Abstract
This study aims to investigate the diagnostic value of integrating multi-parametric magnetic resonance imaging (mpMRI) radiomic features with tumor abnormal protein (TAP) and clinical characteristics for diagnosing prostate cancer. A cohort of 109 patients who underwent both mpMRI and TAP assessments prior to prostate biopsy were enrolled. Radiomic features were meticulously extracted from T2-weighted imaging (T2WI) and the apparent diffusion coefficient (ADC) maps. Feature selection was performed using t-tests and the Least Absolute Shrinkage and Selection Operator (LASSO) regression, followed by model construction using the random forest algorithm. To further enhance the model’s accuracy and predictive performance, this study incorporated clinical factors including age, serum prostate-specific antigen (PSA) levels, and prostate volume. By integrating these clinical indicators with radiomic features, a more comprehensive and precise predictive model was developed. Finally, the model’s performance was quantified by calculating accuracy, sensitivity, specificity, precision, recall, F1 score, and the area under the curve (AUC). From mpMRI sequences of T2WI, dADC(b = 100/1000 s/mm2), and dADC(b = 100/2000 s/mm2), 8, 10, and 13 radiomic features were identified as significantly correlated with prostate cancer, respectively. Random forest models constructed based on these three sets of radiomic features achieved AUCs of 0.83, 0.86, and 0.87, respectively. When integrating all three sets of data to formulate a random forest model, an AUC of 0.84 was obtained. Additionally, a random forest model constructed on TAP and clinical characteristics achieved an AUC of 0.85. Notably, combining mpMRI radiomic features with TAP and clinical characteristics, or integrating dADC (b = 100/2000 s/mm²) sequence with TAP and clinical characteristics to construct random forest models, improved the AUCs to 0.91 and 0.92, respectively. The proposed model, which integrates radiomic features, TAP and clinical characteristics using machine learning, demonstrated high predictive efficiency in diagnosing prostate cancer.
Similar content being viewed by others
Introduction
Prostate cancer is the second most common male cancer globally1. The gold standard for diagnosing prostate cancer remains the prostate biopsy2,3. However, early-stage prostate cancer often presents with subtle clinical manifestations and a covert onset, underscoring the critical importance of early detection and diagnosis.
Although prostate-specific antigen (PSA) is currently widely utilized for prostate cancer screening, relying solely on PSA can lead to over-diagnosis, contributing to ongoing controversies surrounding its use4,5. Tumor abnormal protein (TAP), an emerging biomarker indicative of glycosylation dysregulation, derives its diagnostic value from the aberrant activation of glycosyltransferases during the early stages of malignant transformation. This dysregulation alters the branching structures of glycoprotein sugar chains, leading to the formation of soluble TAP that enters circulation, typically appearing 6–24 months earlier than clinical symptoms or radiological manifestations. Numerous studies have demonstrated that TAP is closely associated with the occurrence, progression, and prognosis of malignant tumors6. Our previous study revealed that TAP is increased in prostate cancer, and has more diagnostic capabilities than other diagnostic methods in prostate cancer7.
Radiomics involves extracting features from medical images and applying machine learning algorithms to transform medical images into high-dimensional data that can be amenable to computational analysis. Multi-parametric magnetic resonance imaging (mpMRI) plays a crucial role in managing prostate cancer and has emerged as a highly promising non-invasive imaging technique for prostate cancer identification8,9. Machine learning enables models to escalate their performance exponentially with increasing amounts and complexity of data10,11, holding the potential for transformative impact within oncology12. Despite this, current radiomics research predominantly focuses on disease prognosis, postoperative pathological prediction, and disease diagnosis in prostate cancer13,14,15,16, with few studies integrating radiomics with clinical data, genomics, or proteomics. Moreover, compared with other types of cancer, research on prostate cancer in this area is relatively scarce.
Consequently, to address these dual challenges, we propose the present study. This study establishes a non-invasive diagnostic framework for prostate cancer, underpinned by MRI-derived radiomic features, complemented by TAP and clinical characteristics.
