Abstract
Saposhnikovia divaricata (S. divaricata) has demonstrated significant efficacy in mitigating arsenic poisoning. However, its protective effects of S. divaricata against NaAsO2-induced neural injury remain unclear. Based on network pharmacology, through intersection analysis, the common targets of the effective ingredients and targets of S. divaricata were selected from the TCMSP database, and the targets related to neural injury and arsenic toxicity were selected from the GeneCards database. Subsequently, protein–protein interaction (PPI), GO and KEGG enrichment analyses were carried out. We validated the PI3K/AKT pathway in vitro by assessing drug effects by CCK8 assays, flow cytometry, Western blot, and RT-qPCR. The PI3K/AKT pathway was screened by using network pharmacology, and AKT1 was identified as a core target via topological analysis using the degree algorithm among 45 potential targets derived from 72 traditional Chinese medicine targets, 9717 nerve injury targets, and 2224 arsenic toxicity targets. Molecular docking revealed that 9 active components of S. divaricata interacted with AKT1. In vitro experiments revealed that S. divaricata effectively mitigated neural injury by increasing cell viability, antioxidant activity and Bcl2 level while decreasing the apoptosis rate and the levels of Bax, Caspase8, Caspase3 and Cleaved-caspase3 in HT22 cells exposed to NaAsO2. The ratios of p-PI3K/PI3K and p-AKT/AKT were further increased by S. divaricata as well as Nrf2 expression. Our findings indicated that S. divaricata exerted multicomponent, multitarget and multipathway protection against NaAsO2-induced neurotoxicity. These effects were related to the antioxidant and antiapoptotic properties of S. divaricata, which might be achieved by activating the PI3K/AKT signaling pathway.
Similar content being viewed by others
Introduction
Arsenic is a nonmetallic element that is present extensively within natural rocks, soils, and aquatic environments1. Research has demonstrated that arsenic can accumulate extensively within the brain via the blood‒brain barrier, leading to mitochondrial membrane instability and subsequent energy depletion, thereby exerting potent neurotoxic effects2,3. Prolonged exposure to high levels of arsenic compounds may induce peripheral neural injury and developmental disorders related to cognitive and behavioral aspects in both humans and animals4,5. In contemporary medical practice, sodium dithio-propyl sulfonate (DMPS) is known for its substantial arsenic-detoxifying properties and is frequently employed as a specific therapeutic agent for arsenic intoxication6,7. Despite its utility, the treatment is associated with adverse effects such as nausea, tachycardia, and vertigo, along with allergic reactions, including rashes, shivering, fever, and even anaphylactic shock and exfoliative dermatitis, prompting a search for safer and more healthful substitutes8. As a traditional Chinese medicine, Saposhnikovia divaricata (S. divaricata) has pronounced anti-inflammatory, immunomodulatory, and antiviral properties and is clinically applied for the management of liver and cardiac injuries induced by arsenic poisoning9,10,11. Notably, there are no reports in the literature on the protection of S. divaricata against arsenic-induced neuronal damage.
Network pharmacology represents an emerging academic field that elucidates the intricate interplay among drugs, their targets, biochemical pathways, and associated diseases, which is instrumental in the discovery of novel therapeutics, the prediction of therapeutic targets, and the elucidation of pharmacological mechanisms of action12,13.
In the present investigation, network pharmacology was used to identify the pivotal targets associated with S. divaricata intervention and NaAsO2-induced neural injury in HT22 cells. A model of arsenic exposure and S. divaricata intervention was subsequently established in vitro, and the effects of NaAsO2 and S. divaricata on HT22 cells were assessed. Finally, the involved signaling pathways were further explored via the CCK8 assay, Western blot analysis, and flow cytometry techniques.
Materials and methods
Bioinformatics research
Acquisition of Saposhnikovia divaricata targets
Using the TCMSP (https://old.tcmsp-e.com/tcmsp.php) database14, we applied screening criteria of oral bioavailability (OB) ≥ 30% and compound drug-likeness (DL) ≥ 0.18 as screening conditions to identify active compounds, including (2R, 3R) 3-(4-hydroxy-3-methoxy-phenyl)-5-methoxy-2-methyl-2,3-dihydropyrano [5,6-h][1,4]benzodioxin-9-one (chloramphenicol A), 11-hydroxysec-o-beta-d-glucosylhamaudol_qt, anomalin, divaricatacid, divaricatol, Ammidin, ledebouriellol, phelloptorin, 5-O-Methylvisamminol, Phellopterin, sitosterol, wogonin, beta sitosterol, Mandenol, isoimperatorin, Prangenidin, methyl icosa-11,14-dienoate, and Decursin from S. divaricata. The identified active ingredients were subsequently imported into the PubChem database (https://pubchem.ncbi.nlm.nih.gov/)15 to retrieve their SMILES numbers or chemical structural formulas. SMILES numbers or chemical structural formulas of active ingredients were imported into the SwissTarget prediction database (http://www.swisstargetprediction.ch/)16 to ascertain potential targets associated with traditional Chinese medicine drugs.
Acquisition of genes related to arsenic poisoning and nerve injury
We employed “arsenic” and “nerve injury” as search terms within the GeneCards database (https://www.genecards.org/)17 to extract genes related to arsenic poisoning and nerve injury.
