain tumors using transfer learning. Appl. Sci.. 12 (11), 5645. https://doi.org/10.3390/APP12115645 (2022).
Basthikodi, M., Chaithrashree, M., Ahamed Shafeeq, B. M. & Gurpur, A. P. Enhancing multiclass brain tumor diagnosis using SVM and innovative feature extraction techniques. Sci. Rep. 14(1), 1–15. https://doi.org/10.1038/s41598-024-77243-7 (2024).
Halder, T. K., Sarkar, K., Mandal, A. & Sarkar, S. A novel histogram feature for brain tumor detection. Int. J. Inform. Technol. (Singapore). 14 (4), 1883–1892. https://doi.org/10.1007/S41870-022-00917-W/METRICS (2022).
Medicine, S. K. C. in B and, Multi-level feature fusion network for kidney disease detection. ElsevierSUR KhanComputers in Biology and Medicine, 2025•Elsevier. Accessed May 22, 2025. (2025). https://www.sciencedirect.com/science/article/pii/S0010482525005657.
Amin, J. et al. A new model for brain tumor detection using ensemble transfer learning and quantum variational classifier. Comput. Intell. Neurosci. 2022 (1), 3236305. https://doi.org/10.1155/2022/3236305 (2022).
Saleh, A., Sukaik, R. & Abu-Naser, S. S. Brain tumor classification using deep learning. In Proceedings – 2020 International Conference on Assistive and Rehabilitation Technologies, iCareTech 2020. Published online August 1,:131–136. (2020). https://doi.org/10.1109/ICARETECH49914.2020.00032.
Bilal, O., Asif, S., Zhao, M., Khan, S. & Electrical, Y. L. C. An amalgamation of deep neural networks optimized with Salp swarm algorithm for cervical cancer detection. Comput. Electr. Eng.. Accessed May 22, 2025. (2025).https://www.sciencedirect.com/science/article/pii/S0045790625000497.
Khan, S. U. R., Asif, S. & Bilal, O. Ensemble Architecture of Vision Transformer and CNNs for Breast Cancer Tumor Detection From Mammograms. Int. J. Imaging Syst. Technol. 35(3). https://doi.org/10.1002/IMA.70090 (2025).
Özkaraca, O. et al. Multiple brain tumor classification with dense CNN architecture using brain MRI images. Life. 13 (2), 349. https://doi.org/10.3390/LIFE13020349 (2023).
Khan, S. U. R., Zhao, M. & Li, Y. Detection of MRI brain tumor using residual skip block based modified MobileNet model. Computing 28(4). https://doi.org/10.1007/S10586-024-04940-3 (2025).
Khan, S. U. R., Asif, S., Zhao, M., Zou, W. & Li, Y. Optimize brain tumor multiclass classification with manta ray foraging and improved residual block techniques. Multimed. Syst. 31(1). https://doi.org/10.1007/S00530-025-01670-3 (2025).
Hekmat, A., Zuping, Z., Bilal, O. & Khan, S. U. R. Differential evolution-driven optimized ensemble network for brain tumor detection. Int. J. Mach. Learn. Cybern.. Published online 2025. https://doi.org/10.1007/S13042-025-02629-6.
Khan, S., Asif, S. & Zhao, M. Optimized deep learning model for comprehensive medical image analysis across multiple modalities. Neurocomputing. Accessed May 22, (2025). https://www.sciencedirect.com/science/article/pii/S0925231224019532.
Brain Tumor, M. R. I. & Dataset Accessed April 8, (2025). https://www.kaggle.com/datasets/masoudnickparvar/brain-tumor-mri-dataset?select=Training.
Mathivanan, S. K. et al. A secure hybrid deep learning framework for brain tumor detection and classification. J. Big Data. 12 (1), 1–35. https://doi.org/10.1186/S40537-025-01117-6/FIGURES/11 (2025).
Hun, W. J. et al. Brain Tumor Detection via Transfer Learning and Ensemble Learning.In IEEE 12th Conference on Systems, Process & Control (ICSPC). Published online December 7, 2024:95–100. https://doi.org/10.1109/ICSPC63060.2024.10862279 (2024).