Method and materials
Patient and data collection
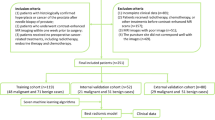
A retrospective analysis was conducted on 109 consecutive patients who underwent mpMRI, TAP testing, and prostate biopsy at the Second Affiliated Hospital of Soochow University between December 2017 and September 2022. An extensive array of clinical data were collected including patient age, PSA levels, prostate volume, Gleason scores(GS), and Prostate Imaging Reporting and Data System (PI-RADS) scores. Patients were included in this study if they met the following criteria: (1) Presence of a well-defined prostate lesion on mpMRI, as classified by the Prostate Imaging Reporting and Data System Version 2.1 (PI-RADS v2.1); (2) Availability of a complete clinical dataset including patient age, PSA levels, prostate volume and Gleason scores; (3) Voluntary provision of consent for blood tests for TAP; (4) Underwent their first prostate biopsy.
Exclusion criteria included the following: (1) History of prior prostate biopsy. (2) Diagnosis or treatment of extraprostatic malignancies. (3) Presence of TAP-positive confounding factors, including active rheumatoid arthritis, uncontrolled diabetes mellitus, recent fractures, autoimmune disorders, or tuberculosis. (4) Lack of complete clinical data, precluding statistical analysis due to incomplete documentation. The patient selection process and characteristics of those meeting inclusion criteria and no exclusion criteria are summarized in Fig. 1.
MRI acquisition
The imaging was performed on a Philips 3.0 Tesla Ingenia MRI scanner, following a meticulously designed scanning protocol that encompassed sagittal T2-weighted imaging (T2WI), axial T2WI, Diffusion-Weighted Imaging (DWI), and Dynamic Contrast-Enhanced MRI (DCE-MRI). Regarding the DWI sequence, b-values of 100/1000 s/mm² and 100/2000 s/mm² (s/mm²) were selected. Subsequent to the scanning process, the Apparent Diffusion Coefficient (ADC) map was automatically generated through the integrated post-processing software. For the DCE-MRI component, a total of 64 time points were acquired to capture the dynamic enhancement patterns.
Tumor segmentation and extraction of imaging features
The process was carried out by an experienced clinical surgeon with over five years of experience in image interpretation, having handled over 1000 s/mm² cases, in collaboration with a radiologist possessing a 15 year tenure in image interpretation. They carefully described the region of interest (ROI) of the lesion based on prostate magnetic resonance imaging (MRI) reports, validated by pathological findings from biopsy procedures. In cases of disagreement, discrepancies were resolved through joint discussion to achieve consensus. Using the 3D Slicer software platform, the tumor ROI was precisely delineated on both T2WI and ADC sequences. Subsequently, radiomic features were systematically extracted from the ROI within each patient’s T2WI and ADC sequences. These features were categorized into three distinct classes: 1.Shape-based features. 2. First-order features. 3. Texture features, including Gray Level Run Length Matrix (GLRLM), Gray Level Co-occurrence Matrix (GLCM), Gray Level Size Zone Matrix (GLSZM), Neighborhood Gray Tone Difference Matrix (NGTDM), and Gray Level Dependency Matrix (GLDM). A total of 390 features were extracted, and these extracted features conformed to the Initiative for Biomarker Standardization in Medical Imaging (IBSI)17. Figure 2 provides a representative illustration of the lesion segmentation workflow applied to prostate cancer patients.
Feature selection and construction of radioproteomics models
An independent t-test was used to evaluate the differences among all radiomic features between the benign and malignant cohorts, with the aim of eliminating those radiomic features that did not exhibit statistically significant associations with the diagnostic outcome. Subsequently, the radiomic features that demonstrated significant differences were subjected to the Least Absolute Shrinkage and Selection Operator (LASSO) regression analysis. Through a rigorous 10-fold cross-validation process, the optimal lambda (λ) value was determined, which facilitated the identification of the most efficient selection of radiomic features along with their corresponding coefficients, thereby achieving feature screening and dimensionality reduction. Using the random forest algorithm, the radiomics features selected through the above methods were integrated into radioproteomics models for each patient based on T2WI, dADC(b = 100/1000 s/mm2), dADC(b = 100/2000 s/mm2), and the sequence containing all three mpMRI features.