Construction of the component-target disease network
The targets related to the S. divaricata, along with those linked to arsenic and nerve injury, were input into the Venny website (https://bioinfogp.cnb.csic.es/tools/venny/)18 for common targets. The interaction targets were constructed via the String database (https://cn.string-db.org/)19 alongside Cytoscape version 3.7.2 software for protein‒protein interaction analysis; this facilitated the extraction of significant indicators for subsequent experiments through PPI network scoring analysis in Cytoscape 3.7.2 software while employing the NetworkAnalyzer plugin for topological assessment, yielding metrics such as degree centrality and betweenness centrality values.
Enrichment analysis of active ingredients-disease intersection targets
The clusterProfiler package was used to perform gene ontology (GO) enrichment analyses on intersection targets between active ingredients and differentially expressed genes across three levels: biological process (BP), cellular component (CC), and molecular function (MF), which also included Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment assessments.
Construction of a protein interaction network diagram for PI3K/AKT pathway analysis
Cytoscape 3.7.2 software was used to generate a PPI network diagram illustrating protein interactions; additionally, the CytoHubba tool was used for degree value calculations, leading us to identify core target proteins of this pathway.
Molecular docking
This project aimed to link with the PubChem database to download 3D structures corresponding to effective components derived from S. divaricata; candidate target protein structures along with ligand configurations were sourced from the PDB database (https://www.rcsb.org/)20. Subsequently, molecular docking simulations were performed via AutoDock Tools software, followed by visualization via PyMOL software to effectively demonstrate the docking results.
Experimental verification
Medications and reagents
Saposhnikovia divaricata (HPLC purity > 98%) was commercially purchased from Beijing Tongrentang Pharmaceutical Co., Ltd. (Beijing, China). Classification and identification were conducted by Professor Shaohuan Liu (School of Pharmacy, Guizhou Medical University) through macroscopic and microscopic characterization. The voucher specimen (No.2021110701) has been stored in the Herbarium of the School of Basic Medicine, Guizhou Medical University (Room 5–9 of the Physiology Laboratory), and was obtained through licensed commercial channels; sodium arsenite NaAsO2 (HPLC > 98%) was acquired from Shanghai Sigma High Tech Co., Ltd. (Shanghai, China); fetal bovine serum, DMEM and penicillin/streptomycin were purchased from Thermo Fisher Scientific (Massachusetts, USA); polyclonal antibodies against PI3K, p-PI3K, AKT, p-AKT, Caspase3, Cleaved-caspase3, Caspase8, Bcl2, and Bax and GAPDH polyclonal antibodies were sourced from Wuhan Sanying Biotechnology Co., Ltd. (Wuhan, China); a cell apoptosis detection kit was provided by Kaiji Biological Company (Nanjing, China); a reactive oxygen species detection kit was obtained from Biyuntian Biotechnology Co., Ltd. (Shanghai, China); and a trace reduced glutathione (GSH) determination kit was obtained from Nanjing Jiancheng Biotechnology Research Institute (Nanjing, China).
Cell culture
The mouse hippocampal neuron cell line HT22 was obtained from Wuhan Punosai Biotechnology Co., Ltd. (Wuhan, China). Subsequently, the HT22 cells were cultivated in low-glucose medium supplemented with 10% fetal bovine serum and 1% penicillin‒streptomycin and placed in a 37 °C, 5% CO2 incubator. The culture medium was changed daily, and the cells were passaged or cryopreserved when the confluence reached approximately 80–90%.
CCK8 assay
When a standard 96-well plate was used, 8000 cells were inoculated into each well with 100 µL of culture medium, and the culture plate was subsequently placed in a culture incubator (37 °C, 5% CO2) for 12–24 h. Following cell adhesion to the plate surface, different treatments were administered for an additional 24 h. Ten microliters of CCK8 reagent was added to each well and incubated for 2 h before the absorbance was measured at a wavelength of 450 nm.
Reactive oxygen species (ROS) assay
Well-cultured HT22 cells were inoculated in a six-well plate; when the cell density reached approximately 70-80%, different treatments were applied for 24 h: control, NaAsO2 (12.5 µM) and NaAsO2 + S. divaricata (12.5 µM + 400 µg/mL). The culture medium was removed after 24 h, and the cells were washed twice with PBS. One milliliter of serum-free culture medium was added to each well, 1 µL of DCFH-DA probe was added to each well, and the mixture was incubated for 20 min in a dark incubator. Then, the cells were collected, and 300 µL of PBS was added, after which the mixture was resuspended and tested by flow cytometry. The experiment was repeated three times for different batches of cells.
Glutathione (GSH) assay
After the cells were washed twice with PBS, they were collected via low-speed centrifugation and resuspended in 0.3–0.5 L of isotonic PBS buffer (0.1 M, pH 7.4). Following cell lysis by ultrasonication, the test mixture was prepared according to the reagent kit instructions, and the absorbance was measured at 405 nm via an enzyme-linked immunosorbent assay.
Assay of the apoptosis rate
Well-cultured HT22 cells were inoculated in a six-well plate. When the cell density reached approximately 70−80%, different treatments were carried out for 24 h: a control group, a NaAsO2 group (12.5 µM), and a NaAsO2 + S. divaricata group (12.5 µM + 400 µg/mL). After centrifugation at 1200 rpm for 5 min, each well was washed with 1 mL of PBS to remove residual media. The supernatant was discarded, followed by the addition of 50 µL of bin-containing buffer and 2.5 µL of FITC; this mixture was incubated in the dark for 5 min at room temperature before 5 µL of PI and another 200 µL of PBS were added. Each experiment was repeated three times using different batches of cells.
