Abstract
Wound healing is a dynamic process involving tissue formation, debris removal and ultimately remodeling to restore skin integrity. Although wound healing is generally successful, this process can eventually fail, leading to chronic wounds like pressure ulcers (PUs), whose presence/absence has been considered by WHO as good indicator of patient’s wellbeing and care quality. PUs are stratified into grades I to IV grades based on their severity, however, the existence of systemic markers predicting their clinical progression remains unexplored. Here, we performed a serum proteomic and transcriptomic profiling of 54 patients with PUs ranging from grade II to grade III-IV. Unsupervised clustering identified a distinctive immune-related proteomic and transcriptomic blood profile in high-grade PUs. Specifically, pathways controlled by inflammatory-linked genes such as IER3, TSLP, and TNFAIP6 (TSG-6) were found to be upregulated in high-grade PUs, together with a reduction in the levels of potent immunomodulators such as IL-10, IFNγ, MCP-2/CCL8, and CXCL-10 in serum from grade III-IV PUs patients. All together, indicating an altered inflammatory state in advanced PUs. This study provides novel insights regarding the use of omic approaches to find potential systemic biomarkers for the prediction of severity in PUs and could help to understand the molecular mechanisms underlying the chronic progression of this pathology.
Similar content being viewed by others
Introduction
Wound healing is a concept encompassing both the physiological processes of new tissue formation and debris removal. This process is often divided into four overlapping and continuous phases, lasting approximately three months including haemostasis, inflammation, proliferation and remodeling1. The healing process is mediated by the action of a plethora of molecules including cytokines, chemokines and growth factors, with direct steps leading to the restoration of the skin’s physiological barrier function2. Combined with impaired cellular and systemic host responses to stress, local tissue hypoxia, repetitive trauma and heavy bacterial burden, have been described to effectively disrupt the wound healing process, causing tissue damage and ultimately promoting a persistent inflammatory state1.
The term ‘chronic wounds’ includes a heterogeneous group of skin lesions that do not follow the normal course of healing, and includes diabetic foot ulcers, venous and arterial ulcers and pressure ulcers (PUs), among others3. In this work we will focus on PUs. PUs are clinically defined as localized damage affecting the skin and/or the underlying tissue, as a consequence of pressure with or without the combination of shear4. PUs are usually developed on bony prominences, however, their occurrence can also be related due to an overuse of medical devices4.
WHO considers the presence of PUs as a good indicator of the quality of care received by the patient. PUs significantly affect both patient’s quality of life and healthcare resource utilization5,6. The reported worldwide prevalence of PUs, according to WHO, ranges from 5 to 12%, however, their true incidence is difficult to determine, as many cases are treated at home and go unreported. PUs are mostly caused by external pressure applied to an area of the body, mainly bony prominences, leading to blockage of capillaries and causing ischemia, hypoxia, edema, inflammation and ultimately necrosis and ulcer formation. PUs are generally classified into grades I-IV based on the international National Pressure Ulcer Advisory Pane/European Pressure Ulcer Advisory Panel (NPUAP/EPUAP) system, established in 20194. Grades I-II PUs include incipient ulcers that can easily regenerate if the antipathogenic factors are alleviated. In case underlying causes persist, the natural progression leads to ulcers becoming more severe and potentially irreversible, with involvement of injured subcutaneous tissues including in some cases muscle or bone (Grades III-IV PUs)7.
Patients suffering from chronic ulcers are heterogeneous cohorts, usually composed of elderly people with multiple comorbid diseases, and under diverse treatments8. Thus, this clinical situation makes extremely needed the identification of systemic molecular biomarkers, which could easily predict disease severity outcomes, with a future aim of improving patient’s guidance for the selection of better and more effective interventions23. From the molecular perspective, normal wound repair usually involves several processes such as cytokine release, rearrangement of adhesion molecules/cytoskeletal components, as well as the alteration in the expression of tissue remodeling molecules such as matrix metalloproteases (MMPs). Particularly, non-healing wounds commonly manifest the presence of a prolonged inflammation status, deregulation of protease levels, a reduced growth factor activity, stem cell dysfunction and cellular senescence9,10,11,12. In addition, some cellular events have been also associated with the wound healing process, such as the release of tumor necrosis factor alpha (TNF-α) and different growth factors, involving platelets, neutrophils, macrophages and fibroblasts12. To clarify the complex role exerted by all these mediators in this process, the combination of omic approaches such as transcriptomics and proteomics can provide a holistic understanding of cellular mechanisms at a molecular level. This multi-omic approach could enable a personalized medicine with the ability to validate biomarkers for diagnosis and ultimately help to describe potential therapeutic targets to prevent wound healing worsening12.
In this work, we propose that PUs progression to irreversible stages (Grades III-IV) lies in a systemic and chronic inflammatory process, elucidated by specific transcriptomic and proteomic profiling changes. Considering most of the research conducted on ulceration uses skin/tissue-derived samples, this work also aimed to identify non-invasive and easy to detect systemic markers able to classify patients with grade I-II and III/IV (irreversible) PUs, lately shedding a light into the understanding of the molecular/cellular mechanisms governing the chronic progression of this pathology.
Results
Patient phenotypes and characteristics
Patient information was thoroughly analyzed to ensure that significant differences in the transcriptomic and proteomic analyses were solely and exclusively due to ulcer-related factors.