Construction of clinical models and joint models of radiomic features and clinical characteristics
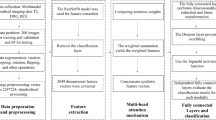
In the present study, we compiled a dataset integrating TAP and a range of clinical variables, including patient age, PSA levels, prostate volume and Gleason scores. Using the Least Absolute Shrinkage and Selection Operator (LASSO) regression, we identified and selected independent risk factors for prostate cancer. Subsequently, a clinical prediction model was constructed based on these identified risk factors. Using the random forest algorithm, we developed several predictive models: one focusing exclusively on clinical characteristics that included TAP, and two integrated models combining radiomic features with clinical data. Specifically, these integrated models were constructed using T2WI coupled with dADC (b = 100/1000 s/mm²) and dADC(b = 100/2000 s/mm²) respectively, along with the corresponding clinical data. We compared the predictive capabilities among these developed models. A schematic workflow of the model development process is illustrated in Fig. 3.
Statistical analysis
Statistical analyses were performed using SPSS software. For count data comparison, the chi-square test was applied. To assess the normality of the measurement data, Shapiro-Wilk test were conducted beforehand. Thereafter, either independent samples t-tests or non-parametric Mann-Whitney U test were conducted, depending on whether normality assumptions were met. To assess the predictive efficacy of the investigated models, several metrics were calculated, including Receiver Operating Characteristic (ROC) curves and the Area Under the Curve (AUC), sensitivity, specificity, accuracy recall and F1 score. Statistical significance was defined as p < 0.05.
Result
Clinical data
Based on the inclusion criteria, a total of 109 patients were enrolled, comprising 57 patients with benign prostatic hyperplasia and 52 patients with prostate cancer. The dataset included prostate mpMRI images, and clinical data, including TAP, age, PSA levels, Gleason scores, and PI-RADS scores. A comprehensive overview of the cohort’s clinical characteristics is provided in Table 1.
Selection of features and extraction of radiomics features
A total of 390 radiomic features were extracted from the ROI of each patient’s T2WI, dADC(b = 100/1000 s/mm²), and dADC(b = 100/2000 s/mm2) images. Following rigorous feature selection and dimensionality reduction processes using the Student’s t-test and LASSO regression, 8, 10, and 13 radiomic features were identified as significant and selected from the T2WI, dADC(b = 100/1000 s/mm2), and dADC(b = 100/2000 s/mm2). These refined features are detailed in Table 2, providing insights into their discriminatory power. For visualization, the selected radiomic features of each imaging modality are graphically depicted in Fig. 4.
(A, B, and C) represent the effectiveness curves for T2WI, dADC (b = 100/1000 s/mm2) and dADC100/2000 s/mm2), respectively. (D, E, and F) depict the convergence graphs of the corresponding sequence feature coefficients, where Lasso model was employed along with cross-validation for feature selection.
Using the mpMRI sequences of T2WI, dADC(b = 100/1000 s/mm2), and dADC(b = 100/2000 s/mm2), 8, 10, and 13 radiomic features were identified as significantly associated with prostate cancer, respectively. Individual random forest models were constructed using each set of these features, resulting in AUC values of 0.83 (sensitivity: 0.813, specificity: 0.588), 0.86 (sensitivity: 0.750, specificity: 0.822), and 0.87 (sensitivity: 0.750, specificity: 0.941). Subsequently, by integration of these three datasets into a consolidated random forest model, the AUC becomes 0.84 (sensitivity: 0.750, specificity: 0.941), demonstrating enhanced diagnostic performance when multiple mpMRI-derived biomarker sets are combined.
