Abstract
The ongoing biodiversity crisis calls for a complete biodiversity inventory of marine and terrestrial ecosystems. The task is particularly challenging for fragmented island territories, where baseline biodiversity information is often difficult to procure. By centralising information from different sources (museums, research institutions, citizen scientists), ‘big-data’ platforms provide an opportunity to evaluate species biodiversity information of understudied regions. Using data primarily sourced from the Global Biodiversity Information Facility (GBIF), and complemented by a review of 56 potential data sources—of which nine provided unique, non-redundant records—we curated the first biogeographic dataset for both marine and terrestrial animal species in French Polynesia, a large territory composed of 124 islands and atolls that belongs to the Central Pacific region, a marine biodiversity hotspot facing conservation challenges. The dataset revealed heterogeneous species richness across archipelagos and islands, prompting an investigation into potential sampling biases (institutional, taxonomic, spatial) as well as an assessment of island-specific accessibility biases. We estimated that the archipelagos and islands had an inventory completeness rate that ranges from 1.9 to 98.4%, suggesting that a large proportion of the studied area remains poorly documented. Spatial and temporal sampling biases were partly explained by accessibility constraints (proximity to airports, roads or ports), and inventory completeness was higher for marine than terrestrial species. The biases quantified here challenge our ability to conduct biogeographic analyses that integrate the land-sea meta-ecosystem. Our database allows identifying taxa and sampling locations that require urgent attention, as well as comprehensively recorded species that can serve as indicators for environmental degradation. Explicitly acknowledging the inherent biases of biodiversity datasets is the first step towards a more comprehensive characterization of species diversity across fragmented territories. This information is crucial for guiding sound adaptive-management and conservation planning strategies.
Similar content being viewed by others
Introduction
Humans are driving an unprecedented erosion of marine and terrestrial biodiversity, fundamentally altering the structure and functioning of ecosystems, and in return threatening the beneficial contributions that nature provides1,2,3. Implementing conservation actions to confront this crisis requires comprehensive and spatially explicit baseline information on species diversity across the planet4. Ultimately, these data are essential for guiding conservation management based on a sound understanding of the ecological and evolutionary processes that drive spatial and temporal patterns of species distribution across ecosystems5,6.
Thanks to the concerted efforts from museums, research institutions, citizen scientists, and ‘big-data’ platforms facilitating the integration of information, biodiversity records are increasingly available7,8,9. Over the last two decades, many initiatives to centralise species occurrence data have emerged, notably some online repositories including the Global Biodiversity Information Facility (GBIF, https://www.gbif.org/) and the Ocean Biodiversity Information System (OBIS, https://obis.org/). By adhering to the FAIR principles (Findability, Accessibility, Interoperability, and Reusability) and the metadata-sharing standards, such as the Darwin Core (DwC)10, Ecological Metadata Language (EML)11, and BioCASe12, these intergovernmental research infrastructures promise to expedite the study of biodiversity across ecosystems. GBIF and OBIS are the largest open-access occurrence data portals for terrestrial and marine species, both being routinely used to inform resource management and conservation programs (e.g13,14,15,16,17).
Despite their growing popularity, open-access biodiversity databases have been criticised on the grounds of poor data quality, potentially limiting their scope and applicability18. Important shortfalls that are often cited include standardisation issues during sampling19,20, incomplete and/or incorrect records (e.g., species misidentification) and sampling biases, either spatial/temporal (i.e., unbalanced sampling efforts across space/time), taxonomic (i.e., skewed sampling favouring certain taxa), or both20,21,22,23. While cleaning and filtering methods allow readily correcting for incomplete and/or incorrect entries, sampling biases are difficult to diagnose and require special attention24. The spatial sampling bias, considered one of the main challenges limiting our comprehensive understanding of large-scale biodiversity patterns25, can be partly explained by socio-economic reasons (e.g., wealthy zones are more likely to be surveyed26), a scientific bias towards certain taxa21, differences in sampling standards27, and/or by logistical difficulties to access certain locations28,29. Notably, implementing standardized sampling methods in future research is essential, as it enhances comparability and integration, increases the reproducibility of findings, and improves data quality and reliability, while also saving time and resources30. These shared data collection protocols should be adopted and facilitated for both scientific and non-scientific personnel31.
Remote oceanic islands are likely to show sampling gaps due to their geographical isolation, which ultimately results in patchy and poorly representative data for the study region. The difficulties and high costs associated with organising monitoring campaigns further exacerbate these biases. As a result, some islands are underrepresented in long-term monitoring schemes32 and, aside from a few exceptions (e.g.,33), comprehensive biodiversity studies across widespread archipelagos remain rare. This paucity of information for islands and atolls is particularly detrimental because they are a priori highly vulnerable ecosystems that potentially harbour high levels of endemism due to their isolation34,35. Additionally, fragmented archipelagos are unique natural laboratories that provide opportunities for studying the ecological and evolutionary processes driving biodiversity patterns, dispersal potential, endemism and extinction rates, for both marine and terrestrial organisms. However, a proper understanding of these biogeographical processes first requires robust baseline information on species distribution36,37.
With 124 high islands and atolls spread across five archipelagos covering 4.8 million km238,39, French Polynesia represents the epitome of a fragmented territory. The large number of islands, their relative isolation, and the sheer variation in geomorphological characteristics they exhibit complicate efforts to survey the entire region or avoid sampling biases. Indeed, the marine and terrestrial biogeography of French Polynesia has only been partly studied, with a remarkable skew towards specific taxonomic groups. In the marine realm, targeted investigations have mainly focused on marine molluscs, brown seaweeds (Phaeophyceae), and reef fishes40,41,42,43,44,45,46,47,48. In the terrestrial realm, data compilations include a checklist of the recorded land and fresh-water arthropods49, a biogeographic atlas of birds50, and an inventory of the vascular flora51,52,53, as well as some rare studies focusing on the phylogeographic origins of specific terrestrial biota (e.g.,54,55). Overall, the lack of a centralised, complete, and unbiased dataset for the region prevents an exhaustive analysis of the biogeographical status of marine and terrestrial species across French Polynesia. As a model of a highly fragmented island system, improving our fundamental understanding of French Polynesian biogeography is not only critical for cataloguing the existing fauna of the region, but also for contributing to our general comprehension of the ecological processes driving the current biodiversity crisis in isolated systems35,56.
Using data originally downloaded from open-access portals (GBIF, OBIS) and 56 additional occurrence data sources, we compiled and curated the first biogeographic dataset for both marine and terrestrial animal species in French Polynesia. We used these data to: (1) provide a baseline characterization of the number of species in the region; (2) identify taxonomic groups that might require further investigation, as well as comprehensively recorded species that can serve as indicators for environmental degradation; (3) identify poorly- and well-surveyed islands; and (4) quantify island-specific accessibility biases leading to heterogeneous sampling efforts.
Materials and methods
Data collection
We downloaded occurrence data from the GBIF portal (http//gbif.org; https://doi.org/10.15468/dl.gaxgr7) on May 24, 2023, covering French Polynesia (polygon spanning between 5°S and 30°S, and 134°W and 155°W). To identify any additional biodiversity records not included in GBIF, we reviewed 56 data sources, including local reference guides, expedition reports, and repositories (Table S2). Nine sources contained unique georeferenced species records that were integrated into our final dataset. The remaining sources were used to cross-check metadata, validate completeness, or support contextual interpretation. Species occurrences are defined as records of a particular species (or other taxonomic rank), with a geographic location and timestamp. These raw data were treated following the Darwin Core10, Ecological Metadata Language11, and BioCASe12 standards. A pre-filtration of the data was done to exclude records missing geographic location and/or taxonomic classification (e.g., not available or zeros), yielding 343,780 records (Fig. 1). Because GBIF and OBIS signed a data-sharing agreement which was effective at the time we downloaded the data, the marine data from OBIS was also contained in our GBIF data. The coastline shapefiles used to analyse the region included 120 geographical structures, most of which were atolls and high islands. Hereafter, we refer to all geographical structures as “islands”. Each record retrieved from the GBIF dataset was assigned to its nearest island based on geographic distances estimated using the function st_nearest_feature available in the sf package v.1.0-1557 in R58.
Sankey diagram illustrating the data filtering and quality-control steps. To obtain the final marine, terrestrial and mixed-habitat animal dataset, we downloaded data from GBIF/OBIS data portals and from additional occurrence data sources (Table S2), we removed: records earlier than 1950 (20,967 occurrences), absence data (< 1000 occurrences), occurrences based on invalid recording methods (< 1000 occurrences), invalid taxonomical information (58,490 occurrences), and true duplicates (77,651 occurrences). Finally, data with unavailable habitats were excluded from subsequent analyses (< 1000 occurrences). The cleaned dataset is available in SEANOE (https://www.seanoe.org/data/00878/99018/).