Strika, Z., Petkovic, K., Likic, R. & Batenburg, R. Bridging healthcare gaps: a scoping review on the role of artificial intelligence, deep learning, and large Language models in alleviating problems in medical deserts. Postgrad. Med. J. 101 (1191), 4–16. https://doi.org/10.1093/POSTMJ/QGAE122 (2024).
Wang, J., Wang, S. & Zhang, Y. Deep learning on medical image analysis. CAAI Trans. Intell. Technol. 10 (1), 1–35 (2024).
Tehsin, S., Nasir, I. M., Damaševičius, R. & GATransformer: A graph attention Network-Based transformer model to generate explainable attentions for brain tumor detection. Algorithms. 18 (2), 89. https://doi.org/10.3390/A18020089 (2025).
Sachdeva, J., Sharma, D. & Ahuja, C. K. Comparative analysis of different deep convolutional neural network architectures for classification of brain tumor on magnetic resonance images. Arch. Comput. Methods Eng. 31 (4), 1959–1978. https://doi.org/10.1007/S11831-023-10041-Y/METRICS (2024).
Mathivanan, S. K. et al. Employing deep learning and transfer learning for accurate brain tumor detection. Sci. Rep. 14(1), 1–15. https://doi.org/10.1038/s41598-024-57970-7 (2024).
Ahmed, M. M. et al. Brain tumor detection and classification in MRI using hybrid ViT and GRU model with explainable AI in Southern Bangladesh. Sci. Rep. 14(1), 1–16. https://doi.org/10.1038/s41598-024-71893-3 (2024).
Krishnan, P. T. et al. Enhancing brain tumor detection in MRI with a rotation invariant vision transformer. Front. Neuroinformatics. 18, 1414925. https://doi.org/10.3389/FNINF.2024.1414925/BIBTEX (2024).
Islam, N. et al. Deep Fusion Model for Brain Tumor Classification Using Fine-Grained Gradient Preservation. Published online June 28, 2024. Accessed April 2, (2025). https://arxiv.org/abs/2406.19690v1.
Aamir, M. et al. Brain tumor detection and classification using an optimized convolutional neural network. Diagnostics. 14 (16), 1714. https://doi.org/10.3390/DIAGNOSTICS14161714 (2024).
Rao, K. N. et al. An efficient brain tumor detection and classification using pre-trained convolutional neural network models. Heliyon 10 (17), e36773. https://doi.org/10.1016/J.HELIYON.2024.E36773 (2024).
Tin, M. T. A., Khin, E. E. & Win, L. L. Y. Improving Brain Cancer Detection and Classification Using Xception Deep Convolutional Neural Network. In 5th International Conference on Advanced Information Technologies, ICAIT 2024. Published online 2024. Published online 2024. https://doi.org/10.1109/ICAIT65209.2024.10754910 (2024).
Natha, S. et al. Automated brain tumor identification in biomedical radiology images: A Multi-Model ensemble deep learning approach. Appl. Sci.. 14 (5), 2210. https://doi.org/10.3390/APP14052210 (2024).
Güler, M. & Namlı, E. Brain tumor detection with deep learning methods’ classifier optimization using medical images. Appl. Sci.. 14(2), 642. https://doi.org/10.3390/APP14020642 (2024).
Verma, V. & Aggarwal, A. Exploring Deep Learning for Efficient Data Management in Brain Tumor Classification: An Overview. In 11th International Conference on Reliability, Infocom Technologies and Optimization (Trends and Future Directions), ICRITO 2024. Published online 2024. Published online 2024. (2024). https://doi.org/10.1109/ICRITO61523.2024.10522149.
Shamshad, N. et al. Enhancing brain tumor classification by a comprehensive study on transfer learning techniques and model efficiency using MRI datasets. IEEE Access. 12, 100407–100418. https://doi.org/10.1109/ACCESS.2024.3430109 (2024).