Cellular thermal shift assay (CETSA)
HT22 cells were cultured in 10-cm culture dishes for 24 h and then centrifuged at 12,000 rpm for 5 min at 37 °C. The cells were subsequently resuspended in 1 mL of PBS containing 20 mM Tris-HCl, 100 mM NaCl, 5 mM EDTA, 2 mM phenylmethylsulfonyl fluoride (PMSF), 10 ng/mL leupeptin, and 10 µg/mL aprotinin. Three freeze–thaw cycles were performed on the cells. The cells were exposed to liquid nitrogen for 3 min each cycle and then heated at 25 °C for 3 min. After centrifugation at 12,000 rpm at 4 °C for half an hour, the supernatants from each group were collected. For the S. divaricata-treated groups, S. divaricata at 400 µg/mL and NaAsO2 at 12.5 µM were coadministered for 24 h. For the control groups, NaAsO2 (12.5 µM) was added for 24 h. All the groups were heated at 25 °C for an hour. The pairs in the control and treatment groups were subsequently heated for 3 min at 46, 49, 52, 55, 58, and 61 °C. Finally, the samples were analyzed by Western blotting.
Western blot assay
After total protein was extracted from the cells via the BCA quantitative method, the initial electrophoresis step was performed at 80 V for 30 min, followed by a transition to 120 V for 90 min; the proteins were subsequently transferred to the membrane at 300 mA for 120 min. After the samples were blocked with 5% skim milk powder for 120 min, they were incubated with the primary antibody overnight on a shaking table, followed by incubation with the secondary antibody for 2 h at room temperature, after which electrochemiluminescence was carried out.
RT-qPCR analysis
Real-time quantitative PCR was used to assess total RNA levels after culture medium removal whereupon 1 mL of TRIzol lysis solution was introduced per well—the mixture was thoroughly mixed and then allowed to stand for 5 min on ice prior to the addition of 0.2 mL of chloroform, which necessitated thorough mixing again before the mixture was briefly resuspended and then centrifuged for 15 min at 4 °C and 12,000 rpm. The top aqueous phase was transferred to a new tube, incubated with an equal volume of isopropanol on ice for 15 min, and centrifuged at 4 °C and 12,000 rpm to promote precipitation. The resulting supernatant was discarded, 1 mL of 70% ethanol was added, and the mixture was mixed well, with similar centrifugation steps. The mixture was rolled over filter paper until the ethanol on the wall of the EP tube was air-dried, allowing subsequent dissolution within 20 µL of DEPC-treated water, which was subsequently stored frozen (− 80 °C). Micro-UV spectrophotometry was used to measure the RNA concentration. The reverse transcription reaction system adhered strictly as outlined within the Reagent Kit protocols, yielding a total reaction volume of 20 µL aimed toward first-strand cDNA synthesis, whereas real-time PCR amplification systems comprised 10 µL configured precisely as instructed therein with the respective reagent boxes employed.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 8.0 (GraphPad Software, San Diego, CA) and SPSS Statistics 19.0 (IBM Corp., Armonk, NY). All values were expressed as mean ± standard deviation. Student’s t-tests were performed to compare the means between two groups, whereas one-way ANOVA was used to compare the mean of three or more groups. Significance threshold was P < 0.05.
Results
Obtaining Saposhnikovia divaricata targets
According to the TCMSP database results, 18 active ingredients for S. divaricata were obtained, as shown in Table 1, including (2R, 3R)-3-(4-hydroxy-3-methoxy-phenyl) -5-methoxy-2-methyl-2,3-dihydropyrano [5,6-h][1,4] benzodioxin-9-one (chloramphenicol A), 11-hydroxysec-o-beta-d-glucosylhamaudol_qt, anomalin, divaricatacid, divaricatol, Ammidin, ledebouriellol, phelloptorin, 5-O-Methylvisamminol, Phellopterin, sitosterol, wogonin, betasitosterol, Mandenol, isoimperatorin, Prangenidin, methyl icosa-11,14-dienoate, and Decursin. Based on databases such as PubChem and SwissTarget Prediction, we obtained 209 S. divaricata targets. Through integration and matching, 137 duplicate targets were removed, and 72 target areas were ultimately identified.
Construction of a network of effective ingredients for S. divaricata, targets, and arsenic poisoning
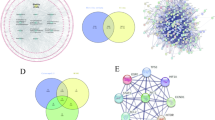
Through the GeneCards database, we obtained a total of 9717 genes related to nerve damage, with 2224 genes related to arsenic poisoning. By crossing 72 effective ingredient targets for S. divaricata with 9717 genes related to nerve damage and 2224 genes related to arsenic poisoning, 45 effective ingredient‒disease interaction targets were obtained, as shown in Fig. 1A. A STRING database was used to perform protein interaction analysis on 45 active ingredient targets, and Cytoscape 3.6.0 was used to perform “active ingredient‒disease target” network analysis. The results are shown in Fig. 1B, where AKT1, CASP3, IL6, and TP53 were key ingredient targets.