Validation of the taxonomic information
To clean, homogenise, and validate the taxonomic information in the dataset, we assumed that misidentifications would occur at the species level. To ensure taxonomic reliability we validated each species name using ad hoc taxonomic data repositories. We first validated the species name of each recognized taxon with WoRMS (World Register of Marine Species, https://www.marinespecies.org/) using the wm_records_name function from the R package worrms59. We then assigned a taxonomic status (i.e., “accepted”, “doubtful”, “synonym”) to each record following the criteria outlined by the GBIF Backbone taxonomy (see https://doi.org/10.15468/39omei, https://hosted-datasets.gbif.org/datasets/backbone/). Taxa that were assigned as either “doubtful” or “synonym” were replaced by the updated taxonomic name provided by WoRMS. Taxa not recognized by the WoRMS repository were further examined using the gna_verifier function from the R package taxize60, which provides a means to validate species names by accessing several additional repositories via specific Application Programming Interfaces (e.g., ITIS: Integrated Taxonomic Information System, https://www.itis.gov/; CoL: Catalogue of Life, https://www.catalogueoflife.org/; bold: Barcode of Living Data, https://www.boldsystems.org/). Taxa that were not recognized neither by WoRMS nor Taxize were submitted to TAXREF (taxonomic reference curated by the French National Museum of Natural History, https://inpn.mnhn.fr/programme/referentiel-taxonomique-taxref) using the rt_taxa_search function from the rtaxref R package61. A final manual check was done for records that could not be identified in the aforementioned taxonomic repositories.
Habitat classification and biogeographical status
Habitat classification for marine and terrestrial species were verified using WoRMS and Taxref, respectively. Habitat information was split into four categories (i.e., marine, brackish, freshwater and terrestrial) according to the classification scheme favoured by WoRMS. Missing habitat information was completed using the TaxRef database. For our analyses of terrestrial and marine ecosystems, we focused on species that were classified as exclusively “marine” or exclusively “terrestrial”. Species classified as amphibious or those inhabiting both terrestrial and marine environments at different life stages (e.g., seabirds like Gygis alba or Sula sula) or during specific phases of their life cycle (e.g., insects with aquatic larval stages) were included in the cleaned dataset (labeled as “Mixed” in Fig. 1) but excluded from further analyses.
Data filtration sequence
Because geographic, taxonomic and accuracy standards have changed over time20,62, and notorious errors were detected in older records, we retained entries dating from 1950 onwards and excluded those without timestamps (Fig. 1). Subsequently, we removed all absence data to rule out potential biases due to false negatives and no-observation data63. We then restricted occurrences to those described in the basisOfRecords column as: “human observation”, “machine observation”, “material sample”, “material citation” and “preserved specimen” according to recommendations by Smith et al. 201864. Cross-checking values corrected real duplicates in decimalLatitude, decimalLongitude, ScientificName, Year, Month and Day categories. Finally, records lacking species names and Habitat information were removed from the dataset (Fig. 1).
Taxonomic biases: identifying under- and over-represented groups
We estimated the taxonomic bias at the Class level based on its over- or under-representation, relative to an “ideal sampling effort index”. The ideal number of records for a given class was estimated based on the hypothetical scenario where each species received the same number of records, and therefore each class received a number of records directly proportional to its number of species21 according to:
Ideal = Nrec * (Nsp_group/ Nsp_tot)
where Nrec = total number of records, Nsp_group = number of distinct species within the taxonomic group, and Nsp_tot = total number of species present in the whole dataset. Taxonomic bias was assessed based on the difference between the ideal and observed sampling efforts, calculated for each class with more than 100 records in marine habitats and more than ten records in terrestrial habitats. To highlight values that deviated significantly from the ideal, we applied an inverse hyperbolic sine transformation to the data. We also identified the top ten most representative species for each habitat.
Spatial and temporal heterogeneity in the sampling effort
To examine spatial heterogeneity in the sampling effort, we first mapped the number of records and species estimated for each island within each archipelago. The agreement between the number of records and species per island (log10-transformed) was evaluated based on the Pearson correlation coefficients and its statistical significance. Additionally, to quantify the prevalence of heterogeneous sampling effort across space, we assessed the proportion of species recorded only once at each island, a common method in biodiversity studies to evaluate sampling completeness and detect potential under-sampling biases65. The parameter uniqueness—species that have only been collected once—is widely recognized as an indicator of incomplete sampling66,67, allowing researchers to infer the adequacy of the sampling effort and identify areas that may require further investigation. We considered qk as the number of species documented in k sampling-effort units, so that the number of species observed in a single sampling-effort unit is q1 (i.e., unique), the number of duplicates is q2, and so on.
Inventory completeness
To investigate the degree of inventory completeness in the dataset for both marine and terrestrial ecosystems at the scale of the archipelago and the island, we estimated the species inventory completeness percentage (C), calculated as:
C(i) = (Sobs(i)/ Sest(i)) * 100
where i = each island or archipelago, Sobs = number of species observed, and Sest = number of species estimated at each archipelago and island68. To estimate Sest, we used the Species Accumulation Curve (SAC) approach that describes the relationship between species richness and sampling effort, i.e., the number of records available in a grid cell69. To derive the SACs, we split the area in 0.05° (~ 25 km2) grid cells. We described the SACs using the specaccuml function (method = “exact”) available in the R package vegan v.2.6–470. We fitted the Michaelis-Menten model with the fitspecaccum function (method = “michaelis-menten”) to provide estimates of the number of species likely to be present (i.e., Sest, which corresponds to the asymptotic richness, or parameter Vm in the Michaelis-Menten equation) and the number of records required to capture 50% (K) of the estimated number of species predicted by the model71,72,73. Using the poolspecaccum function available in the vegan package, we also compared this expected number of species with the nonparametric richness estimators, Chao1 and Jackknife 2 estimators that are recommended when the data contain a high number of unseen species74. Because biodiversity assessments can be biased by grid cells with extremely low species records, we considered a minimum threshold of ten observations to run the SACs, as was done in previous studies68,75.
While we aimed to calculate the SACs for each island and archipelago based on grid cells, as recently done in several studies on macroecology using GBIF datasets75,76,77, only eight islands (i.e., Anaa, Huahine, Moorea, Nuku Hiva, Raiatea-Tahaa, Rangiroa, Tahiti and Ua Pou) were sufficiently large to yield enough cells (greater or equal to 10) to fit the Michaelis-Menten model for terrestrial data. We therefore generated archipelago-scale models based on 5-km grid cells, while for the island-scale models we used the geographic coordinates associated with the species records. A preliminary comparison between these two approaches (0.05-degree resolution grid cells vs. records) revealed a significant correlation between them for the archipelago scale (R2 = 0.96). Therefore, we only presented SACs based on records for both archipelagos and islands. To evaluate inventory completeness, we determined the total number of islands with more than 100 records and C greater or equal to 80%, meaning that at least 80% of the species have been sampled77,78. We then examined the correlation between the number of records and C to test whether these proxies of sampling effort and reliability were associated. We used a Spearman correlation test for non-parametric data. Statistical significance was evaluated based on ⍺ = 0.05.
Sampling bias due to accessibility
To explore the influence of accessibility constraints on these sampling biases, we used a Bayesian approach to estimate how sampling rates vary with proximity to several common anthropic accessibility factors (i.e., rivers, roads, cities, airports, and ports). Using the calculate_bias function from the sampbias R package v. 2.0.079, we estimated the bias weights (w), which quantify the impact of each accessibility factor on sampling rates. These weights are calculated assuming an exponential decline in sampling rates as distance from accessibility factors increases. This package also provides spatially explicit estimates of the number of records (i.e., expected records) using a Poisson sampling process while accounting for the influence of the accessibility factors. Because the geospatial data contained by default in the sampbias package is incomplete for French Polynesia (Natural Earth Data, https://www.naturalearthdata.com/), we manually inputted vector data for rivers, roads, cities with > 1,000 inhabitants, airports, and ports. These data were provided by the French Polynesian agency for marine resources, the Direction des Ressources Marines. We defined a grid (inp_raster parameter) contained within the same polygon used for downloading the GBIF data, with 0.05 degrees resolution (~ 5.5 km). This was done for consistency with the SAC analyses. Each grid cell was assigned to the nearest island based on geographic distances estimated using the function st_nearest_feature available in the sf R package v.1.0-1557.