A random forest model based on TAP and other clinical variables achieved an AUC of 0.85 (sensitivity: 0.750, specificity: 0.765). When combining mpMRI radiomic features with TAP and clinical variables, or When combining mpMRI radiomic features with TAP and clinical variables, or using the dADC (b = 100/2000 s/mm²) sequence with TAP and clinical variables alone, the AUC values were 0.91 (sensitivity: 0.625, specificity: 0.882) and 0.92 (sensitivity: 0.625, specificity: 0.941), respectively. These findings demonstrate that the combined model incorporating radiomic features, TAP, and clinical variables may achieve a superior diagnostic performance in prostate cancer identification, as evidenced by machine learning techniques.
The ROC curves representing the predictive performance of these models are depicted in Fig. 5, while the architecture of the developed random forest models are illustrated in Fig. 6. Table 3 compares the AUC, accuracy, sensitivity, and specificity of the different models for forecasting prostate cancer. Figure 7 displays the distribution of the extracted 31 radiomic features using a heatmap. Additionally, Supplemental Figure S1 presents calibration curves utilized to assess the accuracy of the models’ predicted probabilities.
ROC curves of random forest. (A) ROC curves for radiomics-based T2WI model. (B) ROC curves for radiomics-based dADC(b = 100/1000 s/mm2) model. (C) ROC curves for radiomics-based dADC(b = 100/2000 s/mm2) model. (D) ROC curves for TAP and additional clinical characteristics model. (E) ROC curves for radiomics-based T2WI and dADC model. (F) ROC curves for radiomics-based dADC, TAP and clinical characteristics model. (G) ROC curves for radiomics-based T2WI, dADC, TAP and clinical characteristics model. (Cm: Clinical characteristics model, including TAP. T2: T2WI)
Heatmap for normalized feature value distribution of the extracted 31 features between PCa and BPH. The red regions indicate positive correlations with PCa features, while the blue regions represent negative correlations with nPCa features. The color intensity is proportional to the strength of correlation (darker hues denote strong correlations, whereas lighter hues indicate weak correlations).
Discussion
This study aims to develop a non-invasive diagnosis of prostate cancer through machine learning approaches based on MRI data, an area of growing research interest18. The random forest algorithm, an ensemble machine learning method, aggregates predictions from multiple decision trees, each trained on a randomly selected subset of the entire dataset. By aggregating predictions from all trees, it can reduce noise that may affect individual trees and lead to overfitting, thereby improving accuracy and reducing variance19. Donisi et al. demonstrated using openly accessible datasets that radiomics-based machine learning models can proficiently identify clinically significant prostate cancer (PCa), with the random forest model achieving the highest accuracy20. In the present study, the random forest algorithm showed commendable performance, particularly when applied to ADC sequences integrated with clinical parameters such as TAP, achieving an AUC of 0.915. These findings suggest that our proposed model possesses robust diagnostic capabilities for distinguishing between prostate cancer and benign prostatic hyperplasia.
TAP represents a tumor biomarker identified in recent years, the assessment of which can be used to indirectly reflect the number of transformed cells and the degree of malignancy. It has been widely applied in the screening and prognostic evaluation of gastric cancer, bladder cancer, prostate cancer and other diseases21,22. Over the past few years, there has been significant progress in the diagnostic efficacy of mpMRI of the prostate and its related radiomics research9,23,24,25. Such radiomics studies have predominantly focused on pre-biopsy diagnosis differentiation between indolent and aggressive disease phenotypes, and the diagnosis of clinical significance versus non-clinical significance26,27,28. Currently, TAP is frequently used alone or combined with PSA levels for the diagnosis of prostate cancer7. However, few studies have examined the integration of TAP with alternative diagnostic modalities. Regarding the development of diagnostic models that integrate TAP with radiomics features, there remains a paucity of high-quality research in this domain.