Information was collected on the medical pathologies related to risk factors (sex; age; hospitalizations; admission to ICU; diabetes mellitus; hypercholesterolemia; vascular, neurological and cardiac pathology; antiplatelet, anticoagulant or antibiotic treatment) and on ulceration in question (ulcer location, grade and group) of the 54 patients selected (Supplementary Table 1).
Samples from all the patients in the cohort were used for the proteomic study. For transcriptomics, 10 patients from each group were used. Statistical analysis showed that no significant differences between patients with pressure ulcer grade II and patients with pressure ulcer grade III-IV were found for any of the clinical variables analyzed (Supplementary Table 2). Likewise, we didn’t observe significant statistical differences for any of the clinical variables considering the patients included for the RNA sequencing (Supplementary Table 3).
Our results conclude that cohort of study display homogeneous clinical parameters. Therefore, we continue to analyze the proteomic and transcriptomic profile of these patients.
Patients with grade III-IV Pressure Ulcers display an altered inflammatory response
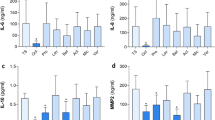
Next, we analyze the proteomic profile in serum samples from all enrolled patients (Supplementary Table 1) with Olink® Target 96 Inflammation panel (Uppsala, Sweden). Anti-inflammatory Interleukin 10 (IL-10), macrophage activation factor gamma Interferon (IFNg), Monocyte Chemotactic Protein (MCP-2 or CCL8) and C-X-C motif chemokine ligand 10 (CXCL10) showed significant differences between patients with grade II PUs and patients with grade III-IV PUs (Fig. 1). The four proteins show a significant decrease in the Grade III-IV group.
IL-10, IFNg, MCP-2 and CXCL10 relative concentration (NPX) comparing patients with grade II PUs (Group 1, G1) and patients with grade III-IV PUs (Group 2, G2) subjects. Mann-Whitney U-test was performed for IL-10, IFNg and MCP-2 and Unpaired T-test for CXCL10. Median with interquartile range was presented. (a) IL-10 (p-value 0.0272); (b) IFNg (p-value 0.0178); (c) MCP-2 (p-value 0.0125); (d) CXCL10 (p-value 0.0494). *: p-value < 0.05.
These results demonstrate a systemic dysregulation of immune mediators related to the inflammatory response taking place during pression ulcers.
Identification of differentially expressed genes between patients with grade II versus grade III/IV pressure ulcers
Additionally, we perform a transcriptomic analysis of the PBMCs (peripheral blood mononuclear cells) to figure out which immune mediators are contributing to the development of the inflammatory response in these patients. This was carried out by RNA sequencing to determine the different transcriptomic profiles of patients with grade II PUs and patients with grade III-IV PUs. In each case, 10 patients were selected to perform RNA sequencing (Supplementary Table 1).
Figure 2 shows distribution of patients with grade II PUs and patients with grade III-IV PUs according to a Principal Component Analysis (PCA) model. From the 44,714 transcripts sequences obtained, 1081 transcripts were identified as significantly differentially expressed between the experimental groups. Differential expression analysis (grade III-IV PUs patients vs. grade II PUs patients) considered as significant transcripts those with p-values < 0.05 and changes in expression Log2 (Fold Change) greater that 2 or lesser than − 2. A total of 117 differentially expressed genes were identified. From all of them, 48 genes were downregulated (Table 1 upside) and 69 were upregulated (Table 1 downside) in patients with grade III-IV PUs vs. grade II PUs. Among overexpressed genes, IER3, TSLP, CD177, DCSTAMP, IGLV3-6, and C2CD4A were directly related to inflammation and immune-linked processes, and MMP27, NSG2 related with tissue remodeling. Among the downregulated genes, we noticed changes in the expression of genes encoding for receptor of cytokines with a widely described role in tissue healing (IL22RA1 and IL17RD) or the proteasome formation (CHGB).
A hierarchical clustering of Z-score normalized expression values was performed with the differentially expressed genes (Fig. 3). Thus, we observed differential gene expression, with genes such as TRAV38-1, FAM106A, IL11, MIR6797, and TRAJ22 showing higher expression levels in patients with grade II ulcers (genes related with T cell receptors and immune response), whereas genes such as EPCAM, MMP27, MIR31B, and CHIT1 exhibited increased expression in patients with grade III–IV ulcers (genes related with matrix extracellular remodeling).
2D hierarchical clustering heatmap of the differentially expressed genes, using Z-score normalized RNA-seq data from patients with grade II PUs (Group 1) compared with patients with grade III-IV PUs (Group 2). Blue bands indicate low gene expression values; red bands indicate high gene expression values.
Our results demonstrate that different stages of PU progression are associated with a specific transcriptomic fingerprint of PBMCs, suggesting a systemic inflammatory response in these patients linked to the progression of the ulcer.