Results
Curated dataset for French Polynesian marine and terrestrial species
From the original 343,780 records included in the dataset, we removed 20,967 records that were either dated before 1950, or which did not have a time stamp (Fig. 1). Then, 58,490 records with no or non-usable species taxonomy were discarded, 86.6% of which sourced from institutions sources (e.g., Museum national d’Histoire naturelle, Smithsonian Institution). A total of 77,651 records were identified as duplicated, 21.5% of which originated from citizen science sources. The number of records accessible per year has increased over time since 1950, reaching maximum values in 2011, 2012, and 2006, with 20,636, 11,263, and 11,039 records, respectively (Fig. S1). This increase in records was mainly explained by the punctual contribution of two out of 130 publishers: OBIS-SEAMAP and UMS PatriNat (OFB-CNRS-MNHN, Paris). The mean number of records per species was 25.8 (median = 3), ranging from 1 to 12,339. Records produced by citizen scientists accounted for 21.7% of the total, corresponding to 40,394 records. Data collected by citizen scientists were also the main source of data (i.e., > 50% of occurrences sourced as citizen science) for 62 islands, and the only source (100%) for two islands (Fangatau, Marutea nord). Human observation, including institutional and citizen science publishers, was the most frequently used recording method, with 75.7% (140,650 records) of total records. Preserved specimen and material sample categories accounted for 20.1% and 3.9% of records, respectively. WoRMS validated the taxonomy of 90.6% of total records and 99.1% of non-terrestrial records. Only 268 species lacked information on their habitat, which we completed manually. The resulting cleaned dataset was composed of 185,758 records including 141,181 marine, 15,940 terrestrial and 28,637 mixed records for 5,953 marine, 1,032 terrestrial and 203 mixed species, collected from 1950 to 2023 (Figs. 1 and S1). The curated dataset is available in SEANOE (https://www.seanoe.org/data/00878/99018/).
Taxonomic composition and biases
The number of recorded species was ~ 5.8 times higher for marine than terrestrial ecosystems, with 5,953 marine and 1,032 terrestrial species, respectively. For marine taxa, the dataset included 18 phyla, with three major groups: Mollusca (2,337 species), Chordata (1,733 species), and Arthropoda (1,148 species), accounting for over 95.4% of marine records (134,734 records). Five classes alone accounted for 78.4% of the observations: Teleostei (76,248 records, 1,547 species), Gastropoda (20,446 records, 2,028 species), Malacostraca (6,788 records, 1,076 species), Bivalvia (4,368 records, 276 species), and Mammalia (2,807 records, 25 species; Fig. 2). The most represented marine species were Carcharhinus amblyrhynchos (grey reef shark, 4,428 records), Carcharhinus melanopterus (blacktip reef shark, 3,987 records), and Triaenodon obesus (whitetip reef shark, 941 records; Fig. 2). A total of 90% of the marine species had 33 or fewer records, and 26% were unique records.
Taxonomic bias assessment. (A) Major class representation in sampling effort (i.e., observed - ideal). Over- and under-representation of each class are illustrated by the green and orange bars respectively. An inverse hyperbolic-sine transformation was used for the x-axis. (B) Number of records for the top 10 most-sampled marine (left) and terrestrial (right) species.
The terrestrial taxa comprised five phyla, including Arthropods (761 species), Mollusca (194 species), Chordata (73 species), Platyhelminthes (3 species), and Nematoda (1 species). The five most recorded classes were Aves (9,061 records, 62 species), Insecta (3,545 records, 687 species), Gastropoda (2,198 records, 194 species), Arachnida (532 records, 62 species), and Squamata (430 records, 10 species), representing 98.9% of all terrestrial species records. A total of 90% of the terrestrial species had 23 records or fewer, and 41% were unique records. Three introduced bird species, Geopelia striata (zebra dove), Acridotheres tristis (common myna), Pycnonotus cafer (red-vented bulbul), were the most recorded terrestrial species, with 1,174, 1,140 and 957 occurrences (Fig. 2), of which 93.1% were provided by the “Cornell Lab of Ornithology”.
Spatial and temporal heterogeneity in sampling effort and the number of recorded species
We observed a significant and strong correlation between the log-10 number of records per island (i.e., a proxy for sampling effort) and the number of species per island for both marine (ρ = 0.984, P < 0.001) and terrestrial (ρ = 0.969, P < 0.001) ecosystems (Fig. S2). This analysis excluded islands that lacked records in both marine and terrestrial habitats.
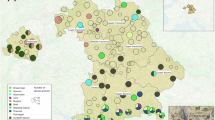
Our dataset included marine species records for 118 out of 124 islands. The number of records per island was heterogeneous (Fig. 3), ranging from 1 to 60,473, with a mean of 1,196 records (median = 77). The number of species present was also highly heterogeneous across space, ranging from 1 to 2,770 species per island, with a mean of 199 species (median = 58) per island. The Society Archipelago (13 islands) held 57.1% of all marine-species records, 75.0% of which were observed in Moorea (60,473 records), Tahiti (9,920 records), and Raiatea-Tahaa (4,213 records; Fig. 4). Considering the other four archipelagos, the islands that exhibited the highest number of records were Rapa (5,797 records) in the Austral islands (11 islands), Fakarava (6,585 records) in the Tuamotu (69 islands), Nuka Hiva (3,356 records) in the Marquesas (17 islands), and Mangareva (2,504 records) in the Gambier (11 islands; Fig. 3). Gambier was the least sampled archipelago, accounting for 2.9% of all marine records, and for only 13.3% of all marine species identified.
Considering the terrestrial habitat, our dataset identified 68 islands with at least one species record, and 52 islands with no records. As for the marine habitat, the number of terrestrial species records per island was heterogeneous (Fig. 3), ranging from 1 to 4,705, with a mean of 234 records per island (median = 8.5). The number of species identified per island ranged from 1 to 384, with a mean of 34 species per island (median = 4.5). The Society Archipelago held 74.4% of all terrestrial species records, 85.2% of which were registered in the trio Moorea (4,314 records, 384 species), Tahiti (4,705 records, 301 species), and Raiatea-Tahaa (1,076 records, 201 species; Fig. 4). Considering the other four archipelagos, the islands showing the highest number of records were Anaa (683 records) in the Tuamotu, Rurutu (441 records) in the Austral, Nuku Hiva (548 records) in the Marquesas, and Mangareva (123 records) in the Gambier (Fig. 4). As for the marine database, the Gambier archipelago had the lowest number of terrestrial records, representing only 4.7% of all terrestrial species identified.
Inventory completeness
Considering the archipelago scale, the SAC analysis showed that the number of species recorded increased with sampling effort. Although the curves for both marine and terrestrial datasets exhibited a plateau, they did not reach a clear saturation point (Fig. S3). Our calculations suggest that marine inventory completeness was comparable among archipelagos, with 76.6%, 74.9%, 75.6%, 79.9% and 76.6% for the Austral, Gambier, Marquesas, Society and Tuamotu Archipelagos, respectively, indicating that at least 70% of the species were detected overall. According to the asymptote values based on the Michaelis-Menten model (Sest), marine species richness was lowest at the Gambier (Sest = 1,055, Chao1 = 1,221, Jackknife 2 = 1,317 species) and the highest at the Society (Sest = 5,230, Chao1 = 6380, Jackknife 2 = 6,969 species) archipelagos. The Austral, Marquesas and Tuamotu Archipelagos showed similar asymptote values of 2,538 (Chao1 = 3,067, Jackknife 2 = 3,291), 2,648 (Chao 1 = 3,497, Jackknife 2 = 2,120), and 2,120 (Chao1 = 2,639, Jackknife 2 = 2,807) expected species, respectively (Table 1).
Inventory completeness for terrestrial species was highly heterogeneous across archipelagos, ranging from 43.0% for the Marquesas, the northernmost and most remote archipelago, to 82.1% in the Society. Inventory completeness for terrestrial species was higher than for marine species in the Society (C = 82.1% versus 79.9%) and Tuamotu Archipelagos (C = 81.5% versus 76.6%), but lower in the Gambier (C = 64.1% versus 74.9%) and Marquesas (C = 43.0% versus 75.6%). For terrestrial species, the asymptote values based on the Michaelis-Menten model (Sest) ranged from 76 (Gambier, Chao1 = 73, Jackknife 2 = 84) to 640 species (Marquesas, Chao 1 = 639, Jackknife 2 = 609), with 484 (Chao 1 = 595, Jackknife = 578), 606 (Chao 1 = 894, Jackknife = 981), and 100 (Chao 1 = 152, Jackknife = 165) species estimated for the Austral islands, Society and Tuamotu, respectively.