In mpMRI, T2WI provides a clear depiction of the prostate gland’s zonal anatomical structure. Meanwhile, ADC values reflect tissue microstructural properties, including cellular density and intracellular complexity. These parameters distinguish malignant lesions by contrasting the higher cell density and more organized microstructure typically found in benign prostate tissue. Parameters derived from ADC values serve as proxies for tumor invasiveness29,30,31,32,33. Additionally, ADC and T2WI have been shown to correlate negatively with the percentage of nuclear or cytoplasmic area measured in histological prostate tissue specimens, and positively correlated with the percentage of lumen space32. As Gleason scores increase, glandular architectural disorganization leads to more chaotic and fragmented lumens34. Consequently, mpMRI integrates morphological, metabolic, and vascular characterization, demonstrating its potential for prostate cancer detection. Based on these fundamental principles, our study was designed and executed using both T2WI and ADC sequences as key imaging modalities.
In this study, we found that the ADC sequence had better diagnostic efficacy compared with the T2WI sequence, consistent with the findings of Hou et al.35. The ADC sequence, based on DWI, accurately measures water molecule diffusion and is sensitive to both intracellular and extracellular water movement. This reflects the microstructural characteristics of tissues and the spatial distribution of water molecules. Conversely, T2WI relies on T2 relaxation time measurements, which assess transverse relaxation characteristics of water molecules. This provides information on the velocity and extent of free water diffusion within tissues. Consequently, the ADC sequence demonstrates higher sensitivity and specificity compared to T2WI for diagnostic purposes.
In our study, the AUC of the random forest model using only the ADC sequence was 0.869, comparable to the AUC of 0.863 reported by Hou et al. for their model employing the same ADC sequence30. Furthermore, we constructed a random forest model integrating ADC sequences combined with TAP and additional clinical features, achieving an improved AUC of 0.915. Interestingly, Hou et al. employed a similar model combining ADC sequences with clinical features, achieving an AUC of 0.912. This indicates that our study comprehensively analyzed clinical and radiomic features while applying optimized machine learning methodologies to construct the predictive model. Importantly, the inclusion of TAP in our model highlights its incremental diagnostic value for differentiating prostate cancer from benign conditions.
However, this study has several limitations. First, the study employed a retrospective design with a single-center dataset, lacking external validation. Second, the small sample size may introduce selection bias and limit the generalizability of results. Clinicians’ assessments of potential lesions depend on individual clinical expertise and experience, introducing subjectivity that leads to variability in interpretations and discrepancies among practitioners. In future studies, it is necessary to address the performance variability, robustness, and reproducibility of radiomics-based models more transparently to enable widespread clinical applications. Large-scale multicenter trials are essential to minimize selection bias and improve model accuracy. Additionally, radiomics-based prognostic assessments for PCa patients should be prioritized to refine clinical decision-making and tailor adjuvant therapies. Compared to existing radiomics studies, our research demonstrates the following methodological advantages.
Firstly, many radiomics studies use nomograms to construct visualizable models36. However, these approaches often lack the computational efficiency to handle large-scale data and complex machine learning operations efficiently. Secondly, compared to Bayesian and K-nearest neighbors (KNN) algorithms, random forest models demonstrate superior performance in handling high-dimensional and imbalanced datasets. Additionally, our study provides a unique visualization of the random forest model’s decision-making process, enhancing transparency and interpretability.
In summary, our study demonstrates that a multimodal model integrating radiomics features, TAP and clinical characteristics presents potential for distinguishing benign from malignant prostate lesions. Our findings underscore the efficacy of mpMRI combined with innovative imageomics methodologies and clinical data for the identification of prostate cancer. Furthermore, integrating automated machine learning into mpMRI frameworks provides a comprehensive approach to enhance cancer detection sensitivity and refine risk stratification strategies.
Data availability
All data generated or analysed during this study are included in this published article.