Network pharmacology analysis. (A) Venn diagram. (B) Interactive PPI network for disease and drug targets. The deeper and larger the node indicated stronger protein-protein connectivity. The top four networks targets analyzed by PPI were AKT1, CASP3, IL6, and TP53, respectively. (C) CC cellular component, (D) BP biological process, (E) MF molecular function. (F) KEGG pathway enrichment analysis. (G) The construction of the protein interaction network diagram on the PI3K/AKT pathway with the AKT1 node the largest and darkest in color. (H) Molecular docking. The main target, AKT1, of the PI3K/AKT signaling pathway was related to the active components such as 5-Methylvisaminol, andromalin, β-Sitosterol, Decursin, isoImperatorin, ledebourielle, Phelloperin, Pragenidin and wogonin.
GO and KEGG enrichment analysis
The 45 effective components of S. divaricata and disease interaction targets involved biological processes (BP), cellular components (CC), and molecular functions (MF), with the top 10 enriched, as shown in Fig. 1C–E. Its mechanism of action includes action on lipopolysaccharides (LPS) and reactive oxygen species (ROS). Figure 1F shows the top 10 pathways with the highest enrichment of 45 active ingredient‒disease interaction targets.
Construction of a protein interaction network diagram of the PI3K/AKT pathway
Because AKT1 is a key component target for arsenic-induced nerve damage and ranks among the top 10 enriched pathways in the PI3K/AKT pathway, the relevant genes in this pathway were analyzed for their protein‒protein interactions through STRING analysis and imported into Cytoscape 3.7.2 to construct an “active ingredient‒disease target” network. The results are shown in Fig. 1G, where AKT1 was the most important key target.
Molecular docking
Using Discovery Studio 2016, the key target AKT1 in the PI3K/AKT signaling pathway was identified, which has binding sites with the active ingredients 5-O-Methylvisamminol, anomalin, beta-sitosterol, Decursin, isoimperatorin, ledebouriellol, Phellopterin, Prangenidin and wogonin of S. divaricata, and their binding energy values are shown in Table 2. Therefore, AKT1 had good binding activity, and the specific binding mode is shown in Fig. 1H.
Saposhnikovia divaricata reversed NaAsO2-induced changes in morphology and cell viability
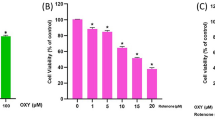
HT22 cells were treated with different concentrations of NaAsO2 (0, 3.125, 6.25, and 12.5 µM) for 24 h. CCK8 analysis revealed that NaAsO2 could reduce HT22 cell viability to varying degrees (P < 0.01) and inhibit cell proliferation. In addition, 12.5 µM NaAsO2 was the optimal condition for the intervention of HT22 cells for 24 h, and S. divaricata resulted in the most significant reversal of NaAsO2 damage to HT22 cells at 400 µg/mL, as shown in Fig. 2A. After 24 h of NaAsO2 and S. divaricata treatment, we observed morphological changes in HT22 cells via optical microscopy and found that, compared with the control group, the NaAsO2 treatment group presented a decreased number of HT22 cells, an increased number of dead cells, and increased transparency. The cell morphology was circular, and the neural processes were significantly shortened. Compared with that in the NaAsO2 treatment group, the morphology of the cells in the NaAsO2 + S. divaricata treatment group was significantly improved, indicating that S. divaricata had a certain protective effect on NaAsO2-induced morphological damage in HT22 cells, as shown in Fig. 2B.
Effects of S. divaricata on the morphological changes, viability, oxidative stress, apoptosis of HT22 cells induced by NaAsO2. (A) Cell viability of HT22 cells in each group. (B) Cell morphology in each group under the microscope. (C) ROS content was assessed by Flow cytometry in each group. (D) ROS content statistical chart (E) GSH content was assessed by ELISA in each group. (F) Apoptosis rates of HT22 cells were assessed by Flow cytometry in each group. (G) Apoptosis rates statistical chart. (H) The relative mRNA levels of Bcl2, Bax, Caspase8, Caspase3, and Cleaved-caspase3 in HT22 cells were assessed by RT-qPCR. (I) The protein expressions of cleaved Bcl2, Bax, Caspase8, Caspase3 and Cleaved-caspase3 were assessed by Western blot. (J) Statistical analysis of the expression intensity of Bcl2, Bax, Caspase8, Caspase3 and Cleaved-caspase3 in HT22 cells. GAPDH served as internal reference. *P < 0.05, **P < 0.01 vs. control group; ^P < 0.05, ^^P < 0.01 vs. 3.125 group; $P < 0.05, $$P < 0.01 vs. 6.25 group, #P < 0.05, ##P < 0.01 vs. NaAsO2 (12.5) group, &P < 0.05 vs. 12.5 + 300 group. The data was displayed as mean ± standard deviation, n = 3.
Saposhnikovia divaricata reversed NaAsO2-induced oxidative stress
To investigate the protective effects of S. divaricata on NaAsO2-induced HT22 cell toxicity, we detected the levels of ROS and GSH synthesis in HT22 cells via flow cytometry and ELISA. The following groups of cells were established: the control group, NaAsO2 (12.5 µM) group, and NaAsO2 + S. divaricata (12.5 µM + 400 µg/mL) group. Compared with that in the control group, the ROS content in the NaAsO2 group increased from 100 ± 55.97% to 421.99 ± 75.12% (P < 0.01). Compared with that in the NaAsO2 group, the ROS content in the NaAsO2 + S. divaricata group decreased from 421.99 ± 75.12% to 166.46 ± 89.49% (P < 0.01), as shown in Fig. 2C,D. Compared with that in the control group, the GSH synthesis in the NaAsO2 treatment group greatly decreased (P < 0.01), whereas compared with that in the NaAsO2 treatment group, GSH synthesis significantly increased in the NaAsO2 + S. divaricata treatment group (P < 0.01), as shown in Fig. 2E, indicating that S. divaricata could reverse NaAsO2-induced oxidative stress in HT22 cells.