At the island scale and for the marine dataset, we fitted SACs for 73 out of 119 islands having at least 10 records (Table 2). Inventory completeness was highly heterogeneous, ranging from 1.9% (Takaroa, Tuamotus, 12 records) to 82.2% (Moorea, Society, 2,826 records), with an average (± SD) of 39.2% (± 20.0%). Assuming a threshold of C ≥ 80% and at least 100 records, only two islands were classified as well-sampled: Moorea (2,826 records, C = 82.2%) and Fakarava (520 records, C = 82.0%). Among the islands with the highest number of records, we identified low to moderate inventory completeness for Tahiti (1,226 records, C = 65.9%), Bora-Bora (521 records, 60.8%) and Raiatea-Tahaa (263 records, C = 28.5%) in the Society, Rapa (385 records, C = 75.6%) and Raivavae (352 records, C = 69.1%) in the Austral Islands, Nuku Hiva (376 records, C = 65.8%) and Hiva Oa (234 records, C = 68.6%) in the Marquesas, Tikehau (360 records, C = 62.6%) and Rangiroa (343 records, C = 52.0%) in the Tuamotu. The correlation between inventory completeness and the number of records per island was moderate (R² = 0.41; P-value < 0.001).
For the terrestrial dataset, 27 islands had sufficient records (> 10 records) to fit SACs (Table 3). Inventory completeness ranged from 27.3% for the Fakarava Atoll (12 records, Tuamotu) to 98.4% for Anaa Atoll (67 records, Tuamotu). Other well-sampled islands (C ≥ 80% and 1 records) included Ua Huka (39 records, C = 87.5%), Hatutaa (23 records, C = 88.7%) and Tahuata (25 records, C = 88.4%) in the Marquesas, Tenararo (26 records, C = 97.5%) in the Tuamotu. Islands with the highest number of records, including Moorea (851 records, C = 76.8%) and Tahiti (1,039 records, C = 68.9%, respectively) were nearly well-sampled. Terrestrial species inventory completeness and sampling effort were not correlated across these islands (R² = 0.20; P-value > 0.05).
Some islands exhibited contrasting patterns between terrestrial and marine inventories. For instance, Anaa was well sampled for terrestrial species (67 records, C = 98.4%) but only moderately sampled for marine species (31 records, C = 17.6%). Fakarava showed the opposite trend, with an almost complete marine inventory (C = 83.20%), while its terrestrial inventory was sparse (12 records, C = 27.3%). Overall, our results indicate that the observed species richness exceeds the Michaelis-Menten estimate by a factor of one to three, suggesting that even our current estimates likely underestimate the true species diversity. This reinforces the notion that species inventories in this region remain incomplete.
Islands for which we were unable to fit SACs were classified as either “neglected islands” (i.e., with no data at all) or “poorly-documented islands” (i.e., with not enough data). For the marine and terrestrial data, we identified three and 52 neglected islands, respectively. The problem of missing data was prevalent across archipelagos, but less important in the Society and Austral archipelagos (Fig. 5). We found 34 and 36 poorly-documented islands for marine and terrestrial ecosystems, respectively. The data scarcity was particularly pronounced in the largest archipelago, the Tuamotu, as well as in the southernmost archipelago, the Gambier (Fig. S4).
Sampling bias due to human accessibility
Sampling effort for marine species was primarily influenced by proximity to roads (w = 0.063), indicating a strong spatial bias towards areas with developed infrastructure. Airports had a moderate effect on sampling distribution (w = 0.008). In contrast, proximity to cities (w = 0.001) and waterbodies (w = 0.0005) had negligible impacts on sampling intensity (Fig. 6; Table S1).
Accessibility bias. Results from the calculate_bias function (Sampbias package) which estimates marine (a) and terrestrial (b) expected occurrences based on accessibility, illustrating the impact of various infrastructure types on species sampling efforts. (Left) The posterior weight shows the relative importance of anthropic accessibility factors: roads, ports/airports, cities, and waterbodies—on sampling efforts in both ecosystems. (Right) The sampling rate, i.e., the expected number of occurrences, as a function of distance from accessibility factors is depicted for marine (a) and terrestrial (b) environments.
Similar results were found for the terrestrial data, where the presence of roads contributed the most to the accessibility bias (w = 0.060). The effect of airports and ports was moderate (w = 0.031) while the influence of cities and water bodies was negligible (cities’ w = 0.004, waterbodies’ w = 0.001) (Fig. 6). The model also revealed a low number of marine and terrestrial records (Table S1), even after correcting for accessibility biases, in the Tuamotu and Gambier Archipelagos, except for Mangareva, Hao, and Arutua Islands. In contrast, most islands in the Society Archipelago were oversampled relative to the overall sampling effort across French Polynesia.
Discussion
Our study compiles the most comprehensive open-source database on animal biodiversity in French Polynesia, illuminating regional- and island-scale biodiversity patterns of marine and terrestrial fauna across this vast and fragmented territory. While our results highlight significant disparities in sampling effort across islands, this work offers valuable quantitative insights into completeness of taxonomic and spatial data throughout French Polynesia. This work also highlights understudied areas and taxonomic groups, providing a practical tool for conservation planners to guide future sampling strategies and enhance biodiversity representativeness. We argue that this integrative approach is essential for explicitly addressing the inherent biases often present in large-scale biodiversity studies23.
Building an accurate open-source biodiversity dataset
While open-source biodiversity datasets offer unique opportunities for studying macroecological processes, global repositories face criticism due to significant variation in data quality and quantity, depending on geographic, temporal, and taxonomic factors22. Ignoring these caveats can lead to erroneous conclusions. However, when carefully considered, they can enhance the utility of open-source data by highlighting critical biodiversity knowledge gaps (e.g80,81,82). Addressing uncertainties in the data first requires acknowledging that open-source biogeographic datasets are likely to be incomplete25, especially in vast and fragmented regions and for specific groups of organisms. Secondly, standardised taxonomic repositories (e.g., WORMS) offer workflows for cleaning data retrieved from open-source platforms while adhering to FAIR data-sharing principles. Here, by applying previously validated filtering protocols63, we enhanced the geographic and taxonomic accuracy of GBIF records for French Polynesia, closely matching recent expert taxonomic assessments.
Our database contains a total of 7,188 species, including 1,893 vertebrates and 5,295 invertebrates. Regarding vertebrates, we found that every known marine mammal (26 out of 26 species) and a large number of birds (126 out of 175 species) previously documented in the region are represented83. Our database includes 2,552 marine molluscs out of 3,022 referenced in a recently published checklist and identification guide46, and the Teleostei class included 1,547 species, which is more than the 1,310 reported in the most complete identification guides for the region84,85. While the taxonomic coverage is reassuring for marine species, it remains relatively limited for terrestrial species. For example, our records include only 757 out of 2,497 insect species (Insecta) and 63 out of 365 spider species (Arachnida) described in the region49. Data scarcity for insects is a global issue, and in some regions, it is partly driven by species extinction rates that outpace discovery rates86,87. Islands, which harbour approximately 20% of the world’s terrestrial biodiversity, are critical reservoirs of fragile and threatened biodiversity56. This highlights the urgent need to document the exceptional biodiversity of insular countries like French Polynesia, where some taxonomic groups, such as ground beetles, contribute significantly to global biodiversity56,88. Our study provides an efficient framework for identifying poorly sampled species, which can be extended to other taxonomic groups in French Polynesia (e.g., plants or algae) and applied more broadly to other regions.
Linnean shortfall
The Linnean shortfall—i.e., only a fraction of the planet’s species has been described—is a major gap in our understanding of biodiversity18, limiting our ability to effectively address the ongoing extinction crisis2. The Linnean shortfall is partly driven by taxonomic sampling biases, where societal preferences influence which groups are more frequently recorded21. This explains why patterns of sampling efforts are often represented by homogeneously-sampled taxonomic groups such as marine mammals82, fishes89 or insects90. Notably, our taxonomic bias analysis revealed a significant under-representation of non-charismatic invertebrate species such as Gastropoda, Malacostraca, Anthozoa, Bivalvia, Polychaeta in the marine environment, as well as Insecta, Gastropoda, Arachnida, Malacostraca, in terrestrial ecosystems. This finding aligns with Troudet et al. (2017)21, who also identified biases against these classes at the global scale. Conversely, vertebrates were well-represented, with the humpback whale (Megaptera novaeangliae) being one of the most frequently recorded species. This discrepancy often stems from the aesthetic appeal of certain species, which influences both public interest and scientific focus91,92,93. Furthermore, studies have effectively shown that visual appeal shapes the perception and prioritisation of species in research and conservation93. To address these biases and enhance biodiversity inventories in French Polynesia, our dataset can help guide future research priorities, focusing on the underrepresented invertebrates and terrestrial species identified. By addressing these gaps, we can move towards a more comprehensive and balanced understanding of biodiversity, which is crucial for developing effective conservation strategies.