PUs progression involves immunological pathways
Gene Set Enrichment Analysis (GSEA) was then performed using all genes to further investigate their biological relevance. Notably, GSEA results indicated that signatures related to inflammation and altered immune responses are significantly enriched in patients with grade III-IV PUs (Group 2) in comparison to grade II PUs patients (Group 1) (Tables 2 and 3; Fig. 4). Concretely, most of the enriched pathways when comparing Groups 2 and 1 of PUs patients indicate immunological alterations, involving a plethora of cellular entities such as T cells, B cells and monocytes/macrophages. Some examples are HAY_BONE_MARROW_FOLLICULAR_B_CELL(FDR q-val < 1.0E-4); GSE22886_NAIVE_BCELL_VS_NEUTROPHIL_DN (FDR q-val < 1.0E-4); GSE22886_ TCELL_VS_BCELL_NAIVE_DN (FDR q-val 5.877E-4); GSE3982_MEMORY_ CD4_TCELL_VS_BCELL_DN (FDR q-val 0.002); GSE25123_WT_VS_ PPARG_KO_MACROPHAGE_UP (FDR q-val 0.002); GSE29618_ MONOCYTE_VS_MDC_UP (FDR q-val 0.002); HAY_BONE_MARROW_NEUTROPHIL (FDR q-val 0.002), which are enriched in Group 2; and to a lesser extend some examples also enriched in Group 1 of less severe PUs patients such as GOCC_T_CELL_RECEPTOR_COMPLEX (FDR q-val 0.0); GSE10325_CD4_ TCELL_VS_MYELOID_UP (FDR q-val 0.02); GSE11057_CD4_EFF_MEM_ VS_PBMC_UP (FDR q-val 0.02).
Finally, an experimental validation in the expression of selected differentially expressed genes assessed by transcriptomics (AP3B2, CD177, COL19A1, CYP26B1, IER3, LIX-1, MMP27, NEGF, OLAH, PGA5, PNMA2, SCN2A,SUMO1P1, TNFAIP6, TSLP) was examined by RT-qPCR. When compared with RNA-seq data, up to 95% of the selected transcripts showed the same expression pattern, thus reinforcing the reliability of the observed changes (Fig. 5). Supplementary Table 5 contains the genes, and the primers sequences used to validate RNA-seq data.
Validation of RNA-seq data by RT-qPCR and 2-ΔΔCT normalization method. RNA-seq (grey or dark blue) compared to RT-qPCR results (dark orange or green). RNA-seq and qPCR results are represented by Log2(FC Group2/Group1; G2/G1). G1: Group 1, corresponding to patients with grade II PUs; G2: Group 2, corresponding to patients with grade III-IV PUs.
Discussion
Pressure Ulcers (PUs) is a complex and multifactorial injury which is becoming a significant and concerning healthcare problem worldwide, especially after COVID pandemic13. As mentioned before, PUs usually affects patients older than 70 years, being developed in more than 80% of hospitalized individuals within the first 5 days of inpatient hospital stay14. PUs progression and chronicity is a dynamic process clinically involving since skin rubbing to blood discharge, and sometimes if become so deep (i.e. stage IV) with profound damage affecting tendons, joints and even the muscle and bone, resulting in life-threatening complications such as infections, malnutrition and anemia15. In this sense, several specialists agreed that prognostic intervention is nowadays the best treatment of PUs, implying the early detection of the injury. Chronic development of PUs are affected by several factors including the advancing age, nutritional status, presence of other chronic comorbidities complicating their detection before they are visually, clinically -and molecularly- irreversible16,17. Majority of PUs are preventable and the availability of biomarkers capable of predicting their onset and progression would represent a significant economic benefit for healthcare systems (e.g., bed availability, treatment duration), while also enhancing patient care and overall wellbeing. The complexity and clinical stratification (grades) of PUs necessitate large-scale studies (omics) to identify biomarkers, enabling big molecular analysis of changes at protein levels and function, as well as describing gene expression alterations to elucidate the cellular events behind their development and chronicity18.
Most of the research performed at describing the development of PUs commonly used skin samples -collected from different depths and location-, with the aim to identify local biomarkers that could help at guiding the selection for the best treatment and predict outcomes19,20,21,22. However, tissue obtained from the surroundings of non-healing wounds (as observed in advanced PU stages) exhibits a hyperproliferative epidermal phenotype, varying levels of fibrosis, and heightened cellular infiltration (mainly leukocytes, macrophages, neutrophils)19,23. In addition, skin sample collection is extremely invasive and painful for patients, involving heterogeneous samples (varying in depth, width, layers of study, etc.), and other less aggressive alternatives need to be considered for routine sampling. Regarding the latter, we wanted here to gain insight into the systemic (instead of local) mechanisms underlying the defective wound healing observed in patients with different stages of PUs, using a multi-omic approach (proteomic and transcriptomic) with the aim to describe blood biomarkers that could help to predict irreversible wound healing in elderly patients. Concretely, we have identified serum alterations related to immune regulation and inflammation when comparing patient samples with low-grade (II) and high-grade (III-IV) PUs. Notably, systemic inflammation has been usually discarded to predict damage for other types of ulceration (e.g. diabetic foot ulcers (DFUs)) due to the insufficient elevation of this signature, mainly influenced by the etiology of the disease (e.g. DFUs are not caused because of immobility but as a result of diabetes complications). We proposed here that a systemic alteration of the inflammatory/immune status is a molecular signature that may help to distinguish severity PUs grades. Transcriptomics have been previously utilized to identify differentially expressed genes at specific stages of wound healing process24,25,26 and mostly focused in DFUs27,28,29,30. Here, a multi-omic approach was performed and transcriptomics were complemented by a proteomic analysis, where we found altered blood levels (decrease in patients with high-grade (III-IV) PUs) of four important immunoregulatory molecules such as IL-10, IFNγ, CXCL10 and MCP-2/CCL8.
Concretely, IL-10 is a widely described immune mediator that can be secreted by T regulatory cells, B regulatory cells and macrophages31, and its alteration has been previously described in the context of pressure ulceration (e.g. DFUs and venous ulcerates)32,33. IL-10 is a pleiotropic factor, controlling inflammation and other varied processes such as M2 polarization of macrophages and wound healing34,35,36. An M2 phenotype downregulation has been previously reported in diabetic foot ulceration32,37,38,39 and could explain here the perpetuation of the inflammatory stage observed in grade III-IV PUs.