Saposhnikovia divaricata downregulated NaAsO2-induced apoptosis
In this study, we explored the effects of S. divaricata on NaAsO2-induced apoptosis in HT22 cells. Compared with that of the control group, the apoptosis rate of the NaAsO2 group significantly increased from 1.83 ± 0.41% to 18.65 ± 1.29% (P < 0.01); compared with that of the NaAsO2 group, the apoptosis rate of the NaAsO2 + S. divaricata group decreased from 18.65 ± 1.29% to 7.53 ± 1.03% (P < 0.01), as shown in Fig. 2F,G. Compared with those in the control group, the protein expression and mRNA levels of Bcl2 in the NaAsO2 group were decreased, whereas the protein expression and mRNA levels of Bax, Caspase8, Caspase3, and Cleaved-caspase3 were increased (P < 0.01). Compared with those in the NaAsO2 group, the protein expression and mRNA levels of Bax, Caspase8, Caspase3 and Cleaved-caspase3 in the NaAsO2 + S. divaricata group were increased, whereas the protein expression and mRNA levels of Bax, Caspase8, Caspase3, and Cleaved-caspase3 were decreased (P < 0.01), as shown in Fig. 2H–J.
The PI3K/AKT pathway May be a key target for Saposhnikovia divaricata
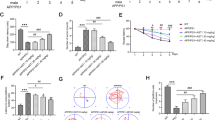
To identify drug targets, we analyzed the binding model between the active ingredients of S. divaricata and AKT1 protein via molecular docking technology, as shown in Fig. 1H. We found that AKT1 could bind to multiple active ingredients of S. divaricata, resulting in a stable binding conformation. The correlation between the S. divaricata and PI3K/AKT ratio was tested via CETSA experiments. Compared with NaAsO2 treatment, S. divaricata treatment increased the stability of PI3K and AKT (Fig. 3A,B). These results indicate that the PI3K/AKT pathway may be a key target for S. divaricata.
Identification of PI3K/AKT as the key target for S. divaricata. (A) The interaction of S. divaricata with PI3K and AKT was assessed by cell thermal displacement measurement in HT22 cells. (B) Quantification of the relative intensity of the PI3K and AKT proteins versus increased temperature. GAPDH served as an internal reference. (C) The protein expressions of p-PI3K, PI3K, p-AKT, AKT and Nrf2 were assessed by Western Blot. (D) Statistical analysis of the ratios of p-PI3K/PI3K, p-AKT/AKT and the expression intensity of Nrf2 in HT22 cells. GAPDH served as an internal reference. *P < 0.05, **P < 0.01 vs. control group, #P < 0.05, ##P < 0.01 vs. NaAsO2 group. The control group was used for comparison. The data was displayed as mean ± standard deviation, n = 3.
Saposhnikovia divaricata activated the PI3K/AKT pathway inhibited by NaAsO2
Finally, to confirm that PI3K/AKT is a target for S. divaricata, we detected the expression of PI3K/AKT pathway proteins and the key downstream signaling molecule Nrf2. Compared with those in the control group, the ratios of p-PI3K/PI3K and p-AKT/AKT and the protein expression of Nrf2 in the NaAsO2 group were lower (P < 0.01). Compared with the NaAsO2 group, the NaAsO2 + S. divaricata group increased the ratios of p-PI3K/PI3K, p-AKT/AKT, and Nrf2 protein expression (P < 0.01), as shown in Fig. 3C,D.
Discussion
Saposhnikovia divaricata, with a spicy, sweet, slightly warm taste, belongs to the bladder, spleen, and liver channels21. As a traditional Chinese medicinal material, S. divaricata is considered a top-grade medicinal herb in the monograph of Shennong Materia Medica22,23. Its active ingredients include mainly chromogenic ketones, polysaccharides, volatile oils, and coumarins, which can eliminate excessive arsenic in the human body and exert strong detoxification effects (Fig. 2)24. Through network pharmacology exploration, a comprehensive dataset was compiled, identifying 78 S. divaricata targets, 9717 nerve injury targets, and 2224 arsenic poisoning targets, with an additional 45 putative targets identified through intersection analysis (Fig. 1A). GO annotations and KEGG pathway analyses of these protein targets revealed significant enrichment in apoptotic processes (Fig. 1C–F). Mochizuki et al. further corroborated that NaAsO2 induces neurotoxicity through apoptotic mechanisms and affects intracellular signaling pathways25. Guided by the results of the enrichment analysis of the top 10 pivotal targets and pathways, the PI3K/AKT signaling pathway was selected as a critical regulatory axis for subsequent investigations26. By utilizing the degree algorithm for topological profiling of the “potential targets”, the most pivotal target, AKT1, was identified (Fig. 1G), followed by molecular docking exercises with the efficacious S. divaricata target (Fig. 1H).