Wallacean shortfall
Another significant gap in our understanding of biodiversity is the incomplete knowledge of species’ geographic distribution, also known as the Wallacean shortfall25,94. Despite extensive efforts, biodiversity sampling remains a resource-intensive, time-consuming and costly process, often resulting in substantial gaps in the spatial coverage of species records. Short-term projects frequently fail to capture the full spectrum of species within an assemblage because many species can be cryptic, rare or elusive, ultimately leading to incomplete assessments of global biodiversity patterns. However, these data gaps and uncertainties can be gauged and possibly mitigated through robust modelling approaches23. In our study, marine inventory completeness was consistently moderate across French Polynesia’s archipelagos, being up to 74% of known species at the regional scale. Furthermore, none of the species accumulation curves for the archipelagos reached saturation, indicating that species richness predictions require more sampling to improve accuracy. Statistical methods to correct these biases (e.g.,66), could be used for comparing community assemblages among archipelagos, as has been recently done with woody plants95. Another strategy is to focus on well-documented groups, with complete inventories, enabling the description of their spatial distribution patterns96.
For terrestrial species, we found that inventory completeness was more variable than that of marine species. The Marquesas Archipelago was especially under-surveyed, as only half of the total estimated animal species have been documented. Owing to their geographical isolation and intricate topography, the Marquesas Islands harbour a high level of floral and faunal endemism, with many native and endemic arthropod species probably yet to be discovered55. Indeed, many studies have highlighted the uniqueness of this archipelago in terms of species assemblages43,97 and genetic diversity98. This biological distinctiveness, combined with the underrepresentation of terrestrial studies compared to marine ones, likely accounts for the discrepancy with other archipelagos, despite the strong interest that scientists have expressed for this biodiversity hotspot99. Prioritising terrestrial biodiversity research in the Marquesas is crucial for establishing reliable comparisons across the land-to-sea continuum in this archipelago. Similarly, a more sustained sampling effort is much needed in the Gambier and Tuamotu Archipelagos, where a significant number of islands remain insufficiently inventoried. This is an urgent call because, while scientific expeditions could potentially discover new species (e.g.,100), other species could become extinct before being documented (e.g101,102).
Sampling effort biases can obscure the true spatial distribution of biodiversity, complicating the identification of biodiversity hotspots and the quantification of biodiversity loss103. Decision-makers rely on data to inform and justify their political choices. However, gaps in biodiversity inventories can hinder conservation efforts and limit our ability to assess their effectiveness. For instance, if the distribution of an endangered species is poorly documented, it becomes difficult to identify and prioritize areas for protection. In Polynesia, a marine mammal observation network has been established, along with sanctuaries on three islands—Rurutu, Tahiti, and Moorea—contributing to the protection of these threatened species. Nevertheless, other areas also merit consideration for protection due to their high marine mammal diversity. Notably, Raiatea-Taha’a stands out, having recorded the highest number of species (23 species) and a significant number of sightings (104 records). Furthermore, Raiatea and Tahaa, which together form the largest lagoon in the Society Archipelago, may host a particularly high level of biodiversity not fully reflected in current GBIF data. This hypothesis is supported by research showing that 26 of the 32 marine sponges recorded across French Polynesia were found in Raiatea-Taha’a104. Similarly, our findings confirmed that the island of Rapa harbours remarkable marine diversity, as evidenced by studies on coral-reef and terrestrial communities, including taxa unique to this island105,106. However, despite being one of the best documented islands in the archipelago (C = 75.6%), Rapa’s inventory completeness remains behind the global threshold of 80%, suggesting that further sampling efforts are necessary to fully capture this island’s biodiversity. Overall, our study contributes to addressing this gap by pinpointing overlooked locations of the Polynesia-Micronesia biodiversity hotspot.
Conservation science is often compelled to assist in decision-making based on limited and incomplete data107. The spatial heterogeneity in sampling effort that we identified for both marine and terrestrial fauna in French Polynesia is considerable, with up to 70% of islands lacking data on their terrestrial environments. This striking data deficiency was also evidenced by another study using GBIF data to analyse species diversity in a remote region63. An additional challenge, particularly for vast and fragmented territories such as French Polynesia, is the need for data at a sufficiently high spatial resolution to capture island-wide variation. We identified 52 islands that either lacked digital data entirely or were poorly documented, likely due to their remoteness. To fill the spatial gaps in biodiversity data for French Polynesia, we recommend that future sampling efforts prioritise these islands, while also considering the disparity in data coverage between marine and terrestrial ecosystems.
The marine-terrestrial sampling bias
Marine and terrestrial ecosystems are often studied separately, partly due to historical, cultural, or practical reasons108,109. However, because the land-sea continuum operates as an integrated meta-ecosystem, this research divide hampers our ability to fully understand and effectively protect interconnected ecosystems103,110. Maintaining a healthy land-sea ecosystem is particularly crucial in small-island territories, where biodiversity is vulnerable to human activities35,56and where the wellbeing of local populations heavily depends on local natural resources, especially through fishing and tourism. French Polynesia is no exception, with tourism as its primary economic activity and fish and invertebrates as staples in the local diet111. Unlike the global trend103our data show that French Polynesian biodiversity is better documented in marine ecosystems than in terrestrial ones. This discrepancy is partly due to the focus of scientific research and exploration on marine environments (e.g., the oldest of the two major ecology research units in French Polynesia, the CRIOBE, is entirely focused on marine environments) and to the inaccessibility of the mountainous regions54 and seamounts112. The gap is also likely influenced by the huge difference in surface area between land (4,167 km²) and sea (2.5*106 km²), which may also explain why the marine habitats host 20 times more species than terrestrial ones. While surface-area differences are a factor to consider, our records indicate that the disparity is also driven by a lack of terrestrial data for over 52 islands, compared to just two islands with missing marine data. The observed imbalance in marine versus terrestrial data coverage is not only due to the inherent differences between these ecosystems but also reflects underlying biases in sampling practices, exacerbated by the accessibility factors.
Sampling bias is partly influenced by accessibility factors
The accessibility bias hypothesis posits that more accessible areas tend to be surveyed more frequently than less accessible zones79. This can significantly impact the global understanding of natural communities103,113. Our database revealed a pronounced geographic bias in species records, with the most accessible islands (i.e., Tahiti and Moorea in the Society Archipelago, Fakarava in the Tuamotu) being heavily sampled. In contrast, less accessible islands (e.g., Tureia, Napuka and Tenarunga in the Tuamotu, Motu One and Motu Nao in the Marquesas) are poorly documented. However, Rapa Island stands out as an exception, having attracted significant attention from the scientific community due to its hosting of several threatened endemic plant and animal species54,105,106,114. The sampling bias in Tahiti and Moorea is also likely related to the presence of local research institutions (e.g., CNRS-EPHE-Université de Perpignan CRIOBE station, Ifremer, IRD, UC-Berkeley Gump station, University of French Polynesia) there. While Tahiti’s international airport contributes to the sampling bias observed in the Society Islands, our accessibility bias analysis indicated that the distance from ‘airports and ports’ was not the main anthropogenic factor explaining the variance in sampling effort at the scale of French Polynesia. Overall, our accessibility bias analysis showed that sampling efforts in both marine and terrestrial datasets are predominantly skewed towards areas near roads and, to a lesser extent, airports/ports. This aggregation pattern around roads is well-documented in the literature for both terrestrial and marine species103,115 particularly in studies based on citizen-science data116.
Accessibility biases can vary depending on geographic and taxonomic contexts116 highlighting the importance of considering situations on a case-by-case basis. For instance, Freitag et al. (1998)117 found that records of smaller species in African terrestrial ecosystems were minimally affected by accessibility biases, whereas larger species were disproportionately represented in protected areas. Similarly, Cardoso et al. (2024)118 identified various accessibility-bias factors for marine species in the western Atlantic Ocean, including proximity to the coastline, research institutions, ports, protected areas, and urban centres. Recognizing and understanding the nuances underlying these various biases is crucial for enhancing the accuracy and comprehensiveness of biodiversity datasets.