Abbreviations
- MRI:
-
Magnetic resonance imaging
- mpMRI:
-
Multi-parametric magnetic resonance imaging
- TAP:
-
Tumor abnormal protein
- T2WI:
-
T2-weighted imaging
- ADC:
-
Apparent diffusion coefficient
- ROC:
-
Receiver operating characteristic
- AUC:
-
The area under the curve
- PSA:
-
Prostate specific antigen
- PI-RADS v2.1:
-
Prostate imaging reporting and data system version 2.1
- DWI:
-
Diffusion-weighted imaging
- DCE:
-
Dynamic contrast-enhanced
- ROI:
-
Region of interest
- GLRLM:
-
Gray level run length matrix
- GLCM:
-
Gray level co-occurrence matrix
- GLSZM:
-
Gray level size zone matrix
- NGTDM:
-
Neighbourhood gray-tone difference matrix
- GLDM:
-
Gray level dependency matrix
- IBSI:
-
Initiative for biomarker standardization in medical imaging
- LASSO:
-
Least absolute shrinkage and selection operator
- SPSS:
-
Statistical product and service solutions
- PCa:
-
Prostatic carcinoma
- Cm:
-
Clinical factor model
- SEN:
-
Sensitivity
- SPE:
-
Specificity
- ACC:
-
Accuracy
- BPH:
-
Benign prostatic hyperplasia
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71(3), 209–249 (2021).
Xiang, J. et al. Transperineal versus transrectal prostate biopsy in the diagnosis of prostate cancer: A systematic review and meta-analysis. World J. Surg. Oncol. 17(1), 31 (2019).
Guo, L. H. et al. Comparison between ultrasound guided transperineal and transrectal prostate biopsy: A prospective, randomized, and controlled trial. Sci. Rep. 5, 16089 (2015).
Carter, H. B. Prostate-specific antigen (PSA) screening for prostate cancer: revisiting the evidence. Jama 319(18), 1866–1868 (2018).
Kim, C. S. et al. Report of the second Asian prostate cancer (A-CaP) study meeting. Prostate Int. 5(3), 95–103 (2017).
Liu, Z. et al. Tumor abnormal protein as a novel biomarker in papillary thyroid carcinoma. Clin. Lab. 63(3), 479–485 (2017).
Fu, L. et al. Clinical application of serum tumor abnormal protein in prostate cancer patients. BMC Cancer 24(1), 665 (2024).
Weinreb, J. C. et al. PI-RADS Prostate Imaging - Reporting and Data System: 2015, Version 2. Eur. Urol. 69(1), 16–40. (2016).
Movahedi, P. et al. Prediction of prostate cancer aggressiveness using (18)F-fluciclovine (FACBC) PET and multisequence multiparametric MRI. Sci. Rep. 10(1), 9407 (2020).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521(7553), 436–444 (2015).
Sherafatmandjoo, H. et al. Prostate cancer diagnosis based on multi-parametric MRI, clinical and pathological factors using deep learning. Sci. Rep. 14 (1), 14951 (2024).
Tran, K. A. et al. Deep learning in cancer diagnosis, prognosis and treatment selection. Genome Med. 13(1), 152 (2021).
Gandaglia, G. et al. Development and internal validation of a novel model to identify the candidates for extended pelvic lymph node dissection in prostate cancer. Eur. Urol. 72(4), 632–640 (2017).
Morlacco, A. et al. The incremental role of magnetic resonance imaging for prostate Cancer staging before radical prostatectomy. Eur. Urol. 71(5), 701–704 (2017).
Ahdoot, M. et al. MRI-targeted, systematic, and combined biopsy for prostate cancer diagnosis. N Engl. J. Med. 382(10), 917–928 (2020).
Cutaia, G. et al. Radiomics and prostate MRI: current role and future applications. J. Imaging 7(2) (2021).
Zwanenburg, A. et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 295(2), 328–338 (2020).
Nketiah, G. A. et al. Utility of T(2)-weighted MRI texture analysis in assessment of peripheral zone prostate cancer aggressiveness: A single-arm, multicenter study. Sci. Rep. 11(1), p2085 (2021).
Chiu, P. K. et al. Enhancement of prostate cancer diagnosis by machine learning techniques: An algorithm development and validation study. Prostate Cancer Prostatic Dis. 25(4), 672–676 (2022).
Donisi, L. et al. A combined radiomics and machine learning approach to distinguish clinically significant prostate lesions on a publicly available MRI dataset. J. Imaging 7(10) (2021).