Arsenic-containing compounds, such as NaAsO2 and As2O3, can disrupt normal neural function by interfering with neurotransmitter synthesis and nerve signal transduction27. Our assessment revealed a gradual decline in viability and cell number along with shortened protrusions and a rounded morphology induced by increasing concentrations of NaAsO2 under the microscope. However, S. divaricata ameliorated these morphological changes, resulting in a substantial increase in cell viability (Fig. 2A). Liu et al. reported that As2O3 could induce oxidative stress, causing oxidative damage to cell membranes and organelles while accelerating neuronal aging and death28. We also observed that NaAsO2 significantly elevated the ROS content within the cells while concurrently decreasing the GSH content, whereas S. divaricata exhibited neuroprotective effects by decreasing the level of ROS and increasing the GSH content (Fig. 2C–E). Apoptosis is tightly regulated by various genes, including those in the Bcl2 and Caspase families29. Bax and Bcl2 are prominent members of the B-cell lymphoma/leukemia-2 (Bcl2) family, and they play opposite roles in NaAsO2-induced apoptosis30,31. Bax is typically regarded as an apoptosis-promoting protein that increases mitochondrial membrane permeability, leading to cytochrome C release along with other proapoptotic factors that initiate apoptotic pathways32,33. Conversely, Bcl2 functions primarily as an antiapoptotic protein, protecting mitochondrial integrity and thereby inhibiting apoptosis34. Cleaved-caspase3, Caspase3 and Caspase8 are key regulatory factors in the process of apoptosis35,36. Caspases 3 and 8 are pivotal for executing and initiating apoptosis, respectively. Caspase8 acts as an initiating protease involved in external apoptotic signaling cascades triggered by receptor-mediated stimuli, whereas activated Caspase3 orchestrates subsequent apoptotic events, including nuclear DNA degradation alongside alterations in cell morphology, culminating in apoptosis itself37,38,39,40. Flow cytometry analysis indicated a marked increase in the apoptosis rate in HT22 cells subjected to NaAsO2 treatment, which was notably reduced by S. divaricata intervention (Fig. 2F,G). Western blot and RT-qPCR analysis further confirmed that S. divaricata decreased the levels of the proapoptotic protein and mRNA of Bax, Caspase8, Caspase3, and Cleaved-caspase3, but increased the level of the antiapoptotic protein Bcl2 in NaAsO₂-treated HT22 cells (Fig. 2H–J). Therefore, these data confirmed our network pharmacological prediction that S. divaricata effectively alleviates NaASO2-induced neuronal apoptosis and related oxidative stress.
Based on the findings from our network pharmacological analysis, we further investigated the roles played by the PI3K/AKT signaling pathway in HT22 cells to elucidate the molecular mechanisms underlying the ability of the S. divaricata to antagonize arsenic neurotoxicity. Studies have shown that the PI3K/AKT pathway plays an important role in arsenic induced neuronal apoptosis41. This pathway comprises three key family members: PI3K, AKT and Nrf2. Stress-activated PI3K and AKT are pivotal in maintaining a balance between cell survival and death42. Through CETSA experiments, we confirmed that the PI3K/AKT pathway represents a potential target for S. divaricata treatment. In addition, the upregulation of the p-PI3K/PI3K and p-AKT/AKT ratios and Nrf2 protein expression were also observed in the S. divaricata group; we hypothesized that this may be due to the interaction between S. divaricata, NaAsO2 and the PI3K/AKT pathway, leading to increased activity of the PI3K/AKT pathway. Acting as both the key downstream regulator in the PI3K/AKT pathway and the oxidative stress transcription factor, Nrf2 may inhibit cell apoptosis by regulating the expressions of Bcl2 and Bax. Our results indicated that NaAsO2 could inhibit the PI3K/AKT pathway, downregulating Nrf2 levels in HT22 cells. Conversely, S. divaricata activated NaAsO2-inhibited signaling and antagonized the NaAsO2-induced downregulation of Nrf2 (Fig. 3).
Therefore, S. divaricata can alleviate NaAsO2-induced cellular apoptosis, thereby reducing associated neurotoxic effects by activating the PI3K/AKT signaling pathway. Considering certain limitations within this study, additional in vitro studies are necessary to validate these mechanisms.
Conclusion
Network pharmacology revealed that the effects of S. divaricata on NaAsO2-induced neurotoxicity are intimately associated with apoptosis and the PI3K/AKT pathway. The experimental findings revealed that S. divaricata exerted therapeutic effects via antioxidant, anti-inflammatory, and anti-apoptotic activities, which may be related to the activation of the PI3K/AKT pathway. Hence, S. divaricata could be employed as a novel therapeutic modality to prevent nerve damage associated with NaAsO2.
Data availability
The data underlying this article will be shared on reasonable request, with the corresponding author.
References
Tabelin, C. B. et al. Arsenic, selenium, boron, lead, cadmium, copper, and zinc in naturally contaminated rocks: A review of their sources, modes of enrichment, mechanisms of release, and mitigation strategies. Sci. Total Environ. 645, 1522–1553. https://doi.org/10.1016/j.scitotenv.2018.07.103 (2018).
Tyler, C. R. & Allan, A. M. The effects of arsenic exposure on neurological and cognitive dysfunction in human and rodent studies: A review. Curr. Environ. Health Rep. 1 (2), 132–147. https://doi.org/10.1007/s40572-014-0012-1 (2014).
Yamauchi, H., Hitomi, T. & Takata, A. Evaluation of arsenic metabolism and tight junction injury after exposure to arsenite and monomethylarsonous acid using a rat in vitro blood-Brain barrier model. PLoS One. 18 (11), e0295154. https://doi.org/10.1371/journal.pone.0295154 (2023).