Institutional bias in open-source databases
While accessibility factors provide important insights into sampling patterns, they are not the sole source of bias impacting our biodiversity records. Institutional biases, particularly those associated with open-source databases, might also play a crucial role. The unevenness in data contributions often stems from disparities in funding, data-sharing policies, and digitization efforts across different regions and institutions. The soaring popularity of GBIF data worldwide is reflected in our dataset for French Polynesia, where the number of records per year increased from 10 in 1950 to 1,866 in 2022. We anticipate that the dataset will continue to grow with the engagement of additional contributors, thereby enhancing its reliability119 if institutions continue to adhere to standardisation protocols10. Interestingly, the surge in data during 2006, 2011, and 2012, which constitutes the bulk of the dataset, was driven by the digitization of the French Museum of Natural History dataset (managed by PatriNat) and a major field sampling campaign by Cornell University (USA). The patchiness in data contributions to global open-source databases can be attributed to differences in funding and data-sharing policies across countries, inadequate efforts in digitalising local and national databases, and the sporadic and spatially heterogeneous nature of formal research campaigns26. Combining GBIF records with national databases can yield more complete inventories, as demonstrated by De Araujo et al. (2022)75 for Amazonian epiphytes. In our study, we applied this approach by not only relying on GBIF as the primary data source but also integrating nine additional local datasets to enhance the completeness of our inventory. This selective integration of external data, including local sources, helped reduce coverage gaps while maintaining data quality, underscoring the importance of leveraging both global and local data sources to mitigate biases in biodiversity records. In the case of French Polynesia, engaging local research institutions, private entities, government agencies and developing a citizen science network to compile and share existing (but often inaccessible) information would significantly reduce biases and strengthen the database. The use and adaptation of existing portals such as FauneFrance (https://www.faune-france.org/) or iNaturalist (https://www.inaturalist.org/) to local flora and fauna could for example be advocated to further centralise and favour the collection and compilation of local naturalist data.
Capitalising from citizen science while reducing biases in open-source datasets
Addressing biases and shortfalls in open-source biodiversity datasets is crucial to ensure their efficiency and accuracy in describing species distribution patterns. Citizen science has been increasingly recognized as an effective method for filling gaps in biodiversity information, especially in areas where formal scientific campaigns are limited or sporadic14,120. In our database for French Polynesia, we observed an increase in species records driven by citizen science initiatives, in agreement with the global trend9. Indeed, a substantial 21.7% of records originated from participatory science efforts. While citizen scientists may not always adhere to standard scientific protocols, their contributions provide valuable insights into broader trends, which can then be rigorously analysed. To minimise taxonomic and geographic biases, the involvement of taxonomic experts remains crucial62.
Conclusions and perspectives
Centralising biodiversity information from museums, research institutions, and citizen scientists into big-data platforms offers a transformative opportunity for evaluating species biodiversity in understudied regions. These platforms enable comprehensive data analysis, facilitate global collaboration, engage the public in science, and ultimately contribute to more informed conservation strategies and biodiversity management. Our study provides significant insights into the biodiversity patterns of both marine and terrestrial fauna across the vast and fragmented territory of French Polynesia. We found that while marine inventory completeness is relatively high, averaging up to 76% of known species at the regional scale, terrestrial biogeography remains underexplored (average of 65%), particularly in the Marquesas and Gambier Archipelagos. The analysis indicates a notable skew in the data toward specific taxonomic groups, highlighting the urgent need for comprehensive surveys to fill these gaps. Furthermore, our findings underscore the value of citizen science initiatives, demonstrating their potential to enhance biodiversity knowledge in regions where formal scientific efforts are limited. Overall, this research not only emphasises the richness of biodiversity in French Polynesia but also calls for collaborative efforts to centralise and analyse biodiversity data. These efforts are crucial for aiding in conservation strategies and improving management of the unique ecosystems in the Indo-Pacific region, a global biodiversity hotspot that includes Micronesia, Polynesia, and Fiji121. By providing a reliable, spatially resolved biodiversity dataset, this study lays the foundations for future macroecological research in French Polynesia that will help respond to both fundamental and applied environmental questions.
Data availability
The analyses scripts are available in GitHub (https://github.com/KilianBARREIRO/biogeography_datadiv). The data are available in SEANOE (https://www.seanoe.org/data/00878/99018/).
References
IPBES. Global Assessment Report on Biodiversity and Ecosystem Services of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services (Bonn, 2019).
Ceballos, G. & Ehrlich, P. R. Mutilation of the tree of life via mass extinction of animal genera. Proc. Natl. Acad. Sci. U. S. A. 120 e2306987120 (2023).
Gorman, C. E. et al. Reconciling climate action with the need for biodiversity protection, restoration and rehabilitation. Sci. Total Environ. 857, 159316 (2023).
Singh, J. S. The biodiversity crisis: A multifaceted review. Curr. Sci. 82, 638–647 (2002).
Newmark, W. D., Jenkins, C. N., Pimm, S. L., McNeally, P. B. & Halley, J. M. Targeted habitat restoration can reduce extinction rates in fragmented forests. Proc. Natl. Acad. Sci. 114, 9635–9640 (2017).
Pilowsky, J. A., Colwell, R. K., Rahbek, C. & Fordham, D. A. Process-explicit models reveal the structure and dynamics of biodiversity patterns. Sci. Adv. 8, eabj2271 (2022).
Farley, S. S., Dawson, A., Goring, S. J. & Williams, J. W. Situating ecology as a Big-Data science: current advances, challenges, and solutions. BioScience 68, 563–576 (2018).
Kays, R., McShea, W. J. & Wikelski, M. Born-digital biodiversity data: millions and billions. Divers. Distrib. 26, 644–648 (2020).
Heberling, J. M., Miller, J. T., Noesgaard, D., Weingart, S. B. & Schigel, D. Data integration enables global biodiversity synthesis. Proc. Natl. Acad. Sci. U. S. A. 118 e2018093118 (2021).
Wieczorek, J. et al. Darwin core: an evolving Community-Developed biodiversity data standard. PLOS ONE. 7, e29715 (2012).
Fegraus, E. H., Andelman, S., Jones, M. B. & Schildhauer, M. Maximizing the value of ecological data with structured metadata: an introduction to ecological metadata Language (EML) and principles for metadata creation. Bull. Ecol. Soc. Am. 86, 158–168 (2005).
Güntsch, A., Berendsohn, W. G. & Mergen, P. The BioCASE Project - a Biological Collections Access Service for Europe. (2007).
Levin, N. et al. Biodiversity data requirements for systematic conservation planning in the mediterranean sea. Mar. Ecol. Prog Ser. 508, 261–281 (2014).
Amano, T., Lamming, J. D. L. & Sutherland, W. J. Spatial gaps in global biodiversity information and the role of citizen science. BioScience 66, 393–400 (2016).
Underwood, E., Taylor, K. & Tucker, G. The use of biodiversity data in Spatial planning and impact assessment in Europe. RIO 4 e28045 (2018).
Lin, H., Caley, M. J. & Sisson, S. A. Estimating global species richness using symbolic data meta-analysis.pdf. Ecography e05617. (2022).
Takashina, N. & Kusumoto, B. A perspective on biodiversity data and applications for spatio-temporally robust Spatial planning for area-based conservation. Discov Sustain. 4, 1 (2023).
Hortal, J. et al. Seven shortfalls that beset Large-Scale knowledge of biodiversity. Annu. Rev. Ecol. Evol. Syst. 46, 523–549 (2015).
Troia, M. J. & McManamay, R. A. Filling in the GAPS: evaluating completeness and coverage of open-access biodiversity databases in the united States. Ecol. Evol. 6, 4654–4669 (2016).
Zizka, A. et al. No one-size-fits-all solution to clean GBIF. PeerJ 8, e9916 (2020).
Troudet, J., Grandcolas, P., Blin, A., Vignes-Lebbe, R. & Legendre, F. Taxonomic bias in biodiversity data and societal preferences. Sci. Rep. 7, 9132 (2017).
García-Roselló, E., González-Dacosta, J. & Lobo, J. M. The biased distribution of existing information on biodiversity hinders its use in conservation, and we need an integrative approach to act urgently. Biol. Conserv. 283, 110118 (2023).
Rocchini, D. et al. A quixotic view of Spatial bias in modelling the distribution of species and their diversity. Npj Biodivers. 2, 10 (2023).
Schiesari, L., Grillitsch, B. & Grillitsch, H. Biogeographic biases in research and their consequences for linking amphibian declines to pollution. Conserv. Biol. 21, 465–471 (2007).
Wüest, R. O. et al. Macroecology in the age of big Data – Where to go from here? J. Biogeogr. 47, 1–12 (2020).
Beck, J., Böller, M., Erhardt, A. & Schwanghart, W. Spatial bias in the GBIF database and its effect on modeling species’ geographic distributions. Ecol. Inf. 19, 10–15 (2014).
König, C. et al. Biodiversity data integration—the significance of data resolution and domain. PLoS Biol. 17, e3000183 (2019).