Regarding the tissue repairing capabilities of IL-10, decrease levels of this cytokine perfectly correlated with the observed downregulation of IL22RA1 and IL17RD in PBMCs of more graded PUs, encoding receptors of cytokines such as IL-22 and IL-17 with a potent and widely described role in promoting wound healing39,40. Despite not being detected among our altered set of molecules, it is worth mentioning that IL-17 secreted by endothelial cells has been already proposed -using transcriptomics- as a promising candidate for the therapeutic treatment of severely developed DFUs41. If these observations can be effectively transferred to other clinically non-related contexts such as altered wound healing occurring at PUs, and together with IL-10, being considered a potential therapy for PUs will require further validation. In addition, research conducted on fluids collected from wounds revealed that non-healing wounds showed alteration of several structural components such as matrix metalloproteinases (MMPs)42. In our research, the only gene encoding for MMPs is MMP27 and resulted overexpressed in patients with grade III-IV PUs, however no reports about its role in ulceration has been published up to date. As observed in other transcriptomic studies of DFUs42, other unexplored genes related to the modulation of keratinocyte migration and wound healing such as ADAMTS2 (encoding for the Metallopeptidase With Thrombospondin Type 1 Motif 2) were also found here to be upregulated in grade III-IV PUs patients, but its importance as a biomarker and contribution to the pathogenesis of chronic PUs wounds will require further exploration.
In addition to IL-10, our proteomic analysis yielded a significant decrease in IFN-γ blood levels in patients with deeper ulcers (grade III-IV PUs), supporting the systemic inflammatory signature described in this study. Taking together the proteomic and transcriptomic data, an alteration of IL-10 and IFN-γ, together with CXCL10 and IL11 is a common feature observed in other inflammatory contexts affecting elderly patient cohorts -usually affected by PUs during hospitalizations- such as COVID19 infections43,44,45. Supporting the systemic inflammation context of grade III-IV PUs, we found that TSLP – which encodes for the alarmin TSLP (Thymic stromal lymphopoietin) –. TSLP is a well-known pro-inflammatory epithelial cell-derived cytokine, which in the context of our observations has been described as a potent inhibitor of IL-10 secretion by T regulatory cells46,47 and key negative regulator of tissue remodeling -via collagen release by fibroblasts- not only in the airways but also in the skin48. Considering collagen deposition is fundamental to the development and resolution of normal wound healing, TSLP levels could play a crucial role in ulcers progression that should not be lightly dismissed and being considered as target of future research in this regard. Finally, regarding the lack of M2 polarization observed in other ulceration processes32,36,37,38, IFN-γ can also influence macrophage polarization towards an M2 phenotype in certain contexts and its decrease levels together with IL-10, supports again the creation of an immune environment inclined to a non-polarization of M2 macrophages. Future studies targeting macrophage function in chronic wound healing will help to clarify this observation.
In connection with systemic inflammation, two genes such as CD177 and TNFAIP6 linked with immune infiltration appeared upregulated in grade III-IV PUs. CD177 is a marker of activated neutrophils which is usually expressed on the surface of these immune cells before migrating to inflamed areas and has been previously described to be also upregulated in invasive S. aureus-infected DFUs49,50. TNFAIP6 (encoding for the protein TSG-6) also modulates neutrophil, and monocyte recruitment wound inflammation, and its gene expression levels has been also described to upregulated in fibroblasts isolated from skin ulcers non-healing DFUs28. In terms of monocyte/macrophage cell infiltration, MCP-2/CCL-8 levels appeared decreased in patients with severe PUs and has been also described as relevant predictor of wound healing in patients with non-healing DFUs51.
Proteomic and transcriptomic data in this study not only intend to help to understand, at a molecular level, how aberrant wound healing processes take place in these ulcers, but also aim to find potential systemic cell-derived biomarkers that could help to predict their severity endpoints. In this sense, a set of less well-known genes and proteins were found to be significantly altered in grade III-IV PUs and could be potentially used to predict their development and pathogenesis. These genes include IER3, DCSTAMP and C2CD4A, and proteins such as MCP-2/CCL8. Concretely, IER3 has a relevant role in immune regulation (via NFκB activation), alteration of blood pressure control, genome stability and more importantly in the context of severe PUs, osteogenic differentiation52,53. IER3 gene alteration was accompanied by an upregulation of genes already described to modulate bone resorption such as DCSTAMP (expressed by dendritic cells and a therapeutic target of inflammatory arthritis)54,55. These observations are of great relevance for the context deeper (grade III-IV/severe) PUs, which can affect not only the skin but also the muscles and bones of the patients. To our knowledge, IER3 and DCSTAMP have not been previously reported to be altered while phenotyping patients the context of ulceration. C2CD4A is a gene involved in the regulation of vascular permeability and, although not been described yet, if related with vessels rupture as observed in deeper ulcers, requires further investigation56.