Gu, C. et al. Quantification of tumor abnormal proteins in the diagnosis and postoperative prognostic evaluation of gastric Cancer. Clin. Med. Insights Oncol. 16, 11795549221104440 (2022).
Zhang, L. et al. Tumor abnormal protein (TAP) examination contributes to primary diagnosis of bladder cancer. Int. J. Clin. Exp. Med. 8(10), 18528–18532 (2015).
Schelb, P. et al. Classification of cancer at prostate MRI: deep learning versus clinical PI-RADS assessment. Radiology 293(3), 607–617 (2019).
Dominguez, I. et al. MRI-Based surrogate imaging markers of aggressiveness in prostate cancer: development of a machine learning model based on radiomic features. Diagnostics (Basel) 13(17) (2023).
Fan, Y. H. et al. The prostate health index aids multi-parametric MRI in diagnosing significant prostate cancer. Sci. Rep. 11(1), 1286 (2021).
Shiradkar, R. et al. Radiomics based targeted radiotherapy planning (Rad-TRaP): A computational framework for prostate cancer treatment planning with MRI. Radiat. Oncol. 11(1), 148 (2016).
Zhang, L. et al. Development and validation of a multiparametric MRI-based radiomics signature for distinguishing between indolent and aggressive prostate cancer. Br. J. Radiol. 95(1131), 20210191 (2022).
Cuocolo, R. et al. Clinically significant prostate cancer detection on MRI: A radiomic shape features study. Eur. J. Radiol. 116, 144–149 (2019).
Wu, R. C. et al. Role of MRI for the detection of prostate cancer. World J. Urol. 39(3), 637–649 (2021).
Demirel, H. C. & Davis, J. W. Multiparametric magnetic resonance imaging: overview of the technique, clinical applications in prostate biopsy and future directions. Turk. J. Urol. 44(2), 93–102 (2018).
Saito, S. et al. Relationship between apparent diffusion coefficient distribution and cancer grade in prostate cancer and benign prostatic hyperplasia. Diagnostics (Basel) 12(2) (2022).
Langer, D. L. et al. Prostate tissue composition and MR measurements: investigating the relationships between ADC, T2, K(trans), v(e), and corresponding histologic features. Radiology 255(2), 485–494 (2010).
Agrotis, G. et al. Diagnostic performance of ADC and adcratio in MRI-based prostate cancer assessment: A systematic review and meta-analysis. Eur. Radiol. 35(1), 404–416 (2025).
Epstein, J. I. An update of the Gleason grading system. J. Urol. 183(2), 433–440 (2010).
He, D. et al. MRI-based radiomics models to assess prostate cancer, extracapsular extension and positive surgical margins. Cancer Imaging 21(1), 46 (2021).
Zhang, H. et al. Diagnostic nomogram based on intralesional and perilesional radiomics features and clinical factors of clinically significant prostate cancer. J. Magn. Reson. Imaging 53(5), 1550–1558 (2021).
Acknowledgements
Not applicable.
Funding
This research was funded by the National Natural Science Foundation of China (No. 81802572 to Yachen Zang) and Gusu Talent Program (GSWS2024023 to Yachen Zang).
Author information
Authors and Affiliations
Contributions
Z.Y., Y.M. and C.Z. concepted and designed the study; Data collection was done by Z.C. and W.Z.; Data analysis was done by C.Z. and Z.W.; Z.C., W.Z. and S.P. drafted the manuscript; Z.Y., Z.J., X.L. and Z.Y. revised the manuscript; All authors were involved in data visualization.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
This study was approved by the Ethics and Research Committee of the second affiliated hospital of Soochow university. Informed consent was obtained from all the patients in the study, and all the procedures were in accordance with the principles of the Declaration of Helsinki.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, C., Wang, Z., Shang, P. et al. Combining multi-parametric MRI radiomics features with tumor abnormal protein to construct a machine learning-based predictive model for prostate cancer. Sci Rep 15, 22816 (2025). https://doi.org/10.1038/s41598-025-05718-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-05718-2