Chu, F. et al. Unveiling the LncRNA-miRNA-mRNA regulatory network in arsenic-induced nerve injury in rats through high-throughput sequencing. Toxics. 11 (12), 953. https://doi.org/10.3390/toxics11120953 (2023).
Weinmuellner, R. et al. Long-term exposure of immortalized keratinocytes to arsenic induces EMT, impairs differentiation in organotypic skin models and mimics aspects of human skin derangements. Arch. Toxicol. 92 (1), 181–194. https://doi.org/10.1007/s00204-017-2034-6 (2018).
Nurchi, V. M. et al. Arsenic toxicity: molecular targets and therapeutic agents. Biomolecules. 10 (2), 235. https://doi.org/10.3390/biom10020235 (2020).
Domingo-Relloso, A. et al. Arsenic exposure, blood DNA methylation, and cardiovascular disease. C Irc Res. 131 (2), e51–e69. https://doi.org/10.1161/CIRCRESAHA.122.320991 (2022).
Bjørklund, G., Mutter, J. & Aaseth, J. Metal chelators and neurotoxicity: lead, mercury, and arsenic. Arch. Toxicol. 91 (12), 3787–3797. https://doi.org/10.1007/s00204-017-2100-0 (2017).
Erdenebileg, S. et al. Saposhnikovia divaricata root and its major components ameliorate inflammation and altered gut microbial diversity and compositions in DSS-induced colitis. Integr. Med. Res. 12 (4), 100998. https://doi.org/10.1016/j.imr.2023.100998 (2023).
Huang, Y. N. Effects of Radix Saposhnikoviae and its Components on ATO Toxicity, and Apoptosis of HL-60/H9c2 Induced ATO[D] (Chinese Academy of Traditional Chinese Medicine, 2014).
Jiang, C. Study on the Extraction of the Effective Components and Protective Effect on Experimental Liver Injury with Radix Saposhnikoviae[D] (Jilin Agricultural University, 2013).
Cui, S., Chen, S., Wu, Q., Chen, T. & Li, S. A network pharmacology approach to investigate the anti-inflammatory mechanism of effective ingredients from Salvia miltiorrhiza. Int. Immunopharmacol. 81, 106040. https://doi.org/10.1016/j.intimp.2019.106040 (2020).
Nogales, C. et al. Network pharmacology: curing causal mechanisms instead of treating symptoms. Trends Pharmacol. Sci. 43 (2), 136–150. https://doi.org/10.1016/j.tips.2021.11.004 (2022).
Ru, J. et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 6, 13. https://doi.org/10.1186/1758-2946-6-13 (2014).
Kim, S. et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 49 (D1), D1388–D1395. https://doi.org/10.1093/nar/gkaa971 (2021).
Gfeller, D., Michielin, O. & Zoete, V. Shaping the interaction landscape of bioactive molecules. Bioinformatics 29 (23), 3073–3079. https://doi.org/10.1093/bioinformatics/btt540 (2013).
Stelzer, G. et al. The GeneCards Suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinf. 54 1.30.1–1.30.33. https://doi.org/10.1002/cpbi.5. (2016).
Li, N. et al. Network pharmacology mechanism of scutellarin to inhibit RGC pyroptosis in diabetic retinopathy. Sci. Rep. 13 (1), 6504. https://doi.org/10.1038/s41598-023-33665-3 (2023).
Szklarczyk, D. et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51 (D1), 5D638–D646. https://doi.org/10.1093/nar/gkac1000 (2023).
Rubach, P., Zajac, S., Jastrzebski, B., Sulkowska, J. I. & Sułkowski, P. Genus for biomolecules. Nucleic Acids Res. 48 (D1), D1129-D1135. https://doi.org/10.1093/nar/gkz845 (2020).
Okuyama, E. et al. Analgesic components of Saposhnikovia root (Saposhnikovia divaricata). Chem. Pharm. Bull. (Tokyo). 49 (2), 154–160. https://doi.org/10.1248/cpb.49.154 (2001).
Li, Z. Y. et al. Neuroprotective effects of total saikosaponins of Bupleurum Yinchowense on corticosterone-induced apoptosis in PC12 cells. J. Ethnopharmacol. 48 (3), 794–803. https://doi.org/10.1016/j.jep.2013.04.057 (2013).
Yang, M. et al. Saposhnikovia divaricata-An ethnopharmacological, phytochemical and pharmacological review. Chin. J. Integr. Med. 26 (11), 873–880. https://doi.org/10.1007/s11655-020-3091-x (2020).
Urbagarova, B. M. et al. Chromones and coumarins from Saposhnikovia divaricata (Turcz.) schischk. Growing in Buryatia and Mongolia and their cytotoxicity. J. Ethnopharmacol. 261, 112517. https://doi.org/10.1016/j.jep.2019.112517 (2020).
Mochizuki, H. Arsenic neurotoxicity in humans. Int. J. Mol. Sci. 20 (14), 3418. https://doi.org/10.3390/ijms20143418 (2019).
He, X. et al. The PI3K/AKT signalling pathway in inflammation, cell death and glial Scar formation after traumatic spinal cord injury: mechanisms and therapeutic opportunities. Cell. Prolif. 55 (9), e13275. https://doi.org/10.1111/cpr.13275 (2022).