In conclusion, in this work we have performed a serum analysis and found relevant proteomic and transcriptomic alterations occurring when comparing patients with grade II and III/VI pressure ulcers. The combination of these omics approaches have allowed us to delve deeper into the molecular mechanisms underlying the wound healing of pressure ulcers. This data supports the hypothesis that PUs development to irreversible stages could be associated with altered inflammatory processes. Despite its limitations, this exploratory work we intended to propose a panel of transcripts (e.g. IL11, TSLP, CD177, DCSTAMP, TNFAIP6, among others) and proteinic (IL-10, IFNγ, CXCL10, MCP-2) blood markers which could be associated with the abnormal development of wound healing in severe ulcers, including PUs. Our data suggests a blockade of the wound healing process in these patients, mainly governed by uncontrolled inflammatory status. This study focused on PUs could open the window of new investigations targeting more specific interventions to treat patients with stage III-IV ulcers (and other types such as DFUs) (e.g., through the application of IL-10 to the wound once developed) or anticipating the development of irreversible ulcers by carrying out a gene study in at-risk patients with ulcers that are still incipient.
Methods
Study design
This project employs a cross-sectional research design and utilizes non-probabilistic, consecutive sampling. All subjects provided written informed consent. The protocol was approved by the Committee of Research and Ethics from Getafe University Hospital (Ref.: CEIM 19/21). All research included here was performed in accordance with relevant guidelines/regulations and informed consent was obtained and signed by all patients. All participants were patients from any medical service evaluated by the plastic surgery service of the Getafe University Hospital during a period of 18 months -starting on 1 July 2019- who attended either on an outpatient basis (emergency or outpatient) or during their hospital stay (admission or discharge).
A total of 54 patients (aged 44–99 years) were recruited and divided into two groups: Group 1 (n = 27 patients with grade II PUs) and Group 2 (n = 27 patients with grade III-IV PUs). Group 1 includes patients with mild ulcers characterized by partial-thickness skin loss with exposed dermis, presenting as a shallow open ulcer with a viable, pink wound bed. Group 2 includes patients with more advanced ulcers, characterized by full-thickness skin loss, with visible subcutaneous fat, and frequently presenting with granulation tissue (epibole) in grade III PUs. For grade IV PUs, the damage may extend to muscle and/or supporting structures such as fascia, tendons, ligaments, cartilage, or bone. All patients included in this study were analyzed by proteomics but only 10 of each group were included in RNA sequencing.
The inclusion criteria required that the patients had the capacity to give informed consent, or, where applicable, a legal representative instead. Following NPUAP/EPUAP guidelines, the patients needed to have developed grade II, III or IV PUs of, at least, 2 cm diameter size, and serum albumin levels > 2 g/dl. Likewise, all individuals with ulcers showing signs of local infection (erythema, heat, purulent discharge or bad odor), active tumour processes, or SARS-CoV2 positive patients were excluded.
Sample collection and processing
Whole blood was collected in BD Vacutainer SST™ II tubes and K3 EDTA BD Vacutainer™ tubes to obtain serum and plasma, respectively. Samples were centrifuged at 2000 g for 10 min to avoid serum recovery. Serum samples were stored at -80ºC until further proteomic analyses. Ficoll-Paque (GE Healthcare™, Chicago, Illinois, USA) density gradient centrifugation gradient was used to separate PBMCs, which were then lysed in Buffer RLT and immediately stored at -80ºC until transcriptomic analyses.
Proteomic study
Protein profiling was obtained by using 1 µl of serum sample at the Proximity Extension Assay (PEA; Olink®, Uppsala, Sweeden) as previously described57,58. The samples were analyzed with the Olink® Target 96 Inflammation panel including 92 proteins59. All samples passed quality control and were randomized prior to analysis on a 96 well plate. Protein levels are reported as Normalized Protein eXpression (NPX) values, a relative quantification unit which is logarithmically related to protein concentration60.
RNA sequencing analysis
RNA was extracted from lysed PBMCs using RNeasy® Mini Kit (Qiagen, Hilden, Germany) with DNase treatment following manufacturer’s recommendations. RNA concentration was determined using a Nano-Drop™ 2000/2000c Spectrophotometer, and its quality was assessed with Experion RNA StdSens analysis kit (RNA quality Indicator; Bio-Rad Laboratories Inc., Hercules, CA, USA), establishing RQI as an indicator of quality. Samples performing an RQI ≥ 8 were selected for the RNA sequencing analysis.
RNA sequencing (RNA-seq) was carried out at the Genomics Unit from CNIC. RNA-seq was performed on DNase I-treated RNA samples (200 ng in 50 ml RNase–free water) with a RIN > 8 (RNA Integrity Number; Agilent; Assessed with the Agilent 2100 Bioanalyzer System; Agilent Technologies, Santa Clara, USA). RNA-seq libraries and sequences were created using the New England BioLabs Next® Ultra II Directional RNA Library Prep Kit at the Illumina HiSeq 2500. All samples were indexed, and multiplex sequencing was conducted on the HiSeq to generate a dataset (minimum of 8 M reads per sample) at 50 nucleotides read length in single-end format (1 × 50). The quality and integrity of sequencing results were monitored for each data collection and sample.
Analysis of RNA-Seq data
FASTQ files obtained after sequencing (GSE230161 on GEO database) were pre-processed first by removing rRNA sequences using SortMeRNA 2.1 and then trimming adapters and low-quality sequences using BBMap version 38.92 and Cutadapt 1.1561. Reads were aligned to GRCh38.p13 (NCBI) using STAR 2.7.10b software62. Samtools 1.13 was used to transform the alignment file into a bam format file and HTSeq 0.6.1p1 (with option -m intersection-nonempty) was used to obtain the reads’ raw counts for each feature (transcript)63,64.