Kadmon, R., Farber, O. & Danin, A. Effect of roadside bias on the accuracy of predictive maps produced by bioclimatic models. Ecol. Appl. 14, 401–413 (2004).
Engemann, K. et al. Limited sampling hampers big data Estimation of species richness in a tropical biodiversity hotspot. Ecol. Evol. 5, 807–820 (2015).
Borges, P. A. V. et al. Global Island monitoring scheme (GIMS): a proposal for the long-term coordinated survey and monitoring of native Island forest biota. Biodivers. Conserv. 27, 2567–2586 (2018).
Alves, C., João Aguuiar, C., Cristina, R., João Pradinho, H. & Ângela, L. Research data management in the field of ecology: an overview. Int. Conf. Dublin Core Metadata Appl. https://doi.org/10.23106/dcmi.952138986 (2018).
Stephenson, P. et al. Priorities for big biodiversity data. Front. Ecol. Environ. 15, 124–125 (2017).
Hachich, N. F. et al. Island biogeography patterns of marine shallow-water organisms in the Atlantic. J. Biogeogr. 42, 1871–1882 (2015).
Simberloff, D. Extinction-proneness of Island species-causes and management implications. Raffles Bull. Zool. 48, 1–9 (2000).
Russell, J. C. & Kueffer, C. Island biodiversity in the anthropocene. Annu. Rev. Environ. Resour. 44, 31–60 (2019).
Warren, B. H. et al. Islands as model systems in ecology and evolution: prospects Fifty years after MacArthur-Wilson. Ecol. Lett. 18, 200–217 (2015).
Whittaker, R. J., Fernández-Palacios, J. M., Matthews, T. J., Borregaard, M. K. & Triantis, K. A. Island biogeography: taking the long view of nature’s laboratories. Science 357, eaam8326 (2017).
Andréfouët, S. & Adjeroud, M. Chapter 38 - French polynesia. In world seas: an environmental evaluation volume II: Indian Ocean to the Pacific. 827–854 (2019).
Galzin, R. & Meyer, J. Y. H. Les 124 Îles de La polynésie française: types, superficies, noms et occupation humaine. Bull. De La. Société Des. Études Océaniennes 123–136 (2024).
Kulbicki, M. Biogeography of reef fishes of the French territories in the South Pacific. Cybium 31, 275–288 (2007).
Salvat, B. Dominant benthic mollusks in closed atolls, French Polynesia. Galaxea J. Coral Reef. Stud. 11, 197–206 (2009).
Tröndlé, J. & Boutet, M. Inventory of marine molluscs of French Polynesia. Atoll Res. Bull. 1–87. https://doi.org/10.5479/si.00775630.570.1 (2009).
Delrieu-Trottin, E. et al. Shore fishes of the Marquesas islands, an updated checklist with new records and new percentage of endemic species. Cl 11, 1758 (2015).
Delrieu-Trottin, E. et al. A DNA barcode reference library of French Polynesian shore fishes. Sci. Data. 6, 114 (2019).
Salvat, B. & Tröndlé, J. Biogéographie des mollusques marins de polynésie Française. Revec 72, 215–257 (2017).
Boutet, M., Gourguet, R. & Letourneux, J. Marine Molluscs of French Polynesia / Mollusques Marins De Polynésie Française (Au Vent Des Iles, 2020).
Vieira, C. et al. Global biogeography and diversification of a group of brown seaweeds (Phaeophyceae) driven by clade-specific evolutionary processes. J. Biogeogr. 48, 703–715 (2021).
Vieira, C. et al. Diversity, systematics and biogeography of French Polynesian Lobophora (Dictyotales, Phaeophyceae). Eur. J. Phycol. 58, 226–253 (2023).
Ramage, T. Checklist of the terrestrial and freshwater arthropods of French Polynesia (Chelicerata; myriapoda; crustacea; Hexapoda). Zoosystema 39, 213 (2017).
Thibault, J. C., Cibois, A. & Lynx birds of Eastern Polynesia: A biogeographic atlas. (Barcelona, 2017).
Florence, J. Flore De La Polynésie Française 1 (IRD édition/MNHN, 1997).
Florence, J. Flore De La Polynésie Française 2 (IRD Éditions/MNHN, 2004).
Chevillotte, H., Ollier, C. & Meyer, J. Y. Base De Données Botaniques Nadeaud De l’Herbier De La Polynésie Française (PAP). Institut Louis Malardé, Délégation À La Recherche, Papeete, Tahiti http://nadeaud.ilm.pf (Institut Louis Malardé, 2019).
Gillespie, R. G., Claridge, E. M. & Goodacre, S. L. Biogeography of the fauna of French polynesia: diversification within and between a series of hot spot archipelagos. Phil Trans. R Soc. B. 363, 3335–3346 (2008).
Hembry, D. H. Evolutionary biogeography of the terrestrial biota of the Marquesas islands, one of the world’s remotest archipelagos. J. Biogeogr. 45, 1713–1726 (2018).
Fernandez-Palacios, J. M. et al. Scientists’ warning – The outstanding biodiversity of Islands is in peril. Global Ecol. Conserv. 31, e01847 (2021).
Pebesma, E. & Bivand, R. Spatial Data Science: with Applications in R (Chapman and Hall/CRC, 2023).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2024).
Chamberlain, S., Vanhoorne, B. & worrms World Register of Marine Species (WoRMS) Client. R package. (2023).
Chamberlain, S. et al. taxize: Taxonomic information from around the web. R package. (2020).
Grenié, M. & Gruson, H. rtaxref: An R Client for TAXREF the French taxonomical reference API. R package. (2022).
Maldonado, C. et al. Estimating species diversity and distribution in the era of B Ig D ata: to what extent can we trust public databases? Glob. Ecol. Biogeogr. 24, 973–984 (2015).
Bonnet-Lebrun, A. S. et al. Opportunities and limitations of large open biodiversity occurrence databases in the context of a marine ecosystem assessment of the Southern ocean. Front. Mar. Sci. 10, 1150603 (2023).
Smith, J. R. et al. A global test of ecoregions. Nat. Ecol. Evol. 2, 1889–1896 (2018).
Lim, G. S., Balke, M. & Meier, R. Determining species boundaries in a world full of rarity: singletons, species delimitation methods. Syst. Biol. 61, 165–169 (2012).
Chao, A. et al. Quantifying sample completeness and comparing diversities among assemblages. Ecol. Res. 35, 292–314 (2020).
Montes, E. et al. Optimizing Large-Scale biodiversity sampling effort: toward an unbalanced survey design. Oceanog 34, 80–91 (2021).
Soberón, J., Jiménez, R., Golubov, J. & Koleff, P. Assessing completeness of biodiversity databases at different Spatial scales. Ecography 30, 152–160 (2007).
Deng, C., Daley, T. & Smith, A. Applications of species accumulation curves in large-scale biological data analysis. Quant. Biol. 3, 135–144 (2015).
Oksanen, J. et al. Vegan: Community ecology package. (2024).
Chao, A. Nonparametric Estimation of the number of classes in a population. Scand. J. Stat. 11, 265–270 (1984).
Chao, A. Estimating the population size for Capture-Recapture data with unequal catchability. Biometrics 43, 783 (1987).
Colwell, R. K. & Coddington, J. A. Estimating terrestrial biodiversity through extrapolation. Phil Trans. R Soc. Lond. B. 345, 101–118 (1994).
Chao, A. & Chun-Huo, C. Species richness: Estimation and Compariso. Wiley StatsRef: Stat. Ref. Online. 1, 26 (2016).
De Araujo, M. L., Quaresma, A. C. & Ramos, F. N. GBIF information is not enough: National database improves the inventory completeness of Amazonian epiphytes. Biodivers. Conserv. 31, 2797–2815 (2022).
Ramírez, F., Sbragaglia, V., Soacha, K., Coll, M. & Piera, J. Challenges for marine ecological assessments: completeness of findable, accessible, interoperable, and reusable biodiversity data in European seas. Front. Mar. Sci. 8, 802235 (2022).
Chanachai, J. et al. What remains to be discovered: A global assessment of tree species inventory completeness. Divers. Distrib. e13862 https://doi.org/10.1111/ddi.13862 (2024).
Soberón, J. & Peterson, T. Biodiversity informatics: managing and applying primary biodiversity data. Phil Trans. R Soc. Lond. B. 359, 689–698 (2004).
Zizka, A., Antonelli, A. & Silvestro, D. sampbias, a method for quantifying geographic sampling biases in species distribution data. Ecography 44, 25–32 (2021).