Gade, V. S. R., Cherian, R. K., Rajarao, B. & Aravind Kumar, M. BMO based improved lite Swin transformer for brain tumor detection using MRI images. Biomed. Signal. Process. Control. 92, 106091. https://doi.org/10.1016/J.BSPC.2024.106091 (2024).
Wageh, M., Amin, K., Algarni, A. D., Hamad, A. M. & Ibrahim, M. Brain tumor detection based on deep features concatenation and machine learning classifiers with genetic selection. IEEE Access. 12, 114923–114939. https://doi.org/10.1109/ACCESS.2024.3446190 (2024).
Rahman, T. & Islam, M. S. MRI brain tumor detection and classification using parallel deep convolutional neural networks. Measurement: Sens. 26, 100694. https://doi.org/10.1016/J.MEASEN.2023.100694 (2023).
Zhang, Z. et al. A novel deep learning model for medical image segmentation with convolutional neural network and transformer. Interdiscip Sci. 15 (4), 663–677. https://doi.org/10.1007/S12539-023-00585-9/METRICS (2023).
Vankdothu, R., Hameed, M. A. & Fatima, H. A brain tumor identification and classification using deep learning based on CNN-LSTM method. Comput. Electr. Eng. 101, 107960. https://doi.org/10.1016/J.COMPELECENG.2022.107960 (2022).
Vankdothu, R. & Hameed, M. A. Brain tumor MRI images identification and classification based on the recurrent convolutional neural network. Measurement: Sens. 24, 100412. https://doi.org/10.1016/J.MEASEN.2022.100412 (2022).
Rasool, M. et al. A hybrid deep learning model for brain tumour classification. Entropy. 24 (6), 799. https://doi.org/10.3390/E24060799 (2022).
Kesav, N. & Jibukumar, M. G. Efficient and low complex architecture for detection and classification of brain tumor using RCNN with two channel CNN. J. King Saud Univ. - Comput. Inform. Sci. 34 (8), 6229–6242. https://doi.org/10.1016/J.JKSUCI.2021.05.008 (2022).
Irsheidat, S. & Duwairi, R. Brain Tumor Detection Using Artificial Convolutional Neural Networks. In 11th International Conference on Information and Communication Systems, ICICS 2020. Published online April 1, 2020:197–203. (2020). https://doi.org/10.1109/ICICS49469.2020.239522.
Choudhury, C. L., Mahanty, C., Kumar, R. & Mishra, B. K. Brain Tumor Detection and Classification Using Convolutional Neural Network and Deep Neural Network. In International Conference on Computer Science, Engineering and Applications, ICCSEA 2020. Published online March 1, 2020. Published online March 1, 2020. (2020). https://doi.org/10.1109/ICCSEA49143.2020.9132874.
Anil, A., Raj, A., Aravind Sarma, H. & Chandran, N. R. Brain tumor detection from brain MRI using deep learning. Int. J. Innovative Res. Appl. Sci. Eng. (IJIRASE). 3 (2), 458–465. https://doi.org/10.29027/IJIRASE.v3.i2.2019 (2019).
Network, K. et al. Convolutional neural network for brain tumor detection. IOP Conf. Ser. Mater. Sci. Eng. 771 (1), 012031. https://doi.org/10.1088/1757-899X/771/1/012031 (2020).
Woźniak, M., Siłka, J. & Wieczorek, M. Deep neural network correlation learning mechanism for CT brain tumor detection. Neural Comput. Appl. 35 (20), 14611–14626. https://doi.org/10.1007/S00521-021-05841-X/FIGURES/17 (2023).
Kalam, R., Thomas, C. & Rahiman, M. A. Brain tumor detection in MRI images using Adaptive-ANFIS classifier with segmentation of tumor and edema. Soft Comput. 27 (5), 2279–2297. https://doi.org/10.1007/S00500-022-07687-4/METRICS (2023).