Data normalization, PCA (considering the 500 top genes with the highest variance) and differential expression analysis were performed using DESeq2 package65. Genes with fewer than 10 counts in the samples were discarded. Finally, genes with a p-value less than 0.05 and a Log2 FC (fold change) greater than 2 (absolute value) were considered as differentially expressed genes.
We used the R package ComplexHeatmap to perform a hierarchical classification of samples and build a heatmap (using Z-scores measurements, “completeness” and “Euclidean distance”)66,67,68.
GSEA Preranked was used to perform gene set enrichment analysis on a pre-ranked gene list, establishing 1000 gene set permutations (enrichment statistic: weighted). Only gene sets with significant enrichment levels were considered (FDR q-value < 0.05 according to GSEA recommendations)69.
Gene expression validation by RT-qPCR
RT-qPCR analysis/validation was performed only for candidate genes previously catalogued as differentially expressed genes (Log2FC ≥ 2 and ≤ -2; p-value < 0.05) when comparing individuals with grades II (n = 10) and III-IV (n = 10) PUs. Briefly, RNA samples (1 µg) were reversely transcribed into a final volume of 20 µl using the High-Capacity RNA to cDNA Kit (Applied Biosystems, Foster City, California, USA). Primers were designed by using OligoArchitect™ (Sigma-Aldrich, San Luis, Missouri, USA), and RT-qPCR was performed using SYBR Green master mix (Takara Kusatsu, Japan) in the equipment Real Time HT 7900 (Applied Biosystems). Reactions were run in triplicates. Expression data were normalized using the 2−ΔΔCT method70, using as housekeeping genes GAPDH and HPRT1.
Statistics
Clinical characteristics were compared between patients with grade II PUs and patients with grade III-IV PUs to determine if there were significant differences in clinical variables. Quantitative variables were analyzed by Shapiro-Wilk test for assessing normality of the data. For normally distributed data, the unpaired Student’s T-test was applied. Otherwise, the non-parametric Mann-Whitney U-test was used. For qualitative variables, Fisher’s exact test was applied. A p-value of 0.05 was considered as a threshold of significance.
For the proteomic approach we compared continuous variables using unpaired Student’s T-test or Mann–Whitney U-test (when data were not normally distributed) between 27 patients with grade II PUs (Group 1) and 27 patients with grade III-IV PUs (Group 2). Data were presented as median with interquartile range (GraphPad Prism 9.2.0). A p-value < 0.05 was considered statistically significant.
Data availability
Raw sequencing data are available in GEO database under accession numbers: [geo] GEO Submission (GSE230161) [NCBI tracking system #23839902]. Please, contact the corresponding author (PFM) for further information if necessary.
References
Ruilong, Z., Liang, H., Clarke, E. & Jackson, C. Meilang, X. Inflammation in chronic wounds. Int. J. Mol. Sci. 17 (12), 2085 (2016).
Jiang, L. et al. Expression of cytokines, growth factors and apoptosis-related signal molecules in chronic pressure ulcer wounds healing. Spinal Cord 52 (2),145–51 (2014).
Stolzenburg-Veeser, L. & Golubnitscha, O. Mini-encyclopaedia of the wound healing—opportunities for integrating multi-omic approaches into medical practice. J. Proteom. 188, 71–84 (2018).
European Pressure Ulcer Advisory Panel, National Pressure Injury Advisory Panel and Pan Pacific Pressure Injury Alliance. Prevention and treatment of pressure ulcers/injuries: Clinical practice guideline. The international guideline 2019. Third edition ed.: Emily Haesler (2019).
Ramos, A. et al. Prevalencia de úlceras Por presión En Un Centro sociosanitario de media-larga estancia. Gerokomos 24 (1), 36–40 (2013).
Tubaishat, A., Papanikolaou, P., Anthony, D. & Habiballah, L. Pressure ulcers prevalence in the acute care setting: A systematic review, 2000–2015. Clin. Nurs. Res. 27 (6), 643–659 (2018).
Kottner, J. et al. Pressure ulcer/injury classification today: An international perspective. J. Tissue Viabil. 29 (3), 197–203 (2020).
Lindley, L. E., Stojadinovic, O., Pastar, I. & Tomic-Canic, M. Biology and biomarkers for wound healing. Plast. Reconstr. Surg. 138 (3), 18S (2016).
Eming, S. A. et al. Differential proteomic analysis distinguishes tissue repair biomarker signatures in wound exudates obtained from chronic wounds and normal healing. J. Proteome Res. 9 (9), 4758–4766 (2010).
Stojadinovic, O. et al. Dysregulation of the epidermal stem cell niche contributes to the pathogenesis of non-healing venous ulcers. Wound Repair. Regen. 22 (2), 220–227 (2014).
Yager, D. R., Zhang, L., Liang, H., Diegelmann, R. F. & Cohen, I. K. Human pressure ulcer wound fluids contain elevated levels of matrix metalloproteinase and activity compared to surgical wound fluids. J. Invest. Dermatol. 107 (5), 743 (1996).
Shah, J. M., Omar, E., Pai, D. R. & Sood, S. Cellular events and biomarkers of wound healing. Indian J. Plast. Surg. 45 (02), 220 (2012).
Gefen, A. & Ousey, K. COVID-19: Pressure ulcers, pain and the cytokine storm. J. Wound Care. 29 (10), 540–542 (2020).
Bansal, C., Scott, R., Stewart, D. & Cockerell, C. J. Decubitus ulcers: A review of the literature. Int. J. Dermatol. 44 (10), 805–810 (2005).