Meyer, C., Weigelt, P. & Kreft, H. Multidimensional biases, gaps and uncertainties in global plant occurrence information. Ecol. Lett. 19, 992–1006 (2016).
Cornwell, W. K., Pearse, W. D., Dalrymple, R. L. & Zanne, A. E. What we (don’t) know about global plant diversity. Ecography 42, 1819–1831 (2019).
Moudrý, V. & Devillers, R. Quality and usability challenges of global marine biodiversity databases: an example for marine mammal data. Ecol. Inf. 56, 101051 (2020).
Clements, J. F. et al. The eBird/Clements checklist of birds of the world. (2024).
Bacchet, P., Zysman, T. & Lefevre, Y. Guide Des poissons de Tahiti et Ses Îles. (Éditions Au Vent des Îles, Tahiti (Polynésie Francaise), 2017).
Siu, G. et al. Shore fishes of French Polynesia. Cybium 41, 245–278 (2017).
Porch, N., Smith, T. R. & Greig, K. Five new Pycnomerus Erichson (Coleoptera: zopheridae: Pycnomerini) from Raivavae. Fr. Polynesia Zootaxa. 4718, 239–250 (2020).
Rocha-Ortega, M., Rodriguez, P. & Córdoba-Aguilar, A. Geographical, Temporal and taxonomic biases in insect GBIF data on biodiversity and extinction. Ecol. Entomol. 46, 718–728 (2021).
Liebherr, J. The first precinctive Carabidae from moorea, society islands: new Mecyclothorax spp. (Coleoptera) from the summit of Mont Tohiea. ZK 224, 37–80 (2012).
Mora, C., Tittensor, D. P. & Myers, R. A. The completeness of taxonomic inventories for describing the global diversity and distribution of marine fishes. Proc. R Soc. B. 275, 149–155 (2008).
Sánchez-Fernández, D., Fox, R., Dennis, R. L. H. & Lobo, J. M. How complete are insect inventories? An assessment of the British butterfly database highlighting the influence of dynamic distribution shifts on sampling completeness. Biodivers. Conserv. 30, 889–902 (2021).
Stokes, D. L. Things we like: human preferences among similar organisms and implications for conservation. Hum. Ecol. 35, 361–369 (2007).
Ducarme, F., Luque, G. M. & Courchamp, F. What are charismatic species for conservation biologists?. BioSci. Master Rev. (2013).
De Pinho, J. R., Grilo, C., Boone, R. B., Galvin, K. A. & Snodgrass, J. G. Influence of aesthetic appreciation of wildlife species on attitudes towards their conservation in Kenyan agropastoralist communities. PLoS ONE. 9, e88842 (2014).
Lomolino, M. V. Conservation biogeography. in Frontiers of Biogeography: New Directions in the Geography of Nature (eds Lomolino, M. V. & Heaney, L. R.) 293–296 (Sinauer Associates, Sunderland, MA, (2004).
Kusumoto, B. et al. Occurrence-based diversity estimation reveals macroecological and conservation knowledge gaps for global woody plants. Sci. Adv. 9 (2023).
Shirey, V., Belitz, M. W., Barve, V. & Guralnick, R. A complete inventory of North American butterfly occurrence data: narrowing data gaps, but increasing bias. Ecography 44, 537–547 (2021).
Biodiversité, T. et Marine des Îles marquises, polynésie française. (Paris, 2016).
Reisser, C. M. O. et al. Population connectivity and genetic assessment of exploited and natural populations of Pearl oysters within a French Polynesian Atoll lagoon. Genes 11, 426 (2020).
Mittermeier, R. A. et al. Hotspots Revisited: Earth’s Biologically Richest and Most Endangered Terrestrial Ecoregions (The University of Chicago Press, 2005).
Williams, J. T., Delrieu-Trottin, E. & Planes, S. A new species of Indo-Pacific fish, Canthigaster criobe, with comments on other Canthigaster (Tetraodontiformes: Tetraodontidae) at the Gambier Archipelago. Zootaxa 3523, (2012).
Zimmermann, G., Gargominy, O. & Fontaine, B. Quatre espèces nouvelles d’endodontidae (Mollusca, Pulmonata) Éteints de Rurutu (Îles australes, polynésie française). Zoosystema 31, 791–805 (2009).
Richling, I. & Bouchet, P. Extinct even before scientific recognition: a remarkable radiation of helicinid snails (Helicinidae) on the gambier islands, French Polynesia. Biodivers. Conserv. 22, 2433–2468 (2013).
Hughes, A. C. et al. Sampling biases shape our view of the natural world. Ecography 44, 1259–1269 (2021).
Hall, K. A. et al. Affinities of sponges (Porifera) of the Marquesas and society islands, French Polynesia. Pac. Sci. 67, 493–511 (2013).
Terrestrial biodiversity of the Austral Islands, french polynesia. (Muséum d’Histoire Naturelle, 2014).
Adjeroud, M. et al. Reefs at the edge: coral community structure around rapa, southernmost French Polynesia. Mar. Ecol. 37, 565–575 (2016).
Soulé, M. E. What is conservation biology?? A new synthetic discipline addresses the dynamics and problems of perturbed species, communities, and ecosystems. BioScience 35, 727–734 (1985).
Raffaelli, D., Solan, M. & Webb, T. J. Do marine and terrestrial ecologists do it differently? Mar. Ecol. Prog. Ser. 304, 283–289 (2005).
Munguia, P. & Ojanguren, A. F. Bridging the gap in marine and terrestrial studies. Ecosphere 6, 1–4 (2015).
Álvarez-Romero, J. G. et al. Integrated Land-Sea conservation planning: the missing links. Annu. Rev. Ecol. Evol. Syst. 42, 381–409 (2011).
Gillett, R. & Tauati, M. I. Fisheries of the Pacific islands. Regional and National information. FAO Fisheries Aquaculture Tech. Paper. 625, 401 (2018).
Hanafi-Portier, M. & Samedi, S. Les monts sous-marins de polynésie française, etat des lieux des connaissances et recommandations scientifiques. https://hal.science/hal-04713244 (2024).
Mangiacotti, M. et al. Assessing the Spatial scale effect of anthropogenic factors on species distribution. PLoS ONE. 8, e67573 (2013).
Barrett, R. L., Taputuarai, R., Meyer, J. Y. H., Bruhl, J. J. & Wilson, K. L. Reassessment of the taxonomic status of Cyperaceae on Rapa iti, Austral islands, French polynesia, with a new combination, Morelotia involuta. Telopea 24, 171–187 (2021).
Reddy, S. & Dávalos, L. M. Geographical sampling bias and its implications for conservation priorities in in Africa. J. Biogeogr. 30, 1719–1727 (2003).
Mair, L. & Ruete, A. Explaining Spatial variation in the recording effort of citizen science data across multiple taxa. PLoS ONE. 11, e0147796 (2016).
Freitag, S., Hobson, C., Biggs, H. C. & Van Jaarsveld, A. Testing for potential survey bias: the effect of roads, urban areas and nature reserves on a Southern African mammal data set. Anim. Conserv. 1, 119–127 (1998).
Cardoso, M. N. M. et al. Causes and effects of sampling bias on marine Western Atlantic biodiversity knowledge. Divers. Distrib. 30, e13839 (2024).
Ivanova, N. V. & Shashkov, M. P. The possibilities of GBIF data use in ecological research. Russ J. Ecol. 52, 1–8 (2021).
Isaac, N. J. B., Van Strien, A. J., August, T. A., De Zeeuw, M. P. & Roy, D. B. Statistics for citizen science: extracting signals of change from noisy ecological data. Methods Ecol. Evol. 5, 1052–1060 (2014).
Fan, H. et al. Conservation priorities for global marine biodiversity across multiple dimensions. Natl. Sci. Rev. 10, nwac241 (2023).
Acknowledgements
We are grateful to Serge Andréfouët and Jean-Yves Hiro Meyer for their insightful comments on the biogeography of French Polynesia and for highlighting relevant prior research in the region.
Author information
Authors and Affiliations
Contributions
All authors have read and approved the final version of the manuscript. K.B. and L.B. designed the study, curated the data, produced the original draft and wrote. C.M. contributed to methodological design the original draft, and writing. S.D., J-.C.G., and J.L.L. contributd to the methodological design and writing. C.J.M. adquired funding, contributed to the methodological design, the original draft and writing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Barreiro, K., Benestan, L., Moritz, C. et al. Species richness variation in marine and terrestrial fauna across widespread, fragmented territories: assessing inherent challenges of data scarcity at local and regional scales. Sci Rep 15, 21043 (2025). https://doi.org/10.1038/s41598-025-06631-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-06631-4