Saeedi, S., Rezayi, S., Keshavarz, H. & Niakan Kalhori, R. MRI-based brain tumor detection using convolutional deep learning methods and chosen machine learning techniques. BMC Med. Inf. Decis. Mak. 23 (1), 1–17. https://doi.org/10.1186/S12911-023-02114-6/TABLES/11 (2023).
Mahmud, M. I., Mamun, M. & Abdelgawad, A. A deep analysis of brain tumor detection from MR images using deep learning networks. Algorithms. 16, (4), 176. https://doi.org/10.3390/A16040176 (2023).
Reyes, D. & Sánchez, J. Performance of convolutional neural networks for the classification of brain tumors using magnetic resonance imaging. Heliyon 10 (3), e25468. https://doi.org/10.1016/J.HELIYON.2024.E25468/ASSET/0650866F-5765-43C5-8267-A0F7C9CF88A7/MAIN.ASSETS/GR016.JPG (2024).
Yazdan, S. A. et al. An Efficient Multi-Scale Convolutional Neural Network Based Multi-Class Brain MRI Classification for SaMD. Tomography 8(4), 1905–1927. https://doi.org/10.3390/TOMOGRAPHY8040161 (2022).
Saxena, P., Maheshwari, A. & Maheshwari, S. Predictive modeling of brain tumor: A deep learning approach. Adv. Intell. Syst. Comput. 1189, 275–285. https://doi.org/10.1007/978-981-15-6067-5_30 (2021).
Asif, S., Zhao, M., Tang, F. & Zhu, Y. An enhanced deep learning method for multi-class brain tumor classification using deep transfer learning. Multimed Tools Appl. 82 (20), 31709–31736. https://doi.org/10.1007/S11042-023-14828-W/METRICS (2023).
Ayadi, W., Charfi, I., Elhamzi, W. & Atri, M. Brain tumor classification based on hybrid approach. Visual Comput. 38 (1), 107–117. https://doi.org/10.1007/S00371-020-02005-1/METRICS (2022).
Mondal, M., Faruk, M. F., Raihan, N. & Ahammed, P. Deep Transfer Learning Based Multi-Class Brain Tumors Classification Using MRI Images. 3rd International Conference on Electrical and Electronic Engineering, ICEEE. Published online 2021:73–76. https://doi.org/10.1109/ICEEE54059.2021.9719003 (2021).
Hashemzehi, R., Mahdavi, S. J. S., Kheirabadi, M. & Kamel, S. R. Detection of brain tumors from MRI images base on deep learning using hybrid model CNN and NADE. Biocybern Biomed. Eng. 40 (3), 1225–1232. https://doi.org/10.1016/J.BBE.2020.06.001 (2020).
Ramaha, N. T. A. et al. Brain pathology classification of MR images using machine learning techniques. Computers. 12 (8), 167. https://doi.org/10.3390/COMPUTERS12080167 (2023).
Deepak, S. & Ameer, P. M. Automated categorization of brain tumor from MRI using CNN features and SVM. J. Ambient Intell. Humaniz. Comput. 12 (8), 8357–8369. https://doi.org/10.1007/S12652-020-02568-W/METRICS (2021).
Mahjoubi, M. A. et al. Improved Multiclass Brain Tumor Detection using Convolutional Neural Networks and Magnetic Resonance Imaging. Int. J. Adv. Comput. Sci. Appl. 14(3). http://www.ijacsa.thesai.org (2023).
Author information
Authors and Affiliations
Contributions
A.A. & N.W.: Draft Preparation, Investigation, Programming; M.R. & A.K.: Conceptualization; Methodology, Draft Preparation; S.P., K.K. & A. Kul: Draft Preparation, Investigation and Reviewing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of this Article contained an error in the author list. Abdullah Al-Danakh was incorrectly listed as an author of the original Article and has subsequently been removed.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Aiya, A.J., Wani, N., Ramani, M. et al. Optimized deep learning for brain tumor detection: a hybrid approach with attention mechanisms and clinical explainability. Sci Rep 15, 31386 (2025). https://doi.org/10.1038/s41598-025-04591-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-04591-3