Teasell, R. & Dittmer, D. K. Complications of immobilization and bed rest. Part 2: Other complications. Can. Fam Phys. 39 (1440-2), 1445–1446 (1993).
Takahashi, P. Y. Pressure ulcers and prognosis: Candid conversations about healing and death. Geriatrics 63 (11), 6–9 (2008).
Qaseem, A., Humphrey, L. L., Forciea, M. A., Starkey, M. & Denberg, T. D. Clinical guidelines committee of the American college of physicians. Treatment of pressure ulcers: a clinical practice guideline from the American college of physicians. Ann. Intern. Med. 162 (5), 370–379 (2015).
Bader, D. & Oomens, C. The potential of biomarkers in the early detection of pressure ulcers. In: (eds Romanelli, M., Clark, M., Gefen, A. & Ciprandi, G.) Science and practice of pressure ulcer management. (Springer, 2018). https://doi.org/10.1007/978-1-4471-7413-4_1.
Utz, E. R. et al. Metalloproteinase expression is associated with traumatic wound failure. J. Surg. Res. 159, 633–639 (2010).
Hawksworth, J. S. et al. Inflammatory biomarkers in combat wound healing. Ann. Surg. 250, 1002–1007 (2009).
Forsberg, J. A. et al. Do inflammatory markers portend heterotopic ossification and wound failure in combat wounds? Clin. Orthop. Relat. Res. 472, 2845–2854 (2014).
Eming, S. A., Martin, P. & Tomic-Canic, M. Wound repair and regeneration: Mechanisms, signaling, and translation. Sci. Transl. Med. 6, 265sr6 (2014).
Stojadinovic, O. et al. Molecular pathogenesis of chronic wounds: The role of beta-catenin and c-myc in the Inhibition of epithelialization and wound healing. Am. J. Pathol. 167, 59–69 (2005).
Leon, C. et al. Transcriptomic analysis of a diabetic Skin-Humanized mouse model dissects molecular pathways underlying the delayed wound healing response. Genes 12 (1), 47 (2021).
Iglesias-Bartolome, R. et al. Transcriptional signature primes human oral mucosa for rapid wound healing. Sci. Transl. Med. 10 (451), eaap8798 (2018).
Wilkinson, H. N., Guinn, B. & Hardman, M. J. Combined metallomics/transcriptomics profiling reveals a major role for metals in wound repair. Front. Cell. Dev. Biol. 9, 788596 (2021).
Rai, V. Transcriptomics revealed differentially expressed transcription factors and MicroRNAs in human diabetic foot ulcers. Proteomes 12, 32 (2024).
Theocharidis, G. et al. Single cell transcriptomic landscape of diabetic foot ulcers. Nat. Commun. 13, 181 (2022).
Wang, Y. et al. An update on potential biomarkers for diagnosing diabetic foot ulcer at early stage. Biomed. Pharmacother. 133, 110991 (2021).
Dangwal, S. et al. Impairment of wound healing in patients with type 2 diabetes mellitus influences circulating MicroRNA patterns via inflammatory cytokines. Arterioscler. Thromb. Vasc Biol. 35, 1480–1488 (2015).
Carlini, V. et al. The multifaceted nature of IL-10: Regulation, role in immunological homeostasis and its relevance to cancer, COVID-19 and post-COVID conditions. Front. Immunol. 8 (14), 1161067 (2023).
Nanda, R., Patel, S., Ghosh, A., Asha, K. S. & Mohapatra, E. A study of Apolipoprotein A1 (ApoA1) and interleukin-10 (IL-10) in diabetes with foot ulcers. Biomed. (Taipei). 12 (1), 30–38 (2022).
Coelho, G. A., Secretan, P. H., Tortolano, L., Charvet, L. & Yagoubi, N. Evolution of the chronic venous leg ulcer microenvironment and its impact on medical devices and wound care therapies. J. Clin. Med. 12 (17), 5605 (2023).
Barrientos, S. et al. Growth factors and cytokines in wound healing. Wound Repair. Regen. 16, 585–601 (2008).
Lauer, G. et al. Expression and prote- olysis of vascular endothelial growth factor is increased in chronic wounds. J. Invest. Dermatol. 115, 12–18 (2000).
Edsberg, L. E. et al. Analysis of the proteomic profile of chronic pressure ulcers. Wound Repair. Regen. 20, 378–401 (2012).
Kotwal, G. J. & Chien, S. Macrophage differentiation in normal and accelerated wound healing. Results Probl. Cell. Differ. 62, 353–364 (2017).
Roy, R. et al. IL-10 dysregulation underlies chemokine insufficiency, delayed macrophage response, and impaired healing in diabetic wounds. J. Invest. Dermatol. 142 (3 Pt A), 692–704 (2022).
Chen, R. & Zou, L. Combined analysis of single-cell sequencing and bulk transcriptome sequencing reveals new mechanisms for non-healing diabetic foot ulcers. PLoS One 19 (7), e0306248 (2024).
Mu, X. et al. IL-17 in wound repair: Bridging acute and chronic responses. Cell. Commun. Signal. 22 (1), 288 (2024).
Lu, Y. et al. Single-cell profiling reveals transcriptomic signatures of vascular endothelial cells in non-healing diabetic foot ulcers. Front. Endocrinol. (Lausanne). 14, 1275612 (2023).