Ariafar, S., Makhdoomi, S. & Mohammadi, M. Arsenic and Tau phosphorylation: a mechanistic review. Biol. Trace Elem. Res. 201 (12), 5708–5720. https://doi.org/10.1007/s12011-023-03634-y (2023).
Liu, Y. et al. Arsenic (III) and/or copper (II) induces oxidative stress in chicken brain and subsequent effects on mitochondrial homeostasis and autophagy. J. Inorg. Biochem. 211, 111201. https://doi.org/10.1016/j.jinorgbio.2020.111201 (2020).
Fan, T. J., Han, L. H., Cong, R. S. & Liang, J. Caspase family proteases and apoptosis, acta biochim biophys sin (Shanghai). 37(11) 719–727. https://doi.org/10.1111/j.1745-7270.2005.00108.x (2005).
Shi, Q. et al. ATF3 promotes Arsenic-Induced apoptosis and oppositely regulates DR5 and Bcl-xL expression in human bronchial epithelial cells. Int. J. Mol. Sci. 22 (8), 4223. https://doi.org/10.3390/ijms22084223 (2021).
Mohammadpour-Gharehbagh, A. et al. Genetic and epigenetic analysis of the BAX and BCL2 in the placenta of pregnant women complicated by preeclampsia. Apoptosis. 24 (3–4), 301–311. https://doi.org/10.1007/s10495-018-1501-8 (2019).
Renault, T. T. et al. Mitochondrial shape governs BAX-induced membrane permeabilization and apoptosis. Mol. Cell. 57 (1), 69–82. https://doi.org/10.1016/j.molcel.2014.10.028 (2015).
Renault, T. T., Floros, K. V. & Chipuk, J. E. BAK/BAX activation and cytochrome c release assays using isolated mitochondria. Methods. 61 (2), 146–155. https://doi.org/10.1016/j.ymeth.2013.03.030 (2013).
Sirtl, S. et al. Hypertonicity-enforced BCL-2 addiction unleashes the cytotoxic potential of death receptors. Oncogene. 37 (30), 4122–4136. https://doi.org/10.1038/s41388-018-0265-5 (2018).
Wang, Y. et al. Chemotherapy drugs induce pyroptosis through caspase–3 cleavage of a gasdermin. Nature 547 (7661), 99–103. https://doi.org/10.1038/nature22393 (2017).
Chen, Q. et al. Discovery of a potent Hedgehog pathway inhibitor capable of activating caspase8-dependent apoptosis. J. Pharmacol. Sci. 137 (3), 256–264. https://doi.org/10.1016/j.jphs.2018.07.001 (2018).
Wang, W. et al. Ropivacaine promotes apoptosis of hepatocellular carcinoma cells through damaging mitochondria and activating caspase–3 activity. Biol. Res. 52 (1), 36. https://doi.org/10.1186/s40659-019-0242-7 (2019).
Mandal, R., Barrón, J. C., Kostova, I., Becker, S. & Strebhardt, K. Caspase–8: the double-edged sword. Biochim. Biophys. Acta Rev. Cancer. 1873 (2), 188357. https://doi.org/10.1016/j.bbcan.2020.188357 (2020).
Powley, I. R., Hughes, M. A., Cain, K. & MacFarlane, M. Caspase–8 tyrosine–380 phosphorylation inhibits CD95 DISC function by preventing procaspase–8 maturation and cycling within the complex. Oncogene. 35 (43), 5629–5640. https://doi.org/10.1038/onc.2016.99 (2016).
Clark, R. S. et al. Caspase–3 mediated neuronal death after traumatic brain injury in rats. J. Neurochem. 74 (2), 740–753. https://doi.org/10.1046/j.1471-4159.2000.740740.x (2000).
Tian, Z. et al. Arsenic trioxide sensitizes pancreatic cancer cells to gemcitabine through downregulation of the TIMP1/PI3K/AKT/mTOR axis. Transl. Res. 255, 66–76. https://doi.org/10.1016/j.trsl.2022.11.007 (2023).
Zhang, Q. et al. Puerarin inhibited oxidative stress and alleviated cerebral ischemia-reperfusion injury through PI3K/Akt/Nrf2 signaling pathway. Front. Pharmacol. 14, 1134380. https://doi.org/10.3389/fphar.2023.1134380 (2023).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (NO. 31960188), Guizhou Provincial College Students’ Innovation and Entrepreneurship Training Program Project (NO. S202210660078, S202210660077), Guizhou Provincial Science and Technology Support Project (No. 2021-General 415) and the Department of Education of Guizhou Province (Guizhou Teaching and Technology [2023]015).
Author information
Authors and Affiliations
Contributions
D.G.: Validation, visualization; D.G., X.-L.C., H.-L.H., M.X.: Writing-original draft, data curation, methodology; J.-G.P., W.Z.: Writing-review and editing, funding acquisition, supervision; D.G., X.-Y.W., Y.-Y.C., W.-R.K.: Formal analysis; G.-H.H., J.-Y.H., X.-L.Z.: Formal analysis, data curation. B.-F.S., X.-H.L.: Funding acquisition. S.-H.L.: Identification of voucher specimens.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gu, D., Chen, XL., He, HL. et al. Mechanisms of Saposhnikovia divaricata in attenuating NaAsO2-induced neural injury via the PI3K/AKT pathway. Sci Rep 15, 33303 (2025). https://doi.org/10.1038/s41598-025-04735-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-04735-5