Richards, S. M. et al. Molecular characterization of chronic cutaneous wounds reveals subregion- and wound type-specific differential gene expression. Int. Wound J. 21 (4), e14447 (2024).
La-Beck, N. M. et al. Clinical characteristics and patterns of immune responses in COVID-19 patients from a rural community hospital. Cureus 16 (6), e61600 (2024).
Mansoor, S. et al. Expression of IFN-Gamma is significantly reduced during severity of covid-19 infection in hospitalized patients. PLoS One 18 (9), e0291332 (2023).
Nery, G. B. et al. Impact of social distancing from the COVID-19 pandemic on the immuno-inflammatory response of older adults. BMC Geriatr. 24 (1), 99 (2024).
Chen, Z. et al. Neutralization of TSLP inhibits airway remodeling in a murine model of allergic asthma induced by chronic exposure to house dust mite. Plos One 8 (1), e51268 (2013).
Nguyen, K. D., Vanichsarn, C. & Nadeau, K. C. TSLP directly impairs pulmonary Treg function: Association with aberrant tolerogenic immunity in asthmatic airway. Allergy Asthma Clin. Immunol. 6 (1), 4 (2010).
Shin, J. U. et al. TSLP is a potential initiator of collagen synthesis and an activator of CXCR4/SDF-1 Axis in keloid pathogenesis. J. Invest. Dermatol. 136 (2), 507–515 (2016).
Agidigbi, T. S. et al. Transcriptomic identification of genes expressed in invasive S. aureus diabetic foot ulcer infection. Front. Cell. Infect. Microbiol. 13, 1198115 (2023).
Bai, M. et al. CD177 modulates human neutrophil migration through activation-mediated integrin and chemoreceptor regulation. Blood 130 (19), 2092–2100 (2017).
Li, J. Y., Wang, Z. J., Deng, A. P. & Li, Y. M. ENA-78 Is a novel predictor of wound healing in patients with diabetic foot ulcers. J. Diabetes Res. 2695436 (2019).
Arlt, A. et al. Immediate early gene-X1 interferes with 26 S proteasome activity by attenuating expression of the 19 S proteasomal components S5a/Rpn10 and S1/Rpn2. Biochem. J. 402 (2), 367–375 (2007).
Küppers, O. et al. Inflammatory priming of human mesenchymal stem cells induces osteogenic differentiation via the early response gene IER3. FASEB J. 38 (19), e70076 (2024).
Miyamoto, T. The dendritic cell-specific transmembrane protein DC-STAMP is essential for osteoclast fusion and osteoclast bone-resorbing activity. Mod. Rheumatol. 16 (6), 341–342 (2006).
Garcia-Hernandez, M. L. et al. Dendritic cell-specific transmembrane protein is required for synovitis and bone resorption in inflammatory arthritis. Front. Immunol. 13, 1026574 (2022).
Warton, K., Fosterm, N. C., Gold, W. A. & Stanley, K. K. A novel gene family induced by acute inflammation in endothelial cells. Gene 342 (1), 85–95 (2004).
Assarsson, E. et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 9 (4), e95192 (2014).
Lundberg, M., Eriksson, A., Tran, B., Assarsson, E. & Fredriksson, S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 39 (15), e102 (2011).
Olink Proteomics. Target 96 Inflammation [white paper]. (2019). https://www.olink.com/resources-support/document-downloadcenter/. Article number 95302.
Olink Proteomics. Data normalization and standardization [white paper]. (2021). https://olink.com/application/data-normalization-and-standardization/
MARTIN, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 1, 10–12 (2011).
Li, B. & Dewey, C. N. RSEM: Accurate transcript quantification from RNA-Seqdata with or without a reference genome. BMC Bioinform. 12, 323 (2011).
Johnson, W. E., Li, C. & Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated Estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550 (2014).
Gu, Z. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32 (18), 2847–2849 (2016).
Gu, Z. Complex Heatmap Visualization. Imeta. 1 (3), e43 (2022).
R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. (2024). https://www.R-project.org/
Subramanian, A. et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 102 (43), 15545–15550 (2005).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (‐Delta Delta C(T)) method. Methods 25, 402–408 (2001).
Acknowledgements
We would like to thank all institutions involved, including Instituto de Medicina Molecular Aplicada – Nemesio Díez (IMMA-ND, Universidad San Pablo-CEU, CEU Universities, Madrid, España), Department of Nursering of Hospital Universitario de Getafe (Getafe, Madrid, España), and emeis Iberia for supporting this work.
Funding
This work was supported by emeis Iberia.
Author information
Authors and Affiliations
Contributions
LTP, PFM, AEP, RAS, LSZM, OGC and CTC analyzed and interpreted laboratory data. AEP, OGC and CTL analyzed the RNA sequencing data. LTP, JBL and XSH had selected and included the patients. LTP, AEP, JCLR and PFM were major contributors in the writing of the manuscript. MME and TCL contributed on the discussion, correction and writing of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The Ethics local Committee of Research and Ethics from Hospital Universitario de Getafe of Madrid, Spain (approval reference: CEIm19/21) approved this study.
Consent for publication
All subjects provided written informed consent.
Competing interests
The authors declare no competing interests.
Informed consent
All research included here was performed in accordance with relevant guidelines/regulations and informed consent was obtained and signed by all patients.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Peña, L.T., Escolar-Peña, A., Solera, R.A. et al. Systemic immune response alteration in patients with severe pressure ulcers. Sci Rep 15, 19579 (2025). https://doi.org/10.1038/s41598-025-04710-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-